Abstract
Use of therapeutic peptides in cancer therapy has been receiving considerable attention in the recent years. Present study describes the development of computational models for predicting and discovering novel anticancer peptides. Preliminary analysis revealed that Cys, Gly, Ile, Lys, and Trp are dominated at various positions in anticancer peptides. Support vector machine models were developed using amino acid composition and binary profiles as input features on main dataset that contains experimentally validated anticancer peptides and random peptides derived from SwissProt database. In addition, models were developed on alternate dataset that contains antimicrobial peptides instead of random peptides. Binary profiles-based model achieved maximum accuracy 91.44% with MCC 0.83. We have developed a webserver, which would be helpful in: (i) predicting minimum mutations required for improving anticancer potency; (ii) virtual screening of peptides for discovering novel anticancer peptides, and (iii) scanning natural proteins for identification of anticancer peptides (http://crdd.osdd.net/raghava/anticp/).
Cancer with leading cause of deaths remains the matter of health concern for both developed and developing countries1. Despite the advances in cancer treatments, mortality rate due to this deadly disease is still very high1. Owing to the development of resistance by cancer cells towards current anti-cancer chemotherapeutic drugs, there is an urgent need to add new weapons in the anti-cancer drug arsenal to fight with this deadly disease. In the last decade, small peptides having anticancer properties have emerged as a potential alternative approach for cancer therapy2. Peptide-based therapy has numerous advantages over small molecules that involve high specificity, low production cost, high tumor penetration, ease of synthesis and modification etc3.
Anticancer peptides (ACPs) are small (5–30 amino acids) peptides, often derived from antimicrobial peptides (AMPs) and are cationic in nature4. Previous studies have demonstrated that many cationic AMPs, which are toxic to bacteria but not to normal cells, show a broad spectrum cytotoxicity against various cancer cells5. Although ACP is a rapidly emerging field, their mechanism of action remains elusive. However, few studies have suggested that there are few differences between the cell membranes of cancer and normal cells and selective killing of cancer cells by certain ACPs could be due to these differences4,5. In this context, electrostatic interactions between cationic amino acids of ACPs and anionic components of cancer cell membranes are suggested to be one of the major contributing factors in the selective killing of cancer cells by ACPs4. Also, high membrane fluidity and high cell-surface area6,7 of cancer cells compared to untransformed cells lead to enhance the lytic activity of ACPs and binding of the increased number of ACPs, respectively. In addition, few ACPs induce apoptosis (program cell death) by disrupting mitochondrial membrane when delivered into the cancer cells8. Many peptide-based therapies to treat various tumor types are currently being evaluated in various phases of preclinical and clinical trials9,10,11,12. The success of these peptides in clinics has open the door for ACPs to reach clinical settings.
Keeping in mind the immense therapeutic importance of ACPs, in the present study, we have made a systematic attempt to develop in silico methods for the prediction and designing of ACPs. Support vector machine (SVM) based models using various features of peptides like amino acid composition, dipeptide composition and binary profile pattern have been developed. In addition, models discriminating ACPs from AMPs have also been developed. Binary profile-based SVM model using NT10 dataset achieved maximum accuracy of 91.44% with MCC and AUC values 0.83 and 0.94 respectively. To assist scientific community, for the first time, a user-friendly webserver, AntiCP, has been developed to predict and design highly efficacious ACPs.
Results
Compositional analysis
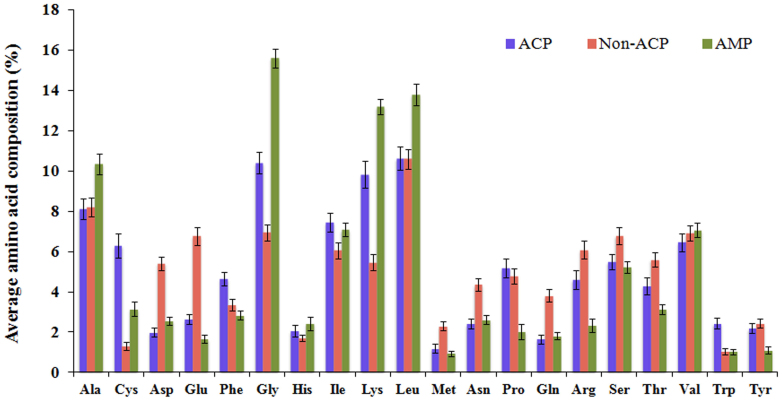
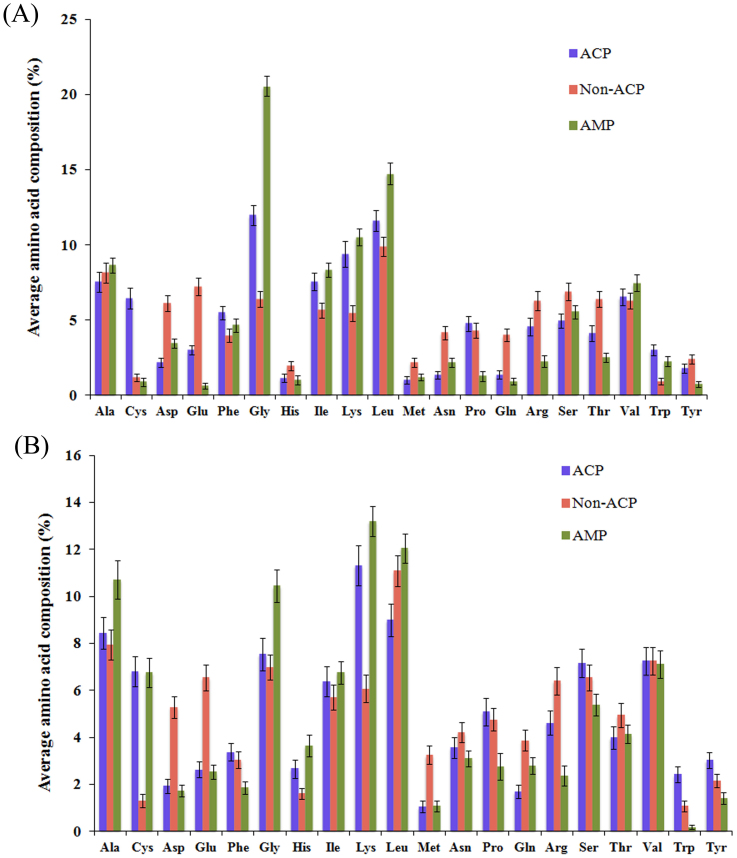
We wanted to develop in silico models, which can differentiate ACPs from non-ACPs, as well as ACPs from AMPs. Therefore, first we sought to determine the frequency of occurrence of all 20 amino acids in these peptides. For this, percent average composition of amino acids in ACPs, non-ACPs (random peptides) and AMPs were calculated and compared. As shown in Figure 1, certain residues, including Gly, Lys, Cys, Phe, Ile, and Trp were found to be abundant in ACPs compared to non-ACPs while Gly, Ala, Lys and Leu were abundant in AMPs compared to ACPs and non-ACPs. Since terminal residues play crucial roles in biological functions of peptides13, we computed and compared the percent average amino acid composition of N-terminal and C-terminal residues (split amino acid composition) in these peptides. As shown in Figure 2A and 2B, average amino acid compositions of terminal residues are more or less similar to whole amino acid composition. However, among N-terminal residues, only Cys was found to be in a higher proportion in ACPs compared to both AMPs and non-ACPs. In C-terminal residue analysis, Tyr and Trp were found to abundant in ACPs compared to both AMPs and non-ACPs (Figure 2B).
Figure 1. Comparison of average whole amino acid composition of anticancer, non-anticancer, and antimicrobial peptides.
Figure 2. Comparison of average amino acid composition of ten (A) N- and (B) C-terminal residues of anticancer, non-anticancer, and antimicrobial peptides.
Residue preference
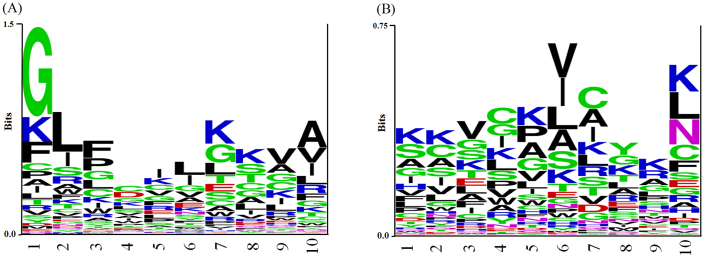
In order to understand residue preference at both termini of peptides, we computed sequence logos. The sequence logos of 10 N-terminal and 10 C-terminal residues are shown in Figure 3A and 3B. As shown, no exclusive preference of residues was observed except Gly at the first position at N-terminus. However, there are few residues like Leu, Lys, Ala and Phe at N-terminus and Val, Cys, Leu and Lys at C-terminus which are also preferred but relatively less preferred than Gly at various positions.
Figure 3. Sequence logo of (A) first ten residues of N-terminus and (B) last ten residues of C-terminus of anticancer peptides where size of residue is proportional to its propensity.
Support vector machine models
SVM models were developed on both realistic datasets (main datasets and alternate datasets) and balanced datasets (balanced dataset-1 and balanced dataset-2) using amino acid composition, dipeptide composition, and binary profiles as input features.
SVM model based on amino acid composition
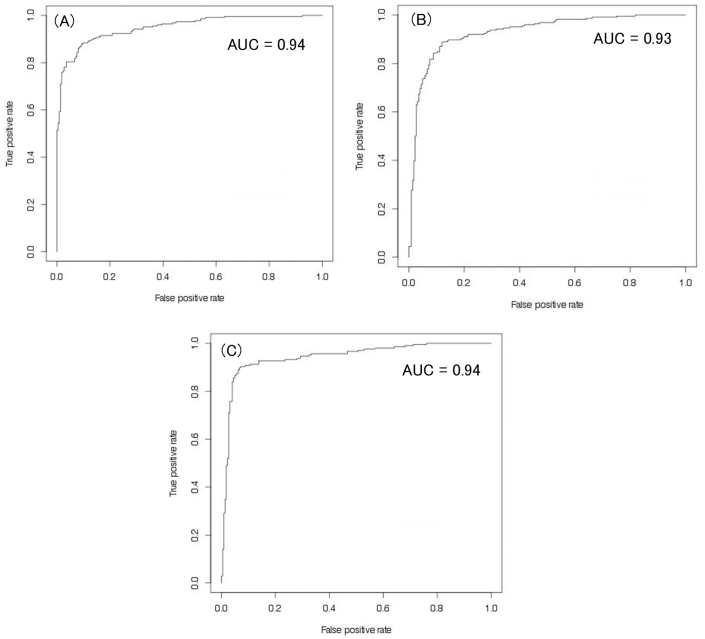
Since certain residues were found to be abundant over others in ACPs and AMPs, ACPs can be discriminated from non-ACPs and AMPs on the basis of their amino acid composition. Therefore, we have developed whole amino acid composition-based SVM models. The performance of whole composition-based SVM models has been shown in Table 1 and 2. The whole composition-based SVM model developed on balanced dataset-1 achieved maximum accuracy of 88.89% with MCC and AUC values 0.78 and 0.94 respectively (Table 1 and Figure 4A). In addition, SVM models based on split amino acid composition (NT5, CT5, NT5CT5, NT10, CT10, and NT10CT10) were also developed. The performance of these models is summarized in Table 1. Model developed with NT10CT10 dataset performed similar to whole composition-based model and achieved maximum accuracy of 88.4% with MCC and AUC values of 0.77 and 0.93, respectively (Table 1). The performance of models developed on main dataset was comparable to models developed on balanced dataset-1 (Table 1).
Table 1. The performance of amino acid composition-based models on main dataset.
| Balanced dataset-1 | |||||
|---|---|---|---|---|---|
| Dataset | Sensitivity | Specificity | Accuracy | MCC | AUC |
| Whole peptide | 88.00 | 89.78 | 88.89 | 0.78 | 0.94 |
| NT5 | 81.33 | 80.44 | 80.89 | 0.62 | 0.86 |
| CT5 | 71.11 | 73.78 | 72.44 | 0.45 | 0.78 |
| NT5CT5 | 82.22 | 83.56 | 82.89 | 0.66 | 0.88 |
| NT10 | 89.37 | 84.34 | 86.91 | 0.74 | 0.92 |
| CT10 | 79.23 | 85.86 | 82.47 | 0.65 | 0.88 |
| NT10CT10 | 89.37 | 87.37 | 88.40 | 0.77 | 0.93 |
| Main dataset | |||||
| Whole peptide | 88.89 | 85.29 | 85.62 | 0.52 | 0.95 |
| NT5 | 73.78 | 88.22 | 86.91 | 0.47 | 0.86 |
| CT5 | 61.78 | 87.64 | 85.29 | 0.38 | 0.80 |
| NT5CT5 | 74.67 | 94.44 | 92.65 | 0.61 | 0.90 |
| NT10 | 82.61 | 92.76 | 91.82 | 0.63 | 0.91 |
| CT10 | 78.26 | 83.80 | 83.29 | 0.43 | 0.89 |
| NT10CT10 | 88.89 | 90.55 | 90.39 | 0.62 | 0.94 |
Table 2. Performances of amino acid composition-based models on alternate dataset.
| Balanced dataset-2 | |||||
|---|---|---|---|---|---|
| Dataset | Sensitivity | Specificity | Accuracy | MCC | AUC |
| Whole peptide | 84.44 | 86.22 | 85.33 | 0.71 | 0.90 |
| NT5 | 84.00 | 84.89 | 84.44 | 0.69 | 0.89 |
| CT5 | 85.33 | 69.78 | 77.56 | 0.56 | 0.83 |
| NT5CT5 | 84.00 | 84.89 | 84.44 | 0.69 | 0.89 |
| NT10 | 81.64 | 86.67 | 84.26 | 0.68 | 0.90 |
| CT10 | 77.29 | 85.78 | 81.71 | 0.63 | 0.87 |
| NT10CT10 | 85.51 | 89.78 | 87.73 | 0.75 | 0.92 |
| Aletrnate dataset | |||||
| Whole peptide | 73.78 | 76.02 | 75.70 | 0.37 | 0.79 |
| NT5 | 68.00 | 62.03 | 62.87 | 0.21 | 0.70 |
| CT5 | 69.08 | 72.25 | 71.83 | 0.30 | 0.76 |
| NT5CT5 | 81.33 | 60.93 | 63.81 | 0.30 | 0.79 |
| NT10 | 69.08 | 72.25 | 71.83 | 0.30 | 0.76 |
| CT10 | 74.88 | 72.69 | 72.98 | 0.34 | 0.79 |
| NT10CT10 | 75.36 | 70.77 | 71.38 | 0.33 | 0.80 |
Figure 4. ROC plot shows performance of models developed using (A) amino acid composition (B) dipetide composition, and (B) binary profiles of patterns (NT10 dataset).
Similarly, SVM models on balanced dataset-2 and alternate dataset were also developed using amino acid composition as input features. The performances of these models are summarized in Table 2. The overall performances of the models developed with balanced dataset-2 were more or less similar to models developed with balanced dataset-1. The whole composition-based SVM model developed on balanced dataset-2 achieved maximum accuracy of 85.33% with MCC and AUC values 0.71 and 0.90 respectively. Similarly, models based on split amino acid composition were also developed (Table 2) and the model developed on NT10CT10 dataset achieved maximum accuracy of 87.73% with MCC and AUC values 0.75 and 0.92 respectively (Table 2). Amino acid composition based models developed on alternate dataset performed poorer than the models developed on balanced dataset-2 (Table 2).
Dipeptide composition-based SVM model
In many previous studies, SVM model based on dipeptide composition has been developed to discriminate different classes of peptides14,15,16. Dipeptide composition is a simple feature, and it encapsulates information of the amino acid fraction as well as local order of amino acids. Therefore, SVM models based on dipeptide composition have been constructed on all the datasets. Performances of dipeptide composition-based models are summarized in Table 3 and 4. Models developed on balanced dataset-1 achieved maximum accuracy of 87.78% with an MCC and AUC values 0.76 and 0.93 respectively (Table 3, Figure 4B). For balanced dataset-2, models developed on whole peptide and NT5CT5 datasets achieved maximum accuracy of 86.89% with MCC and AUC values 0.74 and 0.91 respectively.
Table 3. Performance of dipeptide composition-based models on main dataset.
| Balanced Dataset-1 | |||||
|---|---|---|---|---|---|
| Dataset | Sensitivity | Specificity | Accuracy | MCC | AUC |
| Whole peptide | 88.44 | 87.11 | 87.78 | 0.76 | 0.93 |
| NT5 | 73.33 | 86.67 | 80.00 | 0.61 | 0.86 |
| CT5 | 60.44 | 79.56 | 70.00 | 0.41 | 0.73 |
| NT5CT5 | 78.22 | 88.44 | 83.33 | 0.67 | 0.89 |
| NT10 | 81.16 | 88.89 | 84.94 | 0.70 | 0.91 |
| CT10 | 71.50 | 86.87 | 79.01 | 0.59 | 0.85 |
| NT10CT10 | 83.09 | 88.38 | 85.68 | 0.72 | 0.91 |
| Main Dataset | |||||
| Whole peptide | 90.22 | 84.80 | 85.29 | 0.52 | 0.94 |
| NT5 | 71.11 | 88.89 | 87.27 | 0.46 | 0.85 |
| CT5 | 66.22 | 82.04 | 80.61 | 0.33 | 0.81 |
| NT5CT5 | 80.00 | 85.69 | 85.17 | 0.47 | 0.89 |
| NT10 | 83.09 | 88.63 | 88.11 | 0.54 | 0.92 |
| CT10 | 75.85 | 86.12 | 85.17 | 0.45 | 0.87 |
| NT10CT10 | 84.54 | 85.43 | 85.34 | 0.50 | 0.91 |
Table 4. Performance of dipeptide composition-based model on alternate dataset.
| Balanced dataset-2 | |||||
|---|---|---|---|---|---|
| Dataset | Sensitivity | Specificity | Accuracy | MCC | AUC |
| Whole peptide | 88.89 | 84.89 | 86.89 | 0.74 | 0.91 |
| NT5 | 87.11 | 86.67 | 86.89 | 0.74 | 0.89 |
| CT5 | 76.00 | 75.56 | 75.78 | 0.52 | 0.83 |
| NT5CT5 | 87.56 | 86.22 | 86.89 | 0.74 | 0.91 |
| NT10 | 84.06 | 86.67 | 85.42 | 0.71 | 0.91 |
| CT10 | 84.06 | 75.56 | 79.63 | 0.60 | 0.86 |
| NT10CT10 | 85.51 | 83.56 | 84.49 | 0.69 | 0.89 |
| Alternate dataset | |||||
| Whole peptide | 77.78 | 74.78 | 75.2 | 0.39 | 0.79 |
| NT5 | 74.22 | 62.17 | 63.87 | 0.26 | 0.75 |
| CT5 | 71.50 | 70.70 | 70.81 | 0.30 | 0.78 |
| NT5CT5 | 73.78 | 63.41 | 64.87 | 0.26 | 0.77 |
| NT10 | 71.50 | 70.70 | 70.81 | 0.30 | 0.78 |
| CT10 | 69.57 | 64.58 | 65.24 | 0.24 | 0.74 |
| NT10CT10 | 78.26 | 64.94 | 66.71 | 0.30 | 0.79 |
Binary profile based SVM model
Since apart from composition, order of amino acid is also important feature, therefore, to implement information about frequency as well as the order of residues, we developed models based on binary profiles of peptides. We have used the following three approaches.
N-terminal (NT) approach
For balanced dataset-1, the accuracies of the models developed on NT5 and NT10 datasets were 80.89% and 83.95% with MCC 0.62, 0.68 and AUC 0.87, 0.91 respectively (Table 5). For balanced dataset-2, models developed on NT5 and NT10 datasets achieved maximum accuracies 88.44% and 91.44% with MCC 0.77 and 0.83 and AUC values 0.93 and 0.94 respectively (Table 6 and Figure 4C).
Table 5. Performance of binary profile-based model on main dataset.
| Balanced dataset-1 | |||||
|---|---|---|---|---|---|
| Dataset | Sensitivity | Specificity | Accuracy | MCC | AUC |
| NT5 | 78.67 | 83.11 | 80.89 | 0.62 | 0.87 |
| CT5 | 63.11 | 86.22 | 74.67 | 0.51 | 0.79 |
| NT5CT5 | 81.78 | 86.22 | 84.00 | 0.68 | 0.89 |
| NT10 | 81.16 | 86.67 | 83.95 | 0.68 | 0.91 |
| CT10 | 75.36 | 84.34 | 79.75 | 0.60 | 0.84 |
| NT10CT10 | 81.64 | 88.38 | 84.94 | 0.70 | 0.91 |
| Main dataset | |||||
| NT5 | 80.00 | 87.33 | 86.67 | 0.50 | 0.89 |
| CT5 | 70.67 | 84.76 | 83.47 | 0.40 | 0.83 |
| NT5CT5 | 74.22 | 89.16 | 87.80 | 0.49 | 0.88 |
| NT10 | 80.19 | 92.52 | 91.38 | 0.60 | 0.90 |
| CT10 | 75.85 | 85.33 | 84.45 | 0.44 | 0.87 |
| NT10CT10 | 81.16 | 89.76 | 88.96 | 0.55 | 0.89 |
Table 6. Performance of binary profile-based model on alternate dataset.
| Balanced dataset-2 | |||||
|---|---|---|---|---|---|
| Dataset | Sensitivity | Specificity | Accuracy | MCC | AUC |
| NT5 | 87.11 | 89.78 | 88.44 | 0.77 | 0.93 |
| CT5 | 82.67 | 73.78 | 78.22 | 0.57 | 0.83 |
| NT5CT5 | 88.89 | 89.78 | 89.33 | 0.79 | 0.93 |
| NT10 | 89.37 | 93.33 | 91.44 | 0.83 | 0.94 |
| CT10 | 85.51 | 72.44 | 78.70 | 0.58 | 0.86 |
| NT10CT10 | 85.02 | 96 | 90.74 | 0.82 | 0.94 |
| Alternate dataset | |||||
| NT5 | 67.56 | 73.69 | 72.82 | 0.31 | 0.75 |
| CT5 | 71.56 | 71.43 | 71.45 | 0.31 | 0.75 |
| NT5CT5 | 70.22 | 75.87 | 75.08 | 0.35 | 0.79 |
| NT10 | 71.01 | 71.96 | 71.83 | 0.31 | 0.77 |
| CT10 | 65.22 | 78.08 | 76.38 | 0.33 | 0.77 |
| NT10CT10 | 75.85 | 69.23 | 70.1 | 0.32 | 0.79 |
C-terminal (CT) approach
Similarly, models were developed using 5 and 10 C-terminal residues and performances are summarized in Table 5 and 6. For balanced dataset-1, model developed using 5 and 10 C-terminal residues (CT5 and CT10) achieved accuracies 74.67% and 79.75% with MCC 0.51, 0.60 and AUC 0.79, 0.84 respectively (Table 5). For balanced dataset-2, models developed on CT5 and CT10 datasets achieved maximum accuracies 78.22% and 78.7% with MCC 0.57 and 0.58 and AUC values 0.83 and 0.86 respectively (Table 6).
N + C-terminal (NTCT) approach
Similar strategy, as used in the N- and C-terminal approaches, was applied in this approach also. The comparative performances of SVM model based on N + C terminal residues are shown in Table 5 and 6. For balanced dataset-1, model developed on NT10CT10 datasets achieved maximum accuracy 84.94% with MCC 0.70 and AUC 0.91 (Table 5). For balanced dataset-2, model developed on NT10CT10 dataset achieved maximum accuracy 90.74% with MCC 0.82 and AUC 0.94 (Table 6).
Performance on independent dataset
In order to validate our models, we have evaluated the performances of our best models on an independent dataset. The amino acid composition-based model achieved accuracy 86% with MCC 0.72 while model based on binary profiles (NT10) achieved accuracy 89% with MCC 0.78. These results indicate that our models performed equally well on an independent dataset suggesting that our models are not over trained and may also work in real life. We evaluated the performance of both models (amino acid composition and binary (NT10) based models) using five-fold and ten-fold cross-validation and achieved similar results. In addition, we evaluated the performance of our models 100 times, each time training and testing set of peptides were reshuffled randomly. We computed average performance of these 100 models with standard error, which is summarized in the supplementary information. The average performance of our models indicates that even after repeating 100 times, models performed similarly. This evaluation further demonstrates the reliability of models developed in this study.
Implementation and description of webserver
In order to serve the scientific community, the best SVM-based models were implemented to build a webserver (AntiCP, Figure 5) using a CGI/Perl script. Various tools have been integrated to assist users to design and predict ACPs (Figure 5). Users may submit the peptide, and the server will generate all the possible single substitution mutants of a given peptide. Besides generating mutants, server will also give prediction status as ACP or non-ACP. Along with this, server calculates key physico-chemical properties in a Tabular format. In addition, user can discover novel ACPs by screening multiple peptides at a time. For this, virtual screening tool has been integrated where user has to submit multiple peptide sequences in FASTA format. Another powerful tool is protein scan, which will be useful for the detection of putative ACP regions in the protein. Here, user may submit the protein sequence, and overlapping peptides will be generated by the server, where all the peptides will be clickable. Sorting of results in ascending/descending order of their values is another attractive feature provided with the web server. AntiCP is freely accessible at http://crdd.osdd.net/raghava/anticp.
Figure 5. Schematic representation of AntiCP webserver (developed with scienceslides software, http://www.visiscience.com/) and its various modules.
Discussion
The peptide-based therapeutics is gaining tremendous interest nowadays2,3, which has been reflected in the papers published in the last five years. Many peptides-based strategies for targeting and delivering therapeutics to various tumor types have been used over the years2, and few of them have successfully translated into the clinics. In this context, ACPs have also been emerged as promising candidates for cancer therapy4. Identification and development of novel ACPs in the wet lab is extremely time consuming and labor intensive approach. Therefore, development of in silico methods, which can predict ACPs prior to their synthesis is the need of the hour. Such prediction methods are not only helpful for biologists for designing effective ACPs, but also save money and time. The present study describes an in silico method for designing and predicting ACPs. For the development of SVM models, both positive and negative examples are required. Therefore, we have collected 225 experimentally validated ACPs from literature and from various databases17,18,19. Since, experimentally validated non-ACPs were not reported in the literature, equal number of negative examples were generated randomly from SwissProt proteins and these peptides were assumed to be non-ACPs. This approach has been used in number of previous studies15,16,20 where sufficient amount of negative examples were not available in the literature. As it was observed that most of the ACPs are derived from AMPs, we have collected AMPs without anti-cancer activities (no anti-cancer activities reported in the literature) and developed alternate dataset, which comprises ACPs as positive examples and AMPs without anti-cancer activities as negative examples. The models developed on this dataset discriminated ACPs from AMPs.
A preliminary analysis of amino acid composition has shown that certain residues are dominated in ACPs/AMPs. These differences in amino acid composition between ACPs/AMPs and non-ACPs prompted us to develop SVM models based on amino acid composition and dipeptide composition of peptides. The whole composition-based model performed reasonably well and model developed on balanced dataset-1 performed the best among the rest of the whole composition-based models. However, models developed on split amino acid compositions could not perform better than the whole composition-based models, and it was expected as there was not significant difference observed in amino acid composition between ACPs, non-ACPs and AMPs at N- and C-terminal residues (Figure 1 and 2). We compared the performance of models developed on balanced and realistic datasets and got similar results.
Dipeptide composition is an attractive feature which encapsulates the information of fraction of amino acids as well as their local order. Therefore, we have developed SVM models using dipeptide composition. As shown in the result section, performance of dipeptide composition-based models performed comparable to amino acid composition-based model.
It is well known that peptide's function is strongly related to its residue order. Plethora of studies has suggested that the membrane interaction and insertion of membrane-active peptides (e.g. AMPs, cell penetrating peptides, ACPs, etc.) could be due to their conformation (e.g. helical, β stranded, etc.)21,22, which can be associated to a particular order of amino acids or distribution of residues. Thus, apart from composition of amino acids, order of amino acids is also important feature and might be associated with anti-cancer properties of ACPs. Therefore, to incorporate the order information, binary profiles of the peptides were generated. Binary profiles encapsulate information of both composition and order of amino acids. In many previous studies, binary profiles based models have been used to discriminate various classes of peptide/proteins15,16. In the present study, binary-based models performed reasonably well. In order to provide service to the scientific community, we have implemented best models in a webserver, AntiCP, which is freely available. We hope that our method will provide momentum in the discovery and designing of novel efficient ACPs.
Methods
Datasets
We have extracted 225 experimentally validated anticancer peptides from literature and databases like antimicrobial database (APD, http://aps.unmc.edu/AP/main.php)17, collection of antimicrobial peptides (CAMP, http://www.bicnirrh.res.in/antimicrobial)18, and database of anuran defense peptides (DADP, http://split4.pmfst.hr/dadp/)19. Majority of these peptides are AMPs with a broad spectrum anticancer activities. All these peptides were unique and considered as positive examples. Since there are very few experimentally proved non-anticancer peptides, we derived 2250 random peptides from SwissProt proteins. In this study, we assign these random peptides as non-ACPs (negative examples), though it is possible that some of these random peptides have anticancer properties. We also extracted AMPs from above databases like APD, CAMP, DADP for which no anticancer activity was reported in the literature and considered as non-ACPs. Following datasets were derived from the above data.
Main dataset
This dataset contains 225 experimentally validated anticancer (positive examples) and 2250 random or potential non-anticancer peptides (negative examples).
Alternate dataset
This dataset contains 225 experimentally validated anticancer peptides and 1372 non-anticancer (AMPs without anticancer activities, negative examples).
Balanced datasets
It is a well known fact that classification techniques, particularly machine learning techniques performed best on balanced datasets. Thus, we generated balanced datasets for both main and alternate datasets. Our main balanced dataset contains 225 anticancer and 225 non-anticancer or random peptides (randomly obtained 2250 SwissProt peptides). Similarly, alternate balanced dataset contains 225 anticancer and 225 non-anticancer or AMPs (randomly obtained from 1372 AMPs).
Independent dataset
For developing independent dataset, we collected 50 experimentally validated ACPs from literature and patents and an equal number of random peptides were generated from SwissProt proteins and considered as negative examples. None of the peptides in independent dataset is identical to peptides in training or testing dataset.
Support vector machine
In this study, we developed models for discriminating anticancer and non-anticancer peptides using a highly successful machine learning technique, support vector machine (SVM)23. We developed SVM models using SVMlight Version 6.02 package. Various features, including amino acid composition, dipeptide composition and binary profile of pattern were used as input features.
Residue composition as input features
In order to develop SVM models based on machine learning techniques, one needs fixed length input features. Our dataset contains peptides of variable length; thus we have computed composition profile of peptides. In this study, we computed amino acid and dipeptide composition where information is encapsulated in a vector of 20 and 400 dimensions respectively. The calculation of amino acid and dipeptide composition was described previously15,16.
Binary profile of patterns
Binary profiles is a key feature and has been used in a number of existing methods.
It encapsulates information of both composition and order of amino acid in peptides. Therefore, binary profiles for first 5 and 10 residues from N- and C-terminus were generated for each peptide, where each amino acid is represented by a vector of dimensions of 20 (e.g. Ala by 1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0) as described previously15.
Sequence logos
The sequence logos, which provides information about the position specific frequency of amino acids in peptide, were generated using the WebLogo software24.
Performance measures
The performance of models were evaluated using threshold-dependent and threshold-independent parameters. Sensitivity (Sn), specificity (Sp), accuracy (Ac) and Matthew's correlation coefficient (MCC) were used as threshold-dependent parameters as previously described15. For threshold-independent parameter, ROC (Receiver Operating Characteristic) for all of the models were created in order to evaluate the performance of models.
Cross validation technique
The ten-fold cross validation technique was used to evaluate the performance of various SVM models. In this technique, sequences are randomly divided into ten sets, of which nine sets are used for training and the remaining tenth set for testing. The process is repeated ten times in such a way that each set is used once for testing. Final performance is obtained by averaging the performance of all the ten sets.
Supplementary Material
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 3
Supplementary Dataset 4
Acknowledgments
Authors are thankful to funding agencies Council of Scientific and Industrial Research (project Open Source Drug discovery and GENESIS BSC0121) and Department of Biotechnology (project BTISNET), Govt. of India for financial support.
Footnotes
The authors declare no competing financial interests.
Author Contributions A.T. and P.K. collected the data and created the datasets. A.T., R.K., P.K. and K.C. developed computer programs, implemented SVM and created the back end server. A.T., A.G., P.K. and K.C. developed the front end user interface. A.G. and P.K. analyzed the results and wrote the manuscript. G.P.S.R. conceived and coordinated the project, helped in the interpretation of data, refined the drafted manuscript and gave overall supervision to the project. All of the authors read and approved the final manuscript.
References
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 (2013). [DOI] [PubMed] [Google Scholar]
- Thundimadathil J. Cancer treatment using peptides: current therapies and future prospects. J Amino Acids 2012, 967347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe P., Lisowski V., Martinez J. & Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today 15, 40–56 (2010). [DOI] [PubMed] [Google Scholar]
- Mader J. S. & Hoskin D. W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin Investig Drugs 15, 933–946 (2006). [DOI] [PubMed] [Google Scholar]
- Hoskin D. W. & Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim Biophys Acta 1778, 357–375 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowska K., Nowak J., Kwiatkowski B. & Cichorek M. ESR study of plasmatic membrane of the transplantable melanoma cells in relation to their biological properties. Exp Toxicol Pathol 51, 89–92 (1999). [DOI] [PubMed] [Google Scholar]
- Sok M., Sentjurc M. & Schara M. Membrane fluidity characteristics of human lung cancer. Cancer Lett 139, 215–220 (1999). [DOI] [PubMed] [Google Scholar]
- Ellerby H. M. et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med 5, 1032–1038 (1999). [DOI] [PubMed] [Google Scholar]
- Hariharan S. et al. Assessment of the biological and pharmacological effects of the alpha nu beta3 and alpha nu beta5 integrin receptor antagonist, cilengitide (EMD 121974), in patients with advanced solid tumors. Ann Oncol 18, 1400–1407 (2007). [DOI] [PubMed] [Google Scholar]
- Gregorc V. et al. Phase I study of NGR-hTNF, a selective vascular targeting agent, in combination with cisplatin in refractory solid tumors. Clin Cancer Res 17, 1964–1972 (2011). [DOI] [PubMed] [Google Scholar]
- Khalili P. et al. A non-RGD-based integrin binding peptide (ATN-161) blocks breast cancer growth and metastasis in vivo. Mol Cancer Ther 5, 2271–2280 (2006). [DOI] [PubMed] [Google Scholar]
- Deplanque G. et al. Phase II trial of the antiangiogenic agent IM862 in metastatic renal cell carcinoma. Br J Cancer 91, 1645–1650 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos L. Jr Antibacterial peptides and proteins with multiple cellular targets. J Pept Sci 11, 697–706 (2005). [DOI] [PubMed] [Google Scholar]
- Petrilli P. Classification of protein sequences by their dipeptide composition. Comput Appl Biosci 9, 205–209 (1993). [DOI] [PubMed] [Google Scholar]
- Gautam A. et al. In silico approaches for designing highly effective cell penetrating peptides. J Transl Med 11, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. et al. Computational approach for designing tumor homing peptides. Sci Rep 3, 1607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li X. & Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res 37, D933–937 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Karnik S., Barai R. S., Jayaraman V. K. & Idicula-Thomas S. CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res 38, D774–780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novkovic M., Simunic J., Bojovic V., Tossi A. & Juretic D. DADP: the database of anuran defense peptides. Bioinformatics 28, 1406–1407 (2012). [DOI] [PubMed] [Google Scholar]
- Sanders W. S., Johnston C. I., Bridges S. M., Burgess S. C. & Willeford K. O. Prediction of cell penetrating peptides by support vector machines. PLoS Comput Biol 7, e1002101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. B., Wang X. F., Wang H. Y., Liu Y. & Chen Y. Studies on mechanism of action of anticancer peptides by modulation of hydrophobicity within a defined structural framework. Mol Cancer Ther 10, 416–426 (2011). [DOI] [PubMed] [Google Scholar]
- Eiriksdottir E., Konate K., Langel U., Divita G. & Deshayes S. Secondary structure of cell-penetrating peptides controls membrane interaction and insertion. Biochim Biophys Acta 1798, 1119–1128 (2010). [DOI] [PubMed] [Google Scholar]
- Joachims T. Making large-scale support vector machine learning practical, 169–184 (Scholkopf B., Burges C., & Smola A., Cambridge, MA: MIT Press 1999). [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M. & Brenner S. E. WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 3
Supplementary Dataset 4