Abstract
Background/Aims
Acute liver failure (ALF) is due to severe immune response, resulting in massive apoptosis/necrosis of hepatocytes. The precise mechanism has not been explored yet.
Materials and Methods
The mouse with ALF model was induced by D-GalN/LPS; the hepatic miRNAs expression profile was evaluated by miRNA microarray and verified by RT-PCR. During the ALF in mice, the miR-155 expression was detected in the liver as well as in spleen. Then the correlation between miR-155 and inflammatory cytokines was evaluated. Furthermore, the miR-155 expression in activated Raw264.7 cells and apoptotic hepatocytes was also studied. Finally, the regulatory roles of miR-155 in TNF expression of apoptotic hepatocytes were shown.
Results
It was shown that miRNAs changed in the mice with ALF relating to hepatocytes apoptosis/necrosis; the selected miRNAs were confirmed with RT-PCR. miR-155 was up-regulated, but miR-698, -720, and -329 were down-regulated. Moreover, hepatic miR-155 was up-regulated at all-time points in the liver, but only at 7 h in spleen of mice with ALF. A significant correlation was observed between hepatic miR-155 and TNF/IL-6 in mice with ALF, which was supported by the findings in vitro showing up-regulated miR-155 in Raw264.7 cells and Hepa1-6 cells under LPS or D-GalN+TNF induction, respectively. Moreover, a correlation was observed between miR155 and TNF levels in vivo and in vitro.
Conclusion
These data demonstrate that miR-155 regulates TNF-mediated hepatocyte apoptosis in ALF, which provides some useful information in both basic and clinical researches.
Keywords: miR-155, TNF, acute liver failure, D-GalN, LPS
INTRODUCTION
Acute liver failure (ALF) is a life-threatening syndrome that results from catastrophic liver damage. During ALF, circulating pro-inflammatory cytokines (e.g. TNF, IL-1, IL-6, IL-10, IL-12, and IFN-γ) were regulated via activating of immune cascade following the binding of lipopolysaccharide (LPS) to toll-like receptor 4 (TLR4) (1). Consequently, hepatocyte apoptosis/necrosis occurred resulting from excessive host immune response. Previously, we have shown that TNF plays a critical role in hepatocyte apoptosis/necrosis (2), which was effectively inhibited by anti-TNF-α antibody (3).
MicroRNAs (miRNAs) are negative gene regulators at post-transcriptional level that bind to complementary sequences in downstream mRNAs, resulting in target mRNA silence (4). miRNAs play important roles in a wide variety of biological processes, including proliferation, differentiation, cell fate determination, and apoptosis (5). They also play an important role in innate immune response (6) by regulating pro-inflammatory cytokines production in an autocrine fashion (7). It was demonstrated that over-expression of TNF/IL-6 in monocytes/macrophages was regulated by miR-155 in vitro (8) and in vivo (9). We have previously reported that miR-15b/16 plays a fundamental role in the pathogenesis of TNF-mediated hepatocytes apoptosis (2). However, the role of miR-155 in ALF has not been explored yet.
D-Glucosamine (D-GalN) and LPS-induced ALF in mice is TNF dependent (10); it is the widely used model for study of human ALF (11). In this study, the relationship between miR-155 and TNF was determined in liver tissue of mice with ALF as well as in Raw264.7 cells and Hep1-6 cells induced by LPS and TNF/D-GalN, respectively. The regulatory role of miR-155 in TNF-mediated ALF in vitro was investigated.
MATERIAL AND METHODS
Animal studies
In agreement with animal protocols approved by the Animal Ethics Committee of the Nanjing Medical University, all animals received proper care. Ten-week-old male BALB/c mice (20–22g), obtained from Shanghai SLAC Laboratory Animal Co. Ltd (Shanghai, China), were housed under conventional laboratory conditions with food and water ad libitum. Mice were randomly divided into four groups. In mice model, ALF was induced by intraperitoneal injection of D-GalN (Sigma, USA) (900 mg/kg of body weight) and LPS (Sigma, USA) (10 mg/kg of body weight), as previously described (12), whereas the controls were given D-GalN (900 mg/gram of body weight) or LPS (10 mg/kg of body weight) or saline (0 h) only. The challenged mice were sacrificed at different time points (five per time point). Prior to sacrifice, the serum was collected for biochemical analysis. Liver tissue was stored in liquid nitrogen for miRNA microarray analysis and RT-PCR.
miRNA microarray assay
Microarray assay was performed using a service provider (LC Sciences), as previously described (2). The alteration in the level of miRNAs was considered statistically significant if P-value<0.01.
Total RNA extracted and quantitative real-time RT-PCR
Total RNA from liver tissue or cells was extracted using Trizol regiment (Invitrogen, Paisley, UK) according to the manufacturer’s instructions. The expression levels of miRNAs and mRNAs were detected with SYBR-based quantitative real-time RT-PCR (qPCR). cDNA was synthesized from 0.5 μg of RNA using a reverse transcription kit (Takara); qPCR was performed using the SYBR Green II core kit (Takara) following the manufacturer’s instructions and an ABI 7500 real-time RT-PCR system (Applied Biosystems, Foster City, CA, USA). For each primer set, an optimal dilution was determined, and melting curves were used to determine the specificity of product amplification. Each sample was serially diluted over three orders of magnitude, and all samples were run on the same 96-well plate. qPCR was carried out using primer pairs designed to mouse TNF and IL-6 and housekeeping genes encoding β-actin, to mouse miRNAs and U6 as housekeeping genes, The relative amount of each mRNA/miRNA was measured using the 2−ΔΔCt method (13). All RT-PCR reactions were performed in triplicate, and repeated twice.
Spleen cells isolation
Spleens were collected from the mice 7 h post D-GalN/LPS stimulation. Splenocytes were isolated by passing splenic tissue through 200 mesh. Red blood cells were removed by hypotonic solution, centrifuged at 1500 rpm for 5min, and washed with PBS. Then, the cells were collected, total RNA was extracted, and the expression of miR-155 was detected.
Cell culture and apoptotic induction
Raw264.7 (macrophage cell line) and Hepa1-6 cells (mouse hepatocytes) were maintained in DMEM (Invitrogen) containing 10% FBS (HyClone, South Logan, UT) at 37°C in a 5% CO2 atmosphere. LPS (Sigma) was applied to activate Raw264.7 cells with different concentrations, 50, 100, 500, or 1000 ng/ml, respectively. D-GalN (1 mg/ml) and TNF (100 ng/ml) were used to induce Hepa1-6 cells to apoptosis; and Hochest 33342 staining was applied to detect the cell apoptosis (14). Total RNA was extracted, and the levels of miR-155 and TNF from cells with the stimulations were detected.
miR-155 mimic or inhibitor
Hepa1-6 cells (105) were treated with miR-155-inhibitor (200 nM) or miR-155-mimic (100 nM) (Applied RiboBio), as previously described2. Non-specific miRNA mimic or inhibitor (NSI) (100 nM) was used as negative control. CY3 (100 nM) was used as positive control. The levels of mature miR-155 from different treatments were determined by RT-PCR.
Statistical analysis
The data were presented as means ± SEM, and groups were compared by Student’s t test. Correlation was assessed by Spearman’s r test. p<0.05 was regarded as significant.
RESULTS
Microarray analysis of hepatic miRNAs in ALF in mice
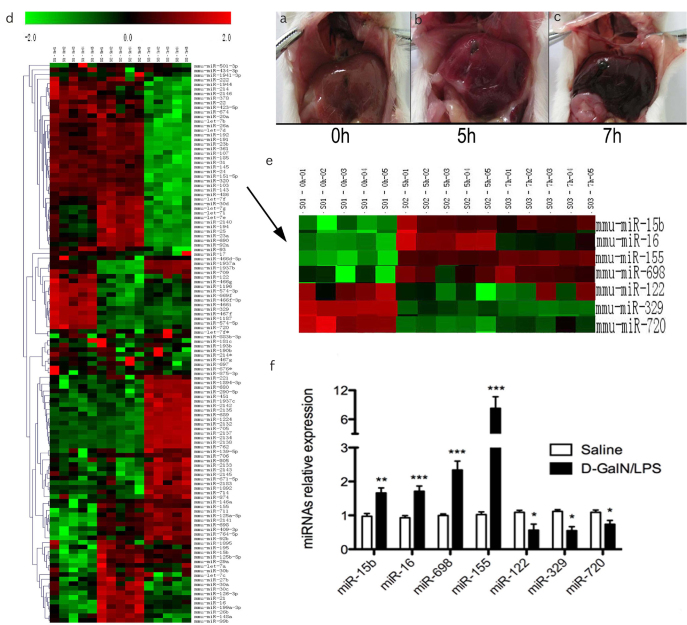
No obvious abnormality was observed in the liver with mock challenge (Figure 1a). Significant swollen and dark red spots (necrosis) were observed in the liver at 5 h post D-GalN/LPS treatment (Figure 1b). Massive dark red necrosis was detected in the liver at 7 h post D-GalN/LPS treatment (Figure 1c). Such macroscopic observation is consistent with our previous study at both macroscopic and microscopic levels (2,12). The miRNA microarray was used to identify modification of miRNAs during the development of ALF; it revealed that there were 122 deregulated hepatic miRNAs (p<0.05) in mice with ALF compared to the control (saline group; 0 h) (Figure 1d). Selected up- or down-regulated miRNAs were verified by RT-PCR. Up-regulation of miR-155 and miR-698, and down-regulation of miR-122, miR-720, and miR-329 were observed (Figure 1e, f).
Figure 1. a–f.
Microarray analysis of hepatic miRNAs expression in ALF
In our previous experiment, we have reported the histopathology changes of liver after D-GalN/LPS induction. Generally, the surface of normal liver looked smooth (A) (n = 10), 5 h after D-GalN/LPS challenged, the color of surface became deepened and it was spotting (B) (n = 10), Extreme damage of liver tissues was checked at 7 h (C) (n = 10), the liver was swelling, and the liver surface had a severe congestion. The expression profile of hepatic miRNA was detected using the LAN-based miRNA microarray in mice at 0, 5 and 7 h post D-GalN/LPS treatment. It was revealed that there were 122 deregulated hepatic miRNAs (P< 0.01) in ALF mice compared to the control (saline group; 0 h) (D) n = 5 for each time point. The miRNAs with more significant changes were showed as the arrow indicated, the top bar represents the signal levels of miRNA expression from −2 (green) to +2 (red) (E), n = 5 for each time point. The up-regulation of miR-15b, miR-16, miR-155 and miR-698 and down-regulation of miR-122, miR-329 and miR-720 were verified by RT-PCR (F) (n = 10).
miR-155 was up-regulated in liver and spleen from mice with ALF
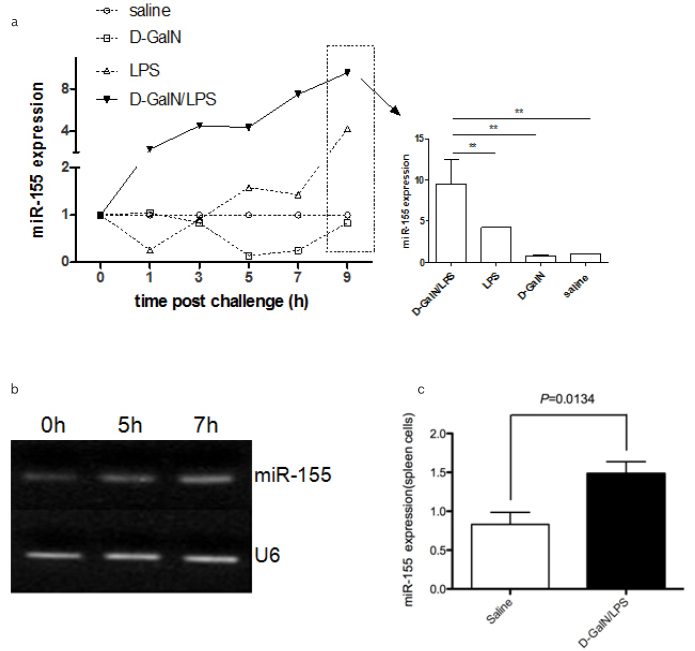
miR-155 was focused due to its highest up-regulation among these deregulated miRNAs. The level of hepatic miR-155 was gradually increased, and it peaked at 7 hrs post D-GalN/LPS treatment, which was ~8-fold or ~2-fold higher than that of saline, D-GaIN and LPS treatment at 9 hrs (p<0.001) (Fig. 2A), respectively. In the LPS group, the level of hepatic miR-155 increased, but it only had a significant difference at 9 hrs. No significant difference of miR-155 was observed among the groups with saline and D-GaIN groups at all-time points. In addition, the results from hepatic miR-155 showed that the significant difference was observed in D-GalN/LPS at 7 hrs post D-GalN/LPS challenge (Figure 2b). Spleen is one of the important organs in the immune system. The expression of miR-155 in splenocytes was determined from the mice with 7 hrs post D-GalN/LPS treatment. Thus, it was only selected to evaluate the expression of miR-155 in the splenocytes with D-GalN/LPS and vehicle groups only, showing that the ~1.5-fold higher from the D-GalN/LPS group than that the vehicle treated group (p<0.05) (Figure 2c).
Figure 2. a–c.
miR-155 was up regulated in both liver and spleen in ALF
The expression of hepatic miR-155 was detected in liver tissue of mice induced by D-GalN/LPS, D-GalN, LPS and saline treatment by RT-PCR respectively (A). There are four groups for this experiment (D-GalN/LPS, D-GalN, LPS and saline), with 6 time points (0, 1, 3, 5, 7, and 9 h) for each group. There were 10 mice per time point for D-GalN, LPS and saline groups respectively and with 100% survival rate for all of the three groups. In D-GalN/LPS group, there were 10, 10, 10, 12, 20 and 35 mice for each time point (0, 1, 3, 5, 7, and 9 h), respectively, prior to challenge. The survival rate was 100% (0 h), 100% (1 h), 100% (3 h), 90% (5 h), 50% (7 h) and 30% (9 h) in D-GalN/LPS challenge group. In addition, the miR-155 PCR productions with D-GalN/LPS induction for 7 h was used to run the gel, U6 was used as the internal control (B). miR-155 expression was detected in spleen tissue in mice with D-GalN/LPS induction for 7 h (C) (n = 10).
The relationship between miR-155 and pro-inflammatory cytokines in the liver from the mice with ALF
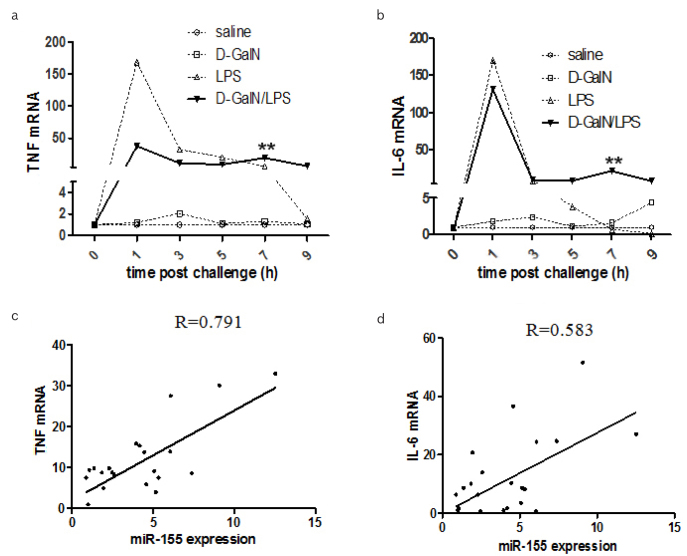
The level of TNF mRNA from the liver of D-GalN/LPS-treated group increased, and it reached at peak at 1 hr (p<0.01), and then persisted till 7 hrs (p<0.01) (Figure 3A), which was ~30-fold higher than the saline- or D-GaIN-treated group. Interestingly, hepatic TNF mRNA was induced by LPS dramatically at 1 hr, which was almost 140-fold higher than that of saline- or D-GalN-treated group. Hepatic TNF mRNA decreased gradually from 3 to 7 hr post LPS challenge. Similar patterns of hepatic IL-6 were observed (Figure 3B) with the different treatments. Furthermore, there was significant correlation between hepatic miR-155 and TNF (Figure 3C) (p<0.05), as well as between miR-155 and IL-6 at 7 hrs post D-GalN/LPS treatment (Figure 3d) (p<0.05).
Figure 3. a–d.
The correlation between miR-155 expression and inflammatory factors
Hepatic TNF and IL-6 mRNA in mice were detected at different time points post D-GalN/LPS, D-GalN, LPS and saline challenged, respectively (A, B) (n = 10). The correlation of miR-155 to TNF and IL-6 respectively were analysed using pearson correlation coefficient analysis (C, D).
miR-155 was up-regulated in both Raw264.7 cells and Hepa1-6 cells
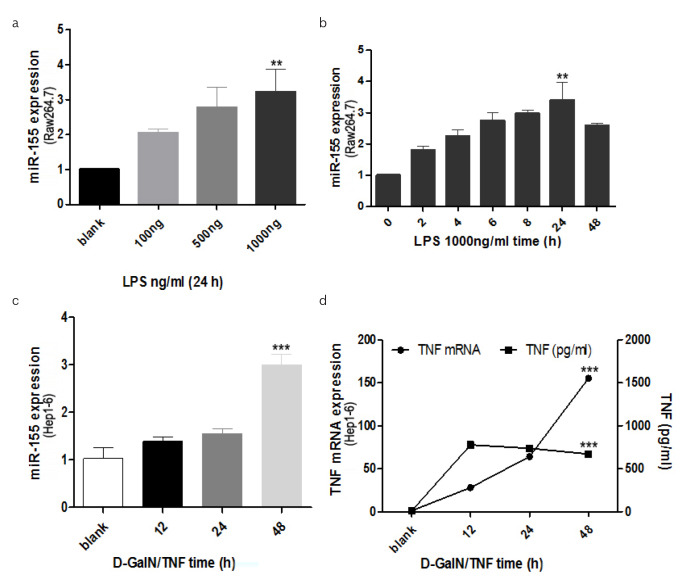
Following the data above, it is of interest to identify the origin of over-expressed miR-155 in liver. A great quantity of macrophages infiltrate in the liver and the activation of Kupffer cells (KCs) plays a key role in ALF (15). Therefore, we supposed miR-155 would over-express in KCs and/or hepatocytes in ALF. Hep1-6 cells apoptosis was induced using D-GalN/TNF in vitro. Morphological apoptotic changes of Hep1-6 cells following DGalN/TNF stimulation were evaluated, that is, shrinkage or rounding of cell shape, and darkened nuclear fluorescence by Hoechst 33342 staining. These apoptotic changes were detectable at 24 h (Figure 4e) and more extensive at 36 h (Figure 4f) compared with blank (mock treated) cells (Figure 4d). According to the ALF condition in vivo, we designed the in vitro study, Raw264.7 cells were induced by LPS with different dose, miR-155 were found with the dose-and time-dependent increase in Raw264.7 cells (Figure 5a, b). Hepatocyte apoptosis in vitro model was also utilized to confirm our in vivo data above. It was observed that miR-155 was time-dependent and peaked at 48 h, which was 3-fold higher than that of mock treated (p<0.05) (Figure 5c). The relationship between TNF mRNA and protein was investigated in the hepatocyte apoptosis in vitro model. It was found that both TNF mRNA and protein were up-regulated, and the over-expression of TNF mRNA was more stable (Figure 5d).
Figure 4. a–f.
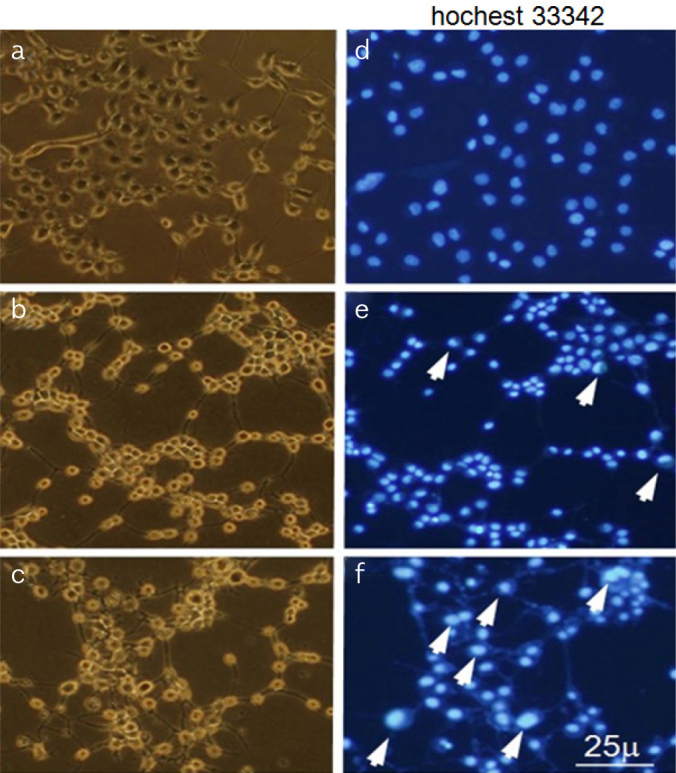
Apoptosis of Hep1-6 cells detection
Apoptosis of Hep1-6 cells with D-GalN/TNF was analysed using fluorescence microscopy (Hoechst 33342 labelled), Healthy cells attached to the bottom of well following mock treatment (A), with intact light blue nuclei (D), D-GalN/TNF (D/T) 24 h treated cells showed slight detachment, rounding in shape (B), with typical apoptotic changes. i.e., shrinkage. Chromatin condensation and margination in the nucleus (white arrow) (E). D-GalN/TNF (D/T) 36 h treated cells showed more severe shrinkage and detachment (C) and more apoptotic changes (white arrow) (F).
Figure 5. a–d.
miR-155 expression in both RAW264.7 and Hep1-6 cells
RAW264.7 cell was induced with LPS, miR-155 was detected with different concentration at 24 h post LPS induction (A) and at different time points with the concentration of 1000ng/ml using RT-PCR (B). miR-155 expression was detected with RT-PCR when the Hep1-6 cells were induced with D-GalN/TNF at several time points (C). TNF mRNA as well as the TNF protein levels in supernatant of Hep1-6 cells with D-GalN/TNF treatment at different time point were detected by RT-PCR and ELISA, respectively (D).
miR-155 regulated TNF in hepatocytes in vitro
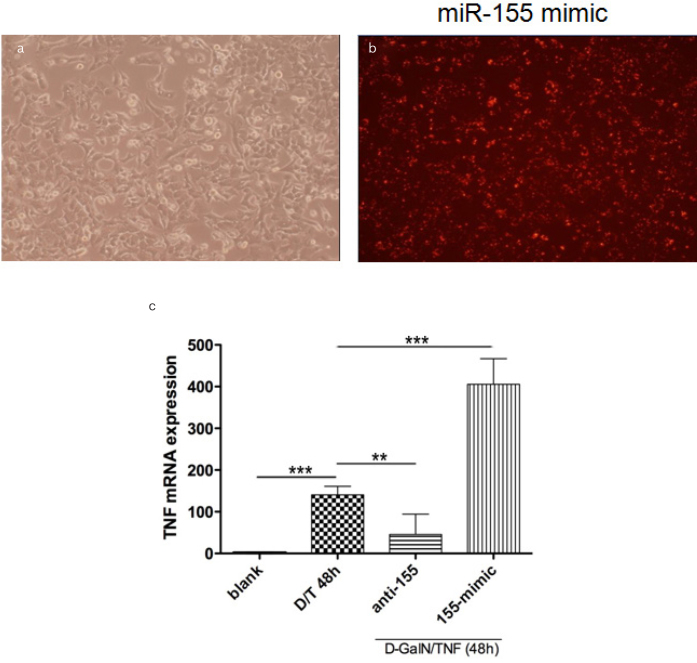
It is important to confirm whether miR-155 can regulate TNF production during hepatocyte apoptosis in vitro. Using D-GalN/TNF induced Hepa1-6 cells apoptosis model, miR-155 mimic (155-mimic) or inhibitor (anti-155) was applied. The transfection efficiency was confirmed using fluorescent cy3 as previously described2 (Figure 6a, b). It was demonstrated that TNF mRNA expression can be suppressed by miR-155 inhibitor, but can also be further up-regulated by miR-155 mimic (Figure 6c).
Figure 6. a–c.
Hep1-6 cells were transfected with miR-155 inhibitor (200nM) or mimic (100nM) (A), dye3 (red fluorescence) was used as positive control (B), the transfection efficiency was verified by RT-PCR. The expression of TNF mRNA in the Hepa1-6 cells transfection of inhibitor of miR-155 (anti-155) or mimic of miR-155 (155-mimic), followed by stimulation of D-GalN/TNF for another 48h, were detected using RT-PCR (C).
DISCUSSION
ALF is due to massive and rapid hepatocyte inflammation, resulting from excessive immune attack. We have shown recently that TNF plays a critical role in the pathogenesis of ALF (3), but the precise underlying mechanisms remain to be clarified. The discovery of miRNAs has revolutionized our understanding of many biological processes, including immunological regulation (16).
In our previous studies, 95 significant deregulated miRNAs were disclosed by microarray (p<0.01), and miR15b/16 was found to regulate the hepatocytes apoptosis in TNF-mediated ALF in mice. Following these studies, 122 changed miRNAs have been identified during the ALF in our study (p<0.05). Among these miRNAs, miR-155 is the highest up-regulated miRNA, over-expressed hepatic miR-155 closely correlated with the severity of liver damage. This finding was supported by the study that miR-155 was implicated as a central regulator of immune response (17) and as the new mediator of inflammation (18).
It is well known that spleen is an important immunological organ, particularly during massive tissue damage. Thus, splenic miR-155 was investigated; and we found that miR-155 was also significantly up-regulated in mice at 7 hrs post D-GalN/LPS challenge, which synchronized the peak of liver damage. However, it is unknown if up-regulated splenic miR-155 results in or from massive hepatic damage, which is currently being investigated.
Based on the findings above, we would like to know whether miR-155 has relationship with pro-inflammatory cytokines during the pathogenesis of ALF. Pro-inflammatory cytokines are produced following the immune response during the ALF (19). In this work, up-regulated circulating pro-inflammatory cytokines TNF and IL-6 in the mice with ALF were verified. Close correlation between miR-155 and TNF, and between miR-155 and IL-6, suggests that miR-155 might contribute to the development of ALF via regulating TNF and IL-6 mediated immunity. This is in line of the findings that miR-155 contributes to the pathogenesis of the rheumatoid arthritis via promoting TNF production (20).
First, it is of interest to identify the origin of over-expressed miR-155 in liver tissue in ALF. KCs are the resident macrophages of the liver; they play a significant role in the pathogenesis of ALF (21), including the phenotype and functions of macrophages, signaling pathways involved in macrophage functional status, and cell-cross talks of KCs with other immune cells. When activated by LPS, they produce and release numerous mediators, including TNF, IL-1, and IL-6 (22); more information on macrophages will promote a better understanding of the cellular molecular mechanisms of ALF and provide new insights for the development of therapeutic targets for ALF. Subsequently, we did the study in mouse macrophage cell line RAW264.7 cells in vitro, and we found that miR-155 was up-regulated with dose- and time-dependent manner after LPS induction, it was consisted with the study that miR-155 levels were increased upon LPS stimulation (8). It was hypothesized that during the ALF, miR-155 over-expresses in KCs and then further augments TNF secretion. Furthermore, to investigate miR-155 expression in the hepatocytes during the ALF, we designed the in vitro study according to the in vivo condition. D-GalN/TNF were used to induce the Hepa1-6 cells to apoptosis; apoptosis of hepatocyte was confirmed using Hoechst 33342 labeling. It was found miR-155 was gradually up-regulated according to the D-GalN/TNF induction. It was supposed that miR-155, at least, was over-expressed in both KCs and hepatocytes during ALF. It was assumed that up-regulated miR-155 from KCs or/and hepatocytes regulate TNF production singly or together during ALF.
Subsequently, our novel observation demonstrated that the levels of TNF mRNA and protein were up-regulated significantly in the Hepa1-6 cells and their supernatant respectively post D-GalN/TNF stimulation, and the levels of TNF mRNA were more stable. Over-expression of miR-155 significantly enhanced the expression level of TNF mRNA, but knocked down of miR-155 can dramatically reduce the expression of TNF mRNA. It indicated that miR-155 stabilizes TNF mRNA, thus increasing the release of this inflammatory mediator during the ALF. It was supported by observation that more TNF transcripts were observed in wild-type mice when compared with miR-155−/− mice (9).
In conclusion, we showed, for the first time, that miR-155 was up-regulated in TNF-mediated mice with ALF and closely related to the liver damage. We provided evidence for the hypothesis that over-expression of miR-155 is produced from active KCs or/and hepatocytes via RAW264.5 and Hepa1-6 cells studies in vitro. We also identified a likely novel mechanism of miR-155 to promote the TNF production and then stimulate and hepatic apoptosis during ALF. Such novel findings may supply the new mechanism of ALF and provide useful information to the development of therapeutic strategy against TNF during ALF. However, further studies are needed to evaluate the specificity and further mechanism of this miRNA in ALF. Generally, our findings suggest that miR-155 contributes to the pathogenesis of ALF by regulating pro-inflammatory cytokine TNF. These data provide useful information for both basic research and clinical therapeutic intervention.
Footnotes
Ethics Committee Approval: This study was approved by the Nanjing Medical University Clinic Institutional Review Board protocols. All applicable Nanjing Medical University Animal Ethics Committee guidelines for the care and use of animals were followed.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Design - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Supervision - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Resources - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Materials - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Data Collection and/or Processing - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Analysis and/or Interpretation - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Literature Search - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Writing Manuscript - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.; Critical Review - C.Y., D.C., G.Z., X.W., Y.Z., Q.Z., F.A.
Conflict of Interest: The authors have no conflicts of interest to declare.
Financial Disclosure: This study was funded by the National Natural Science Foundation of China (81502038, 81773227), the Youth Medical Talent of Jiangsu Province (QNRC2016187), and the Wuxi Medical Innovation Team (CXTD005).
REFERENCES
- 1.Wang X, Ning Q. Immune mediated liver failure. EXCLI J. 2014;13:1131–44. [PMC free article] [PubMed] [Google Scholar]
- 2.An F, Gong B, Wang H, et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis. 2012;17:702–16. doi: 10.1007/s10495-012-0704-7. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Wang H, Bao S, et al. Amelioration of liver injury by continuously targeted intervention against TNFRp55 in rats with acute-on-chronic liver failure. PLoS One. 2013;8:e68757. doi: 10.1371/journal.pone.0068757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gambardella G, Carissimo A, Chen A, et al. The impact of microRNAs on transcriptional heterogeneity and gene co-expression across single embryonic stem cells. Nat Commun. 2017;8:14126. doi: 10.1038/ncomms14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xuan Y, Yang H, Zhao L, et al. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett. 2015;360:89–105. doi: 10.1016/j.canlet.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 6.Lee HM, Kim TS, Jo EK. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. 2016;49:311–8. doi: 10.5483/BMBRep.2016.49.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Zeng Z, Shen X, Wu Z, Dong Y, Cheng JC. MicroRNA-146a-5p Negatively Regulates Pro-Inflammatory Cytokine Secretion and Cell Activation in Lipopolysaccharide Stimulated Human Hepatic Stellate Cells through Inhibition of Toll-Like Receptor 4 Signaling Pathways. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang RS, Hu GQ, Lin B, Lin ZY, Sun CC. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J Investig Med. 2010;58:961–7. doi: 10.2310/JIM.0b013e3181ff46d7. [DOI] [PubMed] [Google Scholar]
- 9.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 10.Shirozu K, Tokuda K, Marutani E, Lefer D, Wang R, Ichinose F. Cystathionine γ-lyase deficiency protects mice from galactosamine/lipopolysaccharide-induced acute liver failure. Antioxid Redox Signal. 2014;20:204–16. doi: 10.1089/ars.2013.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng YJ, Cheng SM, Teng YH, Shyu WC, Chen HL, Lee SD. Cordyceps sinensis prevents apoptosis in mouse liver with D-galactosamine/lipopolysaccharide-induced fulminant hepatic failure. Am J Chin Med. 2014;42:427–41. doi: 10.1142/S0192415X14500281. [DOI] [PubMed] [Google Scholar]
- 12.Yu DS, An FM, Gong BD, et al. The regulatory role of microRNA-1187 in TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int J Mol Med. 2012;29:663–8. doi: 10.3892/ijmm.2012.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng S, Tan H, Ling H, Yuan X. [Detecting overexpression level of HER2 gene in NSCLC by real-time quantitative PCR and the 2[−Delta Delta C(T)] method]. Zhongguo Fei Ai Za Zhi. 2011;14:938–42. doi: 10.3779/j.issn.1009-3419.2011.12.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W, Feng L, Zhang Y, et al. miR-15a induces cell apoptosis by targeting BCL2L2 and BCL2 in HPV-positive hypopharyngeal squamous cell carcinoma. Oncol Rep. 2016;36:2169–76. doi: 10.3892/or.2016.5049. [DOI] [PubMed] [Google Scholar]
- 15.Nati M, Haddad D, Birkenfeld AL, Koch CA, Chavakis T, Chatzigeorgiou A. The role of immune cells in metabolism-related liver inflammation and development of non-alcoholic steatohepatitis (NASH) Rev Endocr Metab Disord. 2016;17:29–39. doi: 10.1007/s11154-016-9339-2. [DOI] [PubMed] [Google Scholar]
- 16.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–57. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 17.Singh UP, Murphy AE, Enos RT, et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143:478–89. doi: 10.1111/imm.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandyopadhyay S, Long ME, Allen LA. Differential expression of microRNAs in Francisella tularensis-infected human macrophages: miR-155-dependent downregulation of MyD88 inhibits the inflammatory response. PLoS One. 2014;9:e109525. doi: 10.1371/journal.pone.0109525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan BZ, Yang BS, Li H, et al. The therapeutic effect of CORM-3 on acute liver failure induced by lipopolysaccharide/D-galactosamine in mice. Hepatobiliary Pancreat Dis Int. 2016;15:73–80. doi: 10.1016/S1499-3872(15)60044-3. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Tian F, Wang F. Rheumatoid arthritis-associated microRNA-155 targets SOCS1 and upregulates TNF-α and IL-1β in PBMCs. Int J Mol Sci. 2013;14:23910–21. doi: 10.3390/ijms141223910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng T, Zhang CL, Xiao M, Yang R, Xie KQ. Critical Roles of Kupffer Cells in the Pathogenesis of Alcoholic Liver Disease: From Basic Science to Clinical Trials. Front Immunol. 2016;7:538. doi: 10.3389/fimmu.2016.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosoian A, Zhang L, Hong F, et al. HIV infection of Kupffer cells results in an amplified proinflammatory response to LPS. J Leukoc Biol. 2016 doi: 10.1189/jlb.3HI0516-242R. [DOI] [PMC free article] [PubMed] [Google Scholar]