Abstract
Rotavirus groups A, B, and C (RVA, RVB, and RVC, respectively) have been the most prevalent and pathogenic in pigs. To date, immunohistochemistry is only available for RVA because of the lack of commercial antibodies for RVB and RVC. We developed a novel in situ hybridization RNA-based chromogenic technique (ISH-RNA) to detect and subtype RVA, RVB, and RVC. We evaluated 33 samples that were reverse-transcription PCR positive for RVA, RVB, and/or RVC. ISH-RNA was able to detect as few as 103 RV RNA copies/mL. The new ISH-RNA test can be useful for routine investigation of rotavirus enteritis in order to guide strategies for control of the infection in pigs, but a full validation study needs to be completed. Pathogenesis studies may be conducted using ISH-RNA based on the identification of replicating virus.
Keywords: Diarrhea, in situ hybridization, rotaviruses, subtyping, swine, viral enteritis
Rotaviruses (RVs, family Reoviridae) are one of the most important causes of diarrhea in young mammals, including children and piglets. RVs are classified into 8 groups (A–H), according to their antigenic characteristics and sequencing of viral protein 6.9,10,14 Rotavirus groups A (RVA), B (RVB), and C (RVC) are distributed endemically in pigs in the United States.6,12 Rotavirus E (RVE) was reported in a single pig sample,2 and rotavirus H (RVH) has been described in piglets with diarrhea in Brazil, Japan, Africa, and the United States.10,11,13,15
Rotaviruses are double-stranded RNA (dsRNA) viruses with 11 genomic segments encoding 6 structural viral proteins (VP1–VP4, VP6, VP7) and 5 or 6 nonstructural proteins (NSP1–NSP6).4 Rotaviruses are commonly detected by PCR in feces from infected piglets. Given the endemic and widespread distribution of RVA, RVB, and RVC in swine herds, more than one RV group is often identified in the same sample in many diagnostic submissions.12 As a result, it is not possible to determine the specific RV group causing atrophic enteritis, which is the histologic lesion typically associated with RV infection. To date, commercial antibodies are available only for RVA, which has been routinely used to detect RVA within histologic lesions using immunohistochemistry. Differential PCR for each RV group in infected tissues only indicates the presence of the virus in the sample but does not allow the evaluation of the actual contribution of each group to the histologic lesions. This information would be extremely important for driving decisions in the field regarding disease prevention of RV strains given that the commercial vaccine only contains RVA, and cross-protection among the other groups does not occur.3
In situ hybridization (ISH) is a method used to detect specific nucleic acid sequences within formalin-fixed, paraffin-embedded (FFPE) tissue sections. Similar to PCR, ISH is based on DNA and RNA sequences but with the ability to identify nucleic acids within histologic lesions. Traditional ISH techniques have good specificity but limited sensitivity as a result of the lower labeling efficiency of short nucleotide sequences.11 A novel ISH-RNA technology (RNAScope, Advanced Cell Diagnostics, Newark, CA) describes single-molecule visualization in a hybridization platform that overcomes the limitations of the classical ISH technique through the amplification of specific hybridization signals,18 with the advantage of targeting messenger RNA (mRNA), which allows the detection of actively replicating microorganisms. Additionally, ISH-RNA can be used to simultaneously detect 2 different targets, offering insight into the contribution of different pathogens associated with histologic lesions. Our objective was to develop and evaluate an ISH-RNA technique in a duplex assay for detection of RVA, RVB, and RVC, by testing 1 probe or 2 probes simultaneously.
FFPE tissues from the University of Minnesota Veterinary Diagnostic Laboratory (Saint Paul, MN) were selected retrospectively based on histologic lesions and PCR results for RVA, RVB, and RVC. The inclusion criteria for ISH testing were the presence of histologic villus tip necrosis (VTN) or villus atrophy (VA), and positive PCR for RVA, RVB, or RVC using reverse-transcription quantitative PCR (RT-qPCR) primers.12
RT-qPCR was performed using 3 freshly sampled 5-cm segments of small intestine from each pig collected from regions adjacent to those submitted for histology and ISH-RNA. RT-qPCR assays were performed as described previously, and samples with cycle threshold (Ct) values <36 were considered positive.12
For ISH-RNA, probes were designed based on the VP6 gene 2-1162 region for RVA (GenBank KR052739), 45-1160 region for RVB (GenBank KF882558), and 2-1317 region for RVC (GenBank KJ814483), according to PCR primers used in qPCR.3 The actual sequence used to design the probe is intellectual property of the provider (Advanced Cell Diagnostics), and, therefore, is not available for public access.
FFPE tissue sections were deparaffinized in xylene and rehydrated through a series of alcohol washes. Rehydrated sections were processed (RNAScope 2.5 HD duplex detection kit, Advanced Cell Diagnostics), according to the manufacturer’s instructions. Briefly, tissues were treated with hydrogen peroxide at room temperature (RT) for 10 min. Then, tissue sections were placed in citric buffer and boiled for 15 min and incubated in protease at 40°C for 30 min. The slides were hybridized with rotavirus-specific probes at 40°C for 2 h in a humidified tray. After a wash step of 2 min in wash buffer, a sequence of amplifiers (amp) was added as follows: amp 1 for 30 min at 40°C; amp 2 for 15 min at 40°C; amp 3 for 30 min at 40°C; amp 4 for 15 min at 40°C; amp 5 for 30 min at RT; amp 6 for 15 min at RT. After amp 6, the red signals (RVB and RVC, peroxidase label) were detected by incubating the slides with a freshly prepared red solution, for 10 min, also at RT. Then, the procedure continued with addition of amp 7 for 15 min at 40°C; amp 8 for 30 min at 40°C; amp 9 for 30 min at RT; and amp 10 for 15 min also at RT. Finally, green signals (RVA, alkaline phosphatase label) were detected by applying a freshly prepared green solution for 10 min at RT. Slides were counterstained with hematoxylin. A wash step of 2 min was performed between amplifiers and/or colorimetric solutions by immersing the slides in the kit wash buffer, with occasional agitation. According to the positive RT-qPCR results, the probes were combined as either RVA and RVB, or RVA and RVC, to detect the RV groups in serial-cut sections of intestine.
To achieve the number of viral copies/mL regardless of the Ct value for each virus in each sample, we used the standard curve established previously.12 Briefly, the standard curve of the RT-qPCR was established based on the results of 7,508 samples, tested for RVA, RVB, and RVC by conventional PCR and by the results of gBlocks gene (Integrated DNA Technologies, Skokie, IL) containing the RVA, RVB, and RVC RT-qPCR targets, serially diluted (4 × 107 to 4 copies per reaction) to determine the curve slope.
Microscopic assessment of histologic lesions (presence, distribution, and intensity of VTN and/or VA), and ISH-RNA results were evaluated by 2 blinded and independent pathologists. Cross-reaction and nonspecific signals were evaluated using samples negative for RVs and positive for other important swine pathogens (porcine epidemic diarrhea virus, transmissible gastroenteritis virus, porcine reproductive and respiratory syndrome virus, and porcine circovirus 2).
A total of 33 RT-qPCR–positive samples for RVA, RVB, and/or RVC were tested (Table 1). ISH-RNA was able to detect VP6 mRNA in samples with PCR Ct values of ⩽31. Based on the standard curve generated in the validation of the RT-qPCR assay,12 a Ct value of 31 represents ~103 RNA copies/mL. Two samples (samples 31 and 32) were positive by ISH and PCR for the 3 RV groups, with Ct values of 22–26 for RVA, 29–31 for RVB, and 26–28 for RVC. RVB was detected by RT-qPCR and ISH-RNA in 2 samples (samples 31 and 32), which were also positive by RT-qPCR and ISH-RNA for RVA and RVC (Fig. 1A). Two samples (samples 31 and 32) were RT-qPCR positive only for RVC (Ct 17 and 20; Fig. 1B), and 2 other samples were positive by RT-PCR and ISH-RNA only for RVA (Ct 20 and 22; Fig. 1C). VA was more frequently associated with RVA infection (25 of 27 RVA-positive samples had VA). VTN was observed in infections of all 3 RV groups (13 of 33). From the samples that were infected by RVC, only 2 did not have histologic lesions (samples 31 and 32). Twelve negative control samples were negative by RT-qPCR and ISH-RNA for all 3 RV groups (data not shown). We found no cross-reaction among the RV groups or other tested swine viruses. The evaluations by the 2 independent pathologists were in agreement.
Table 1.
Ct values for samples positive on RT-qPCR, in situ hybridization results, and type of lesion in intestinal samples from diarrheic piglets.
| Sample ID | RVA |
RVB |
RVC |
Type of lesion | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ct | Viral copies/mL | ISH | Ct | Viral copies/mL | ISH | Ct | Viral copies/mL | ISH | ||
| 1 | 23 | 8.12 × 108 | + | − | NA | NT | 27 | 5.57 × 107 | ++ | VTN |
| 2 | 22 | 1.59 × 109 | ++ | − | NA | NT | 30 | 7.46 × 106 | + | VA |
| 3 | 22 | 1.59 × 109 | + | − | NA | NT | 27 | 5.57 × 107 | + | VTN, VA |
| 4 | 23 | 8.12 × 108 | ++ | − | NA | NT | 30 | 7.46 × 106 | + | VA |
| 5 | 24 | 4.16 × 108 | + | − | NA | NT | 30 | 7.46 × 106 | + | VTN, VA |
| 6 | 27 | 8.12 × 108 | ++ | − | NA | NT | 30 | 7.46 × 106 | + | VA |
| 7 | 23 | 8.12 × 108 | + | − | NA | NT | 31 | 3.82 × 106 | − | VA |
| 8 | 24 | 4.16 × 108 | + | − | NA | NT | 30 | 7.46 × 106 | + | VTN, VA |
| 9 | 24 | 4.16 × 108 | ++ | − | NA | NT | 30 | 7.46 × 106 | + | VA |
| 10 | 23 | 8.12 × 108 | ++ | − | NA | NT | 31 | 3.82 × 106 | + | VA |
| 11 | 25 | 2.13 × 108 | + | − | NA | NT | 31 | 3.82 × 106 | − | VA |
| 12 | 25 | 2.13 × 108 | + | − | NA | NT | 33 | 1.00 × 106 | − | VA |
| 13 | 26 | 1.09 × 108 | ++ | − | NA | NT | 32 | 1.95 × 106 | + | VA |
| 14 | 24 | 4.16 × 108 | ++ | − | NA | NT | 31 | 3.82 × 106 | + | VA |
| 15 | 22 | 1.59 × 109 | ++ | − | NA | NT | 31 | 3.82 × 106 | + | VA |
| 16 | 22 | 1.59 × 109 | + | − | NA | NT | 30 | 7.46 × 106 | − | VA |
| 17 | 23 | 8.12 × 108 | + | − | NA | NT | 30 | 7.46 × 106 | − | VA |
| 18 | 22 | 1.59 × 109 | ++ | − | NA | NT | 31 | 3.82 × 106 | + | VA |
| 19 | 24 | 4.16 × 108 | ++ | − | NA | NT | 28 | 2.85 × 107 | + | VA |
| 20 | 21 | 3.10 × 109 | + | − | NA | NT | 28 | 2.85 × 107 | − | VA |
| 21 | 21 | 3.10 × 109 | + | − | NA | NT | 35 | 2.62 × 105 | − | VTN, VA |
| 22 | 21 | 3.10 × 109 | + | − | NA | NT | 35 | 2.62 × 105 | − | VTN, VA |
| 23 | 20 | 6.06 × 109 | + | − | NA | NT | − | NA | NT | VTN |
| 24 | 22 | 1.59 × 109 | + | − | NA | NT | − | NA | NT | VA |
| 25 | 22 | 1.59 × 109 | + | 31 | 3.82 × 106 | + | 26 | 1.09 × 108 | ++ | VTN |
| 26 | 26 | 1.09 × 108 | ++ | 29 | 1.46 × 107 | + | 28 | 7.46 × 106 | ++ | VTN, VA |
| 27 | 26 | 1.09 × 108 | + | 28 | 7.46 × 106 | − | 24 | 4.16 × 108 | ++ | VTN, VA |
| 28 | 27 | 5.57 × 107 | + | 27 | 5.57 × 107 | − | 27 | 5.57 × 107 | ++ | VTN |
| 29 | − | NA | NT | 30 | 7.46 × 106 | − | 23 | 8.12 × 108 | + | VTN |
| 30 | − | NA | NT | 16 | 8.84 × 1010 | + | − | NA | NT | VA |
| 31 | − | NA | NT | − | NA | NT | 17 | 4.52 × 1010 | + | No lesions |
| 32 | − | NA | NT | − | NA | NT | 20 | 6.06 × 109 | + | No lesions |
| 33 | − | NA | NT | 15 | 1.73 1011 | + | − | NA | NT | VTN, VA |
Ct = cycle threshold; ISH = in situ hybridization; NA = not applicable; NT = not tested; RT-qPCR = reverse-transcription quantitative PCR; RVA, RVB, RVC = rotavirus groups A, B, and C, respectively; VA = villus atrophy; VTN = villus tip necrosis. − = negative; + = positive; ++ = predominant.
Figure 1.
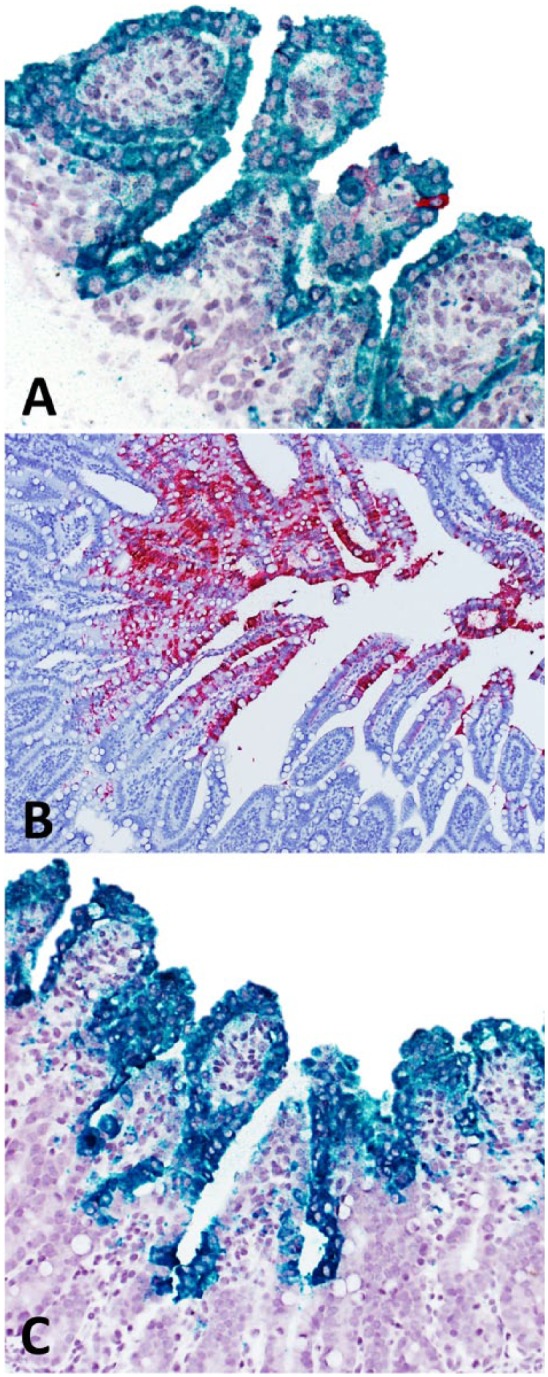
Dual detection of rotavirus A and C (RVA and RVC, respectively) in swine intestinal samples by in situ hybridization RNA-based chromogenic technique (ISH-RNA). A. RVA (green) and RVC (red) in ISH-RNA dual detection. Sample 1. Horseradish peroxidase and alkaline phosphatase. 200×. B. RVC (red) in ISH-RNA dual detection. Sample 26. Horseradish peroxidase. 100×. C. RVA (green) in ISH-RNA dual detection. Sample 18. Alkaline phosphatase. 200×.
The proposed ISH-RNA test is capable of determining RV subtypes within intestinal lesions of piglets previously confirmed by RT-qPCR to be infected by RV, allowing the assessment of the contribution of each RV group within lesions. Although the ISH-RNA test that we developed was used to investigate the relationship of virus presence detected by PCR and the viral presence within lesions characteristic of rotaviral infection, there is potential to establish the technique as one of the services offered to clients by veterinary diagnostic laboratories.
There was a predominance of coinfection of RVA and RVC (23 of 33), but the distribution of these groups was highly variable within the intestinal sections. Although our aim was not to investigate the prevalence of RV groups in field samples, this predominance of RVA and RVC in the RV-positive cases agrees with a published study of distribution of RV groups in pig samples.12
From the 33 samples included in our study, 7 RT-qPCR–positive samples were negative in ISH-RNA. These observations correlate with the segmental distribution of RV lesions commonly observed in infected pigs. It also highlights the importance of systematic collection, including multiple sections of small intestine for histologic evaluation.
With our proposed ISH-RNA test for RV detection in pig tissues, the absence of cross-reaction revealed good specificity of the probes, as well as detection of RV-positive signal demonstrated in tissues with PCR Ct values up to 32. Nevertheless, some of the PCR-positive samples were not positive by ISH-RNA. We suggest 2 possible explanations: 1) RV infection may not be diffuse throughout the intestine, and sampling non-adjacent intestinal segments for PCR and ISH-RNA can result in different outcomes by the 2 techniques; the second explanation, which may be of most concern for diagnosticians, researchers, and pig producers, is the presence of one or more RV subtypes in the pig intestine, without causing lesions. Although the RT-qPCR detects viable and nonviable viral particles, ISH-RNA targets mRNA and only detects actively replicating virus.
We observed VA more frequently in RVA-infected sections; VTN was caused by all 3 RV groups. We also observed RVC ISH-RNA–positive intestinal sections with low Ct values with minimal histologic lesions. VTN and VA have been described as the main microscopic lesions observed in the small intestine of RV-infected piglets.7 Our findings reinforce the advantage of combining PCR detection with ISH-RNA. RT-qPCR screens for RV presence, whereas ISH-RNA allows interpretation of virus in the pathogenesis of the lesions.
ISH-RNA using RNAScope technology has been described to investigate infectious diseases,1,5,8,10,16,17 but to our knowledge, ISH-RNA has not been reported previously for in situ speciation of viral infections. VP6 is an essential viral capsid component. Positive signals generated by hybridization of the VP6 mRNA probe are interpreted as identifying actively replicating virus. The opportunity to subtype and observe in situ transcriptionally active virus also improves the investigation of pathogenesis during natural or experimental infections, not only for pigs, but also for other mammalian species that are susceptible to RVs.
Acknowledgments
We thank all of the employees in the histology and immunohistochemistry laboratories of the University of Minnesota Veterinary Diagnostic Laboratory for their help during the ISH testing, and Lacey Marshal Lund for language assistance.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: TP Resende was supported by CAPES–Brazil. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iD: Talita P. Resende  https://orcid.org/0000-0002-7705-714X
https://orcid.org/0000-0002-7705-714X
References
- 1. Arruda PHE, et al. Detection of a novel sapelovirus in central nervous tissue of pigs with polioencephalomyelitis in the USA. Transbound Emerg Dis 2017;64:311–315. [DOI] [PubMed] [Google Scholar]
- 2. Chasey D, et al. A new type of atypical rotavirus in pigs. Arch Virol 1986;243:235–243. [DOI] [PubMed] [Google Scholar]
- 3. Chattha KS, et al. Strategies for design and application of enteric viral vaccines. Annu Rev Anim Biosci 2015;3:375–395. [DOI] [PubMed] [Google Scholar]
- 4. Estes MK. Rotaviruses. In: Knipe D, et al., eds. Fields’ Virology. Philadelphia, PA: Wolters Kluwer Health/Lippincott Williams & Wilkins, 2007:1917–1974. [Google Scholar]
- 5. Gaynor AM, et al. Localization of bovine papillomavirus nucleic acid in equine sarcoids. Vet Pathol 2016;53:567–573. [DOI] [PubMed] [Google Scholar]
- 6. Homwong N, et al. Three-level mixed-effects logistic regression analysis reveals complex epidemiology of swine rotaviruses in diagnostic samples from North America. PLoS One 2016;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janke BH, et al. Single and mixed infections of neonatal pigs with rotaviruses and enteroviruses: clinical signs and microscopic lesions. Can J Vet Res 1988;52:364–369. [PMC free article] [PubMed] [Google Scholar]
- 8. Joshi LR, et al. Pathogenesis of Senecavirus A infection in finishing pigs. J Gen Virol 2016;97:3267–3279. [DOI] [PubMed] [Google Scholar]
- 9. Kindler E, et al. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect Genet Evol 2013;14:58–67. [DOI] [PubMed] [Google Scholar]
- 10. Luff J, et al. Two canine papillomaviruses associated with metastatic squamous cell carcinoma in two related basenji dogs. Vet Pathol 2016;53:1160–1163. [DOI] [PubMed] [Google Scholar]
- 11. Maes RK, et al. Beyond H & E: integration of nucleic acid–based analyses into diagnostic pathology. Vet Pathol 2014;51:238–256. [DOI] [PubMed] [Google Scholar]
- 12. Marthaler D, et al. Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. J Virol Methods 2014;209:30–34. [DOI] [PubMed] [Google Scholar]
- 13. Marthaler D, et al. Widespread rotavirus H in domesticated pigs, United States. Emerg Infect Dis 2014;20:1195–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matthijnssens J, et al. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 2012;157:1177–1182. [DOI] [PubMed] [Google Scholar]
- 15. Molinari BLD, et al. Species H rotavirus detected in piglets with diarrhea, Brazil, 2012. Emerg Infect Dis 2014;20:2012–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phan TG, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J 2016;13:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Resende TP, et al. A novel RNA-based in situ hybridization to detect Seneca Valley virus in neonatal piglets and sows affected with vesicular disease. PLoS One 2017;12:e0173190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang F, et al. A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagnostics 2012; 4:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]


