Abstract
Obtaining statistically sound numbers of sera from Hendra virus (HeV)-infected horses is problematic because affected individuals usually die or are euthanized before developing a serum antibody response. As a consequence, test validation becomes a challenge. Our approach is an extension of OIE principles for provisional recognition and included 7 validation panels tested across multiple laboratories that provided estimates for test performance characteristics. At a 0.4 S/P cutoff, 16 of 19 sera from HeV-infected horses gave positive results in the HeV soluble G, indirect ELISA (HeVsG iELISA; DSe 84.2% [95% CI: 60.4–96.6%]); 463 of 477 non-infected horse sera tested negative (DSp 97.1% [95% CI: 95.1–98.4%]). The HeVsG iELISA eliminated almost all false-positive results from the previously used HeV iELISA, with marginally decreased relative sensitivity. Assay robustness was evaluated in inter-laboratory and proficiency testing panels. The HeVsG iELISA is considered to be fit for purpose for serosurveillance and international movement of horses when virus neutralization is used for follow-up testing of positive or inconclusive serum samples.
Keywords: Hendra virus serology, horses, network, sensitivity, specificity, validation
Introduction
Since its first description in 1994, Hendra virus (HeV; order Mononegavirales, family Paramyxoviridae, genus Henipavirus, species Hendra henipavirus), a flying fox–transmitted zoonotic agent, has caused rare but deadly losses in humans and horses in Australia.8 Serologic tests have been used in surveillance studies and as part of quarantine procedures for international movement of horses. Traditionally, an indirect ELISA using a Vero cell–derived, detergent-extracted antigen (HeV iELISA) developed at the Australian Animal Health Laboratory (AAHL; Newcomb, Victoria, Australia) was used as a screening test for the detection of antibodies against HeV in horses.4 Although the assay had good sensitivity (Se), its reduced specificity (Sp) resulted in frequent false-positive and nonspecific reactions. To overcome these limitations, a new iELISA for the detection of equine antibody was developed (Colling A, et al. A new networked approach for diagnostic test development and validation when samples from positive reference populations are scarce. Proc 16th Int Symp World Assoc Vet Lab Diagn; Berlin, Germany, 2013), which used a recombinant-expressed HeV soluble G glycoprotein (HeVsG).1 With high fatality rates, affected horses may die or are euthanized before development of a serum antibody response. As a consequence, the number of samples from HeV-infected and seroconverted horses is low, presenting a challenge in test validation.
The World Organization for Animal Health (OIE)’s principle of provisional recognition has been created for situations in which appropriate samples from the target population are scarce and animals are difficult to access, such as for transboundary infectious or wildlife diseases.11 In this approach, critical benchmark parameters such as analytical Se and Sp (ASe, ASp) and repeatability plus initial estimates for diagnostic Se and Sp (DSe, DSp) and reproducibility may allow OIE provisional recognition, pending further validation to fulfill certification requirements. Although our methodology includes analytical and diagnostic aspects, it also explores an alternative to substantiate estimates for test performance characteristics. This included evaluation of different negative horse populations and testing of representative, well-characterized reference samples over the dynamic test range in a network of participating laboratories for assessment of comparative Se and Sp, repeatability, and reproducibility.
Material and methods
Serum samples
Equine sera were sourced from various samples submitted for assessment (disease investigation and export testing) to the laboratories in our study and were grouped into 7 panels according to validation purpose (Table 1). Laboratories from HeV-endemic states (Queensland, n = 265; New South Wales, n = 216) and HeV-free states (AAHL, n = 477; Victoria, n = 200) participated in extended Sp testing. All sera for the extended Sp testing tested prior negative in the HeV iELISA (Table 1, panel 4b).
Table 1.
Validation sample panels: purpose, source of serum, laboratories involved, and assays performed.
| Panel | Purpose | Source of serum | Laboratories/Assays performed |
|---|---|---|---|
| 1 | Relative ASe | One horse, day 8 post-experimental infection with HeV (2008, QLD). | AAHL: HeVsG iELISA; HeV VN |
| 2 | Relative ASp | Eight horses infected with viruses that can cause similar clinical signs to HeV infection: West Nile virus (Kunjin subtype), African horse sickness virus, equine influenza virus (H3N8), eastern, Venezuelan, and western equine encephalitis viruses, and Ross River virus. | AAHL: HeVsG iELISA |
| 3 | DSe (HeV-infected horses) | Convenience samples from outbreak episodes (QLD) and follow-up testing of 15 horses. Thirty-two samples were available from 19 HeV-infected horses. Single samples were available from 8 horses, 2 samples from 10 horses (a–g, i–k), and 4 samples from 1 horse (h). Single samples were generated from repeated bleeds using random selection in Excel resulting in a total of 19 individual samples from 19 infected horses (Table 2, column with an “X”). | AAHL: HeVsG iELISA; HeV iELISA; HeV VN; TaqMan assays; virus isolation |
| 4a | DSp | Healthy, non-infected horses (n = 477); the majority of samples were submitted for export testing quarantine requirements 2011–2012 from QLD, NSW, VIC, and WA. | AAHL: HeVsG iELISA |
| 4b | Extended DSp estimates | Independent intra-laboratory assessment. Healthy horses from NSW (n = 216) and QLD (n = 256), as well as from VIC (n = 200) not exposed to HeV outbreaks. Laboratories in each state had stored horse sera from healthy horses that had tested negative with the HeV iELISA prior to this study. | AAHL, QLDSL, NSWSL, VICSL: HeVsG iELISA |
| 5 | Inter-laboratory robustness study | 10 HeV-positive and 10 HeV-negative horse sera. Positive samples consisted of spiked sera of positive, VN-confirmed sera from 2 naturally infected horses (data not shown). | AAHL, QLDSL, NSWSL, VICSL: HeVsG iELISA |
| 6 | PT | 13 gamma-irradiated sera (2 negative and 13 positive samples at different analyte concentrations) diluted in normal horse serum with detailed instructions for testing. | AAHL, QLDSL, NSWSL, VICSL, WASL: HeVsG iELISA |
| 7 | Excluded samples | 13 sera were identified for which no result was available in the HeV iELISA and/or VN because of nonspecific or toxic reactions. The HeVsG iELISA produced negative results for these samples but results were not included in the analysis because an infection status could not be assigned (data not shown). | AAHL: HeVsG iELISA; HeV iELISA; HeV VN |
AAHL = Australian Animal Health Laboratory; ASe = analytical sensitivity; ASp = analytical specificity; DSe = diagnostic sensitivity; DSp = diagnostic specificity; iELISA = indirect (total antigen) ELISA; NSWSL = New South Wales State Laboratory; PT = proficiency test; QLDSL = Queensland State Laboratory; HeVsG iELISA = HeV soluble G indirect ELISA; VICSL = Victoria State Laboratory; VN = virus neutralization; WASL = Western Australia State Laboratory.
Post-infection sera were obtained from equine HeV infection cases either from experimentally infected horses or field infection events confirmed by agent detection (molecular and/or virus isolation). Intervals between repeated bleeds from the same infected horses were ~1 wk (Table 2).
Table 2.
Diagnostic window of HeVsG iELISA using sera from HeV-infected horses from Queensland and comparison with results from the HeV VN, HeV iELISA, TaqMan M, N, P (genes), PCR, and virus isolation.
| Sample ID | Year | Samples used for DSe | HeVsG iELISA† |
HeV VN‡ |
HeV iELISA§ |
TaqMan‖ M, N, P genes | |
|---|---|---|---|---|---|---|---|
| S/P ratio | Result | Titer | Result | ||||
| 1* | 2006 | X | 0.3 | (+,−) | 1:16 | (+) | P (+) |
| 2 | 2008 | X | 4.7 | (+) | 1:512 | (+) | ND |
| 3 | 2009 | X | 4.2 | (+) | 1:1024 | (+) | N (−), P (−) |
| 4 | 2009 | X | 5 | (+) | 1:16 | (+) | N (−), P (−) |
| 5* | 2011 | X | 0.01 | (−) | 1:16 | (+,−) | M (+), N (+) |
| 6 | 2011 | X | 1.7 | (+) | 1:64 | (+) | M (+,−) |
| 7 | 2012 | X | 0.99 | (+) | 1:4 | (+) | M (+), N (+) |
| 8 | 2012 | X | 0.5 | (+) | 1:16 | (+) | M (+), N (+) |
| 9 (a) | 1994 | 4.7 | (+) | >1:32 | (+) | ND | |
| 10 (a) | 1994 | X | 5 | (+) | 1:512 | (+) | ND |
| 11 (b) | 1994 | X | 5.2 | (+) | >1:32 | (+) | ND |
| 12 (b) | 1994 | 5 | (+) | 1:512 | (+) | ND | |
| 13 (c) | 1994 | 5 | (+) | >1:32 | (+) | ND | |
| 14 (c) | 1994 | X | 5 | (+) | 1:1024 | (+) | ND |
| 15 (d) | 1994 | 0.8 | (+) | >1:32 | (+) | ND | |
| 16 (d) | 1994 | X | 1.5 | (+) | 1:16 | (+) | ND |
| 17 (e) | 1994 | 5.5 | (+) | >1:32 | (+) | ND | |
| 18 (e) | 1994 | X | 4.9 | (+) | 1:512 | (+) | ND |
| 19 (f) | 1994 | 4.6 | (+) | >1:32 | (+) | ND | |
| 20 (f) | 1994 | X | 4.8 | (+) | 1:512 | (+) | ND |
| 21 (g) | 1994 | X | 4.8 | (+) | >1:32 | (+) | ND |
| 22 (g) | 1994 | 5 | (+) | 1:512 | (+) | ND | |
| 23 (h) | 2008 | 4.7 | (+) | 1:2048 | (+) | P (−) | |
| 24 (h) | 2008 | 5.1 | (+) | 1:2048 | (+) | M (+) | |
| 25 (h) | 2008 | X | 4.8 | (+) | 1:2048 | (+) | M (+), P (−) |
| 26 (h) | 2008 | 6.1 | (+) | 1:2048 | (+) | ND | |
| 27 (i) | 2008 | X | 0.6 | (+) | 1:16 | (+) | P (−) |
| 28 (i) | 2008 | 5 | (+) | 1:128 | (+) | N (−), P (−) | |
| 29** (j) | 2009 | 2.1 | (+) | <1:2 | (+) | M (+), P (+) | |
| 30 (j) | 2009 | X | 3.3 | (+) | 1:64 | (+) | N (−), P (−) |
| 31* (k) | 2011 | X | 0.22 | (−) | 1:16 | (+) | M (+), N (+,−), P (−) |
| 32 (k) | 2011 | 0.93 | (+) | 1:32 | (+) | M (−), N (−), P (−) | |
Thirty-two samples were available from 19 Hendra virus (HeV)-infected horses. Single samples were available from 8 horses, 2 samples from 10 horses (a–g, i–k), and 4 samples from 1 horse (h). Intervals between repeated bleeds from the same horse were ~1 wk with the exception of samples 23–26 from horse h, which were taken on 7, 10, 18, and 23 July 2008. Samples marked with an X were used for estimating diagnostic sensitivity (DSe). Sera from repeatedly sampled horses were selected randomly. Virus isolation was only attempted on 3 samples: 1 (+), 25 (−), 27 (+; data not shown).
Molecular results for associated co-collected samples 1, 29 and 31 (k) were positive.
This sample tested negative by VN.
HeV soluble G indirect ELISA (S/P ratio >0.4 = positive (+), 0.25–0.4 = inconclusive (+,−), <0.25 = negative (−).
HeV virus neutralization (titer <1:2 = negative, ≥1:2 = positive).
HeV (total antigen) indirect ELISA: (+) = positive, (+,−) = nonspecific reaction (reactor).
HeV TaqMan assay for M, N, and P genes: (+) = positive, (+,−) = inconclusive, ND = not done).
HeVsG iELISA
The HeVsG iELISA for the detection of antibodies against HeV in horses uses a recombinant-expressed, truncated form of the HeV G envelope glycoprotein antigen.1 Plates (Nunc Maxisorb, Thermo Scientific, Waltham, MA) were coated (37°C for 1 h) with antigen in phosphate-buffered saline (PBS; pH 7.3) at a concentration of 0.073 μg/mL, then blocked with PBS and skimmed milk powder (SMP) for 30 min. Standard ELISA methodology with wash cycles (4×) using PBS and 0.05% Tween 20 (PBST) were followed by incubation steps. Sera originating from a HeV disease investigation were pretreated by dilution 1:5 in PBST and 0.5% Triton X-100, and heating (56°C for 30 min) prior to use. All test and control sera (strong, low, and negative) were prepared to a final 1:100 dilution in PBST and 1% SMP, and were added to the plate and incubated (37°C for 1 h) on a plate shaker. Anti-equine horseradish peroxidase (HRP) conjugate A 6917 (Sigma-Aldrich, Castle Hill, New South Wales, Australia) was used for detection (37°C for 30 min); TMB substrate (3,3’,5,5’-tetramethylbenzidine; T0440, Sigma-Aldrich) was added (7–10 min at ambient temperature); the reaction was stopped with 1 M sulfuric acid and read at 450 nm. After subtraction of background (taken as the optical density [OD] of the negative control serum [NCS]), ODs were transformed to a ratio relative to a low positive (LP) control serum (average OD ~ 0.5), and a signal-to-positive ratio (S/P ratio) was calculated as S/P = (ODTest – ODNCS)/(average ODLP – ODNCS). A negative result gave a S/P ratio of <0.25, and a positive result gave a S/P ratio of >0.4. Results between ≥0.25 to ≤0.4 were regarded as inconclusive. Positive and inconclusive results were followed up with the virus neutralization (VN) test. Using this testing algorithm, a sample was determined positive if confirmed VN-positive.
HeV iELISA
The HeV iELISA for the detection of antibodies against HeV in horse sera was conducted as described previously.12 The assay used detergent-disrupted and inactivated virus antigen derived from whole cell lysates of HeV-infected Vero cells. All sera determined to be positive or nonspecific (mock antigen reactors) were retested by VN.
HeV VN
HeV VN was performed as described previously.2 The test was started as a 1:2 dilution in the first well and performed with a 2-fold dilution across the plate. The titer is expressed as the reciprocal of the highest serum dilution to completely neutralize the virus. Sera with a titer ≥1:2 were considered positive.
TaqMan assays for HeV M, N, and P genes
TaqMan assays for HeV N and M genes were performed as described previously.5,10 TaqMan assay for HeV P gene was performed as an in-house test. This assay was designed to detect both HeV and Nipah virus, but evaluation studies indicated that it was not as sensitive as M and N TaqMan assays for the detection of HeV (Wang J. Evaluation of different PCR assays for detection of Hendra virus in recent outbreak in Queensland. 4th Ann Meet Australian Assoc Vet Lab Diagn, 2008; Brisbane, Qld).
Inter-laboratory comparison
To assess robustness of the HeVsG iELISA, an interlaboratory comparison (ILC) involving 4 laboratories (Queensland, New South Wales, Victoria, and AAHL, Geelong) was conducted between February and May 2012. Each laboratory received antigen; positive and negative control sera; conjugate; consumables; plates; SMP; HRP; TMB substrate; a panel of 10 HeV-positive, spiked (confirmed by VN) sera, and 10 HeV-negative sera; and a protocol with all necessary instructions to perform the HeVsG iELISA. Each laboratory was asked to assess the provided panel of 20 test sera in 3 separate assay runs on different days, with a single operator. In addition, at least 10–15 assay runs, preferably conducted by different operators, should be completed. ILC sera were tested in duplicate. Control samples were tested in quadruplicate (C++, C+, buffer, and C−). Each laboratory was requested to return results in a spreadsheet to AAHL. Any reactive sera in the ELISA were requested to be sent to AAHL for assessment by VN.
Proficiency testing
To further assess reproducibility, a proficiency testing (PT) round was performed with 5 laboratories (Queensland, New South Wales, Victoria, Western Australia, and AAHL, Geelong). The PT panel consisted of 15 gamma-irradiated samples diluted in normal horse serum and included detailed instructions on how to test the samples. The PT panel consisted of 2 negative, 2 weak-positive (assess Se and repeatability), 12 various dilutions of HeV-positive serum (assess efficiency), and 2 identical samples (assess repeatability).
Statistical analysis
Results were stored, and statistical analysis was performed using Excel 2013 (Microsoft, Redmond, WA). Individual samples from repeated bleeds from HeV-infected horses were selected using random sampling function. Ninety-five percent confidence intervals (CIs) for DSe and DSp were calculated using exact binomial methods. Receiver-operating characteristic (ROC) analysis and interactive dot diagram graphs were done with MedCalc v.15.4 (MedCalc Software, Ostend, Belgium). Graphs showing results for ASe and PT round were produced (Prism 5 for Windows v.5.02, GraphPad Software, La Jolla, CA).
Results
Relative ASe and ASp
HeVsG iELISA S/P values associated with a post-infection serum (Table 1, panel 1) were correlated with the VN titer (1:512 or 9.0 as log2 value). A third-order polynomial trend line was fitted (R2 = 1.0) to the plot of S/P ratios against VN titers (Fig. 1). The VN-equivalent titer corresponding to the ELISA S/P threshold (0.4) was 1:2.36 (or ~1 as log2 value), indicating approximately equivalent ASe.
Figure 1.
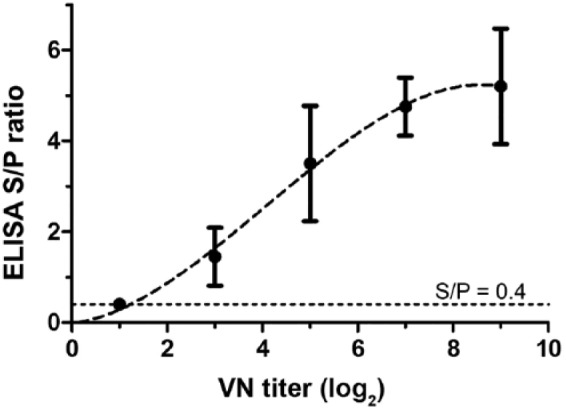
Relative analytical sensitivity: Hendra virus (HeV) virus neutralization (VN) and HeV soluble G (HeVsG) indirect ELISA. A trend line (R2 = 1.0) derived from a third-order polynomial curve fit with 95% confidence interval is shown.
Sera from horses infected with West Nile virus (Kunjin subtype), African horse sickness virus, equine influenza virus (H3N8), eastern, Venezuelan, and western equine encephalitis viruses, and Ross River virus were tested in the HeVsG iELISA (Table 1, panel 2). Results indicated a high level of ASp for HeV antibodies given that all results for non-henipavirus sera were negative (data not shown).
DSe and DSp
Thirty-two samples were available from 19 HeV-infected horses. Single samples were available from 8 horses (samples 1–8), 2 samples from 10 horses (a–g, i–k), and 4 samples from 1 horse (h; Table 2). Single samples were generated from repeated bleeds using random selection resulting in a total of 19 individual samples from 19 infected horses to obtain an estimate for DSe (Table 1, panel 3). Some sera were obtained from serial bleedings at different time intervals after infection (Table 2). For calculation of DSe and DSp, only single samples from individual horses were used. These sera tested positive in the HeV iELISA and were confirmed by VN. At a S/P cutoff of 0.4, 16 of 19 sera from HeV-infected horses tested positive in the HeVsG iELISA, resulting in a DSe of 84.2% (95% CI: 60.4–96.6%; Table 3).
Table 3.
Correlation of positive and negative results of sera from 19 Hendra virus (HeV)-infected and 477 non-infected horses at AAHL, tested using the HeVsG indirect (i)ELISA and virus neutralization (VN).
| HeVsG iELISA | HeV VN |
|
|---|---|---|
| Positive | Negative | |
| Positive | 16 | 14 |
| Negative | 3 | 463 |
| Total | 19 | 477 |
Estimates for DSp (Table 1, panel 4a) were obtained for sera from 477 normal horses. At a 0.4 S/P ratio cutoff, 463 of 477 normal horse sera tested negative in the HeVsG iELISA, resulting in a DSp of 97.1% (95% CI: 95.1–98.4%). The area under the ROC curve (Fig. 2) was 0.974 (95% CI: 0.940–1%), which indicated a highly accurate test.7 Additional analysis shows graphical representation of the spread of data and dependence of false-negative and -positive results on the selected S/P threshold (Fig. 3).
Figure 2.
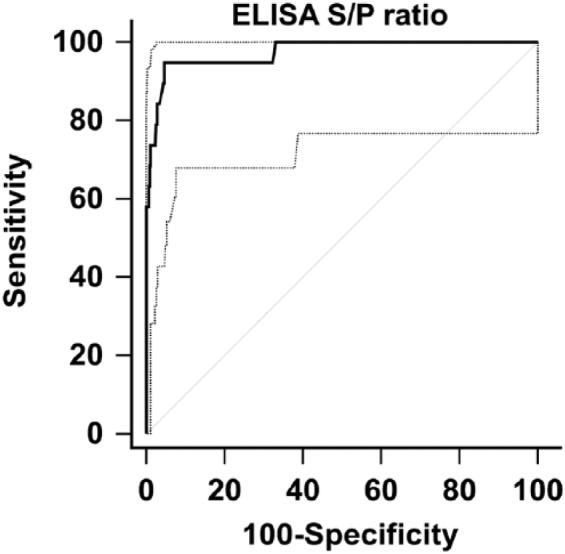
Receiver-operating characteristic curve analysis for Hendra virus soluble G (HeVsG) indirect ELISA using 19 sera from HeV-infected and 477 sera from non-infected horses. The overall area under the curve was 0.974 (95% confidence interval: 0.940–1.0%).
Figure 3.
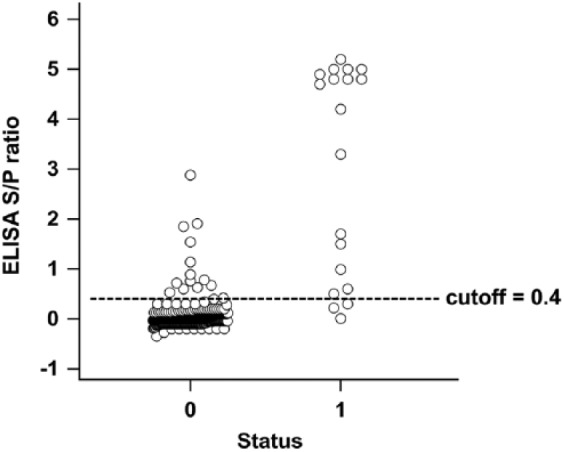
Scatter plot showing results for Hendra virus soluble G (HeVsG) indirect ELISA using 19 sera from HeV-infected (1) and 477 sera from non-infected (0) horses at 0.4 sample-to-positive (S/P) ratio cutoff (dotted line).
To obtain extended SP estimates (Table 1, panel 4b) for the HeVsG iELISA, sera were tested from HeV-affected regions in state laboratories in New South Wales (n = 216) and Queensland (n = 265) and a non-affected state laboratory in Victoria (n = 200). AAHL tested 477 sera variously sourced from affected and unaffected states. Application of a fixed 0.4 S/P ratio cutoff resulted in laboratory-associated DSp estimates of 90%, 81%, and 88.5%, respectively, which were uniformly lower than the 97.1% estimate obtained at AAHL, where the assay was developed.
Results from preliminary comparisons of the HeV iELISA, VN, and HeVsG iELISA indicated a substantially higher relative DSp of the HeVsG iELISA given that it classified all nonspecific reactors as negatives. This included the follow-up of 13 sera (Table 1, panel 7) that were classified as inconclusive by HeV iELISA and VN and negative by HeVsG iELISA (data not shown).
Network approach: ILC
To assess comparative DSe, DSp, repeatability, and reproducibility of the HeVsG iELISA, an ILC involving the 4 laboratories was conducted between February and May 2012 (Table 1, panel 5). Evaluation of qualitative results based on a 0.4 S/P ratio cutoff resulted in 100% agreement of participants for 16 of 20 sera (data not shown). Ten samples were negative, with results close to 0, and 6 samples were positive, with S/P ratio results of 0.7–1.6. The remaining 4 samples had values close to the cutoff, and accordingly lower agreement (64–91%) among laboratories. Assigned status for samples (0.4 S/P ratio cutoff) resulted in 10 negative and 10 positive samples. In addition, repeatability was assessed in each laboratory using strong-positive, positive, negative, and buffer control samples, which were tested repeatedly 14–42 times over a period of 1–6 wk. Coefficient of variation values (mean ÷ standard deviation × 100%) of control samples were within acceptable limits.
Network approach: PT
After completing technology transfer, a PT round was organized to follow up on ILC results and network meeting discussions (Table 1, panel 6). Results for evaluation were obtained from 4 laboratories. Qualitatively, all results were in agreement, apart from 1 false-positive result for sample 4 from the laboratory in Victoria (Fig. 4).
Figure 4.
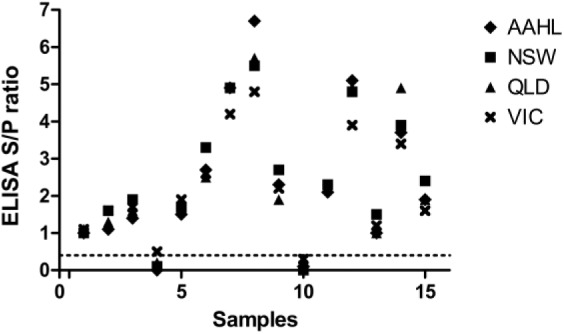
Results from a proficiency test panel of 15 sera tested at AAHL and in state laboratories in Queensland (QLD), New South Wales (NSW), and Victoria (VIC) using the Hendra virus soluble G (HeVsG) indirect ELISA.
Discussion
A review of the literature indicates frequent limitations of field validation and lack of reporting of performance parameters of laboratory tests.6 The situation becomes even more challenging for serologic tests when relatively few animals have been infected and the case fatality rate is high, with death occurring prior to seroconversion, as exemplified by HeV infection in horses. Recognizing these limitations, we introduced a novel HeV equine antibody ELISA into Australian veterinary diagnostic laboratories through a process of systematic validation steps that have included inter-laboratory trials and proficiency testing. Relative ASe and ASp are useful initial metrics of analytical (experimental) performance in assay validation.11 At a positive-to-negative threshold of 0.4 S/P ratio, the HeVsG iELISA–detected levels of antibody were similar to reference VN; additionally, there was no cross-reaction with any sera from animals infected with 8 other viral pathogens of differential diagnostic significance, especially henipaviruses.
DSe and DSp estimates are of more practical value for defining test characteristics. Given that the dynamic immune response to the infectious process particularly affects DSe estimates made relative to the true disease state, the use of VN as a reference test for HeVsG ELISA evaluation is an appropriate approach. Of 19 sera derived from VN-positive, HeV-infected horses, the HeVsG iELISA detected 16 as positive and 1 sample (Table 2, sample 1) as inconclusive, with an S/P ratio of 0.25–0.4. At a threshold level of >0.4 S/P, the DSe was 84.2%. Overall DSe rose to 89.5% (95% CI: 66.9–98.7%) when inconclusive sample 1 was included in the DSe estimate because the established test algorithm required confirmatory testing by VN, which returned a positive result. HeV was isolated or positive molecular detections were recorded for co-collected samples for samples 1, 5, and 31 (k). Therefore, differences can occur between the ELISA and VN detection of acute-stage antibody; this observation supports a multipronged (agent detection and serology) laboratory investigation of all suspected acute HeV cases. Where possible, the collection of follow-up bleeds can confirm seroconversion, as for sample 32 (k) in which the serum tested positive by HeVsG iELISA together with VN and HeV iELISA; co-collected samples were negative in molecular tests suggesting true seroconversion and discontinued virus shedding. In contrast, sample 29 (j) was negative (<1:2) in VN but positive in the molecular tests and both ELISAs, suggesting that the ELISAs sometimes detected antibodies for a period prior to the production of neutralizing antibody.
Serial bleeds from the same infected individual have been regarded as unsuitable to estimate DSe because this approach violates the statistical requirement for independent observations.11 To obtain estimates for DSe, only single samples from individuals were selected randomly when samples from multiple, serial bleeds were available, such as 10 consecutive blood samples from 10 horses and 4 consecutive blood samples from 1 horse (Table 1, panel 3).
For the purpose of assay validation when samples from positive reference animals are scarce, further discussion is needed to redefine meaningful and biological limits for consecutive blood collections from single animals as a potential solution to counterbalance the lack of samples from individually infected animals. Under circumstances of restricted sample availability, a balanced use of repeated bleeds collected at different times from an individual animal may represent an alternative to increase the robustness of Se estimates. As more samples become available, increased robustness and reduced CIs will emerge for the DSe parameter.
A preliminary cutoff using the mean plus 2 standard deviations from 477 negative horses tested (AAHL, S/P ratio 0.51) was further adapted by including DSe estimates using ROC analysis; this established a threshold S/P ratio of 0.4 with associated DSp of 97.1% (95% CI: 95.1–98.4%). By testing and comparing results from negative horse sera from HeV-affected and not-affected states, extended Sp estimates were prepared, although these were uniformly lower (DSp range: 81–90%) than in the developing laboratory (AAHL). Target population, assay robustness, and other external influences are determinants that must necessarily be considered in determining test characteristics, particularly in deploying an assay under validation assessment into other laboratories.
Based on the wide geographic distribution of HeV infection in Australia, the test would need to be robust and rugged enough to be deployed to different laboratories. A laboratory network approach (Laboratories for Emergency Animal Disease Diagnosis and Response [LEADDR]) was used to obtain preliminary estimates for comparative assessments of DSe, DSp, repeatability, and reproducibility. Early results and feedback from participants provided important information for standardization of the HeVsG iELISA, for example water quality and coating conditions. Results from an ILC and a PT round facilitated troubleshooting, changes and adaptation of the protocol, and improved efficiency and continuous monitoring of the technology transfer. Repeatability (robustness) and reproducibility (ruggedness) of the HeVsG iELISA were found to be acceptable after initial discussions and adjustments. Ongoing test performance is presently monitored in real time at various network laboratories using Network Quality Control sera distributed to test laboratories and evaluated in a monthly network meeting. Further improvements are expected once the test is produced under standardized industrial conditions as a kit. The newly developed HeVsG iELISA was found to be fit for purpose as a screening test for surveillance studies for HeV and international movements of horses. Estimates for DSe were similar to the traditional HeV iELISA (using sodium dodecyl sulfate antigen), but the relative Sp of the HeVsG iELISA was substantially higher resulting in fewer false-positive or nonspecific reactions. The test algorithm, a screening test (ELISA) followed by a confirmatory test (VN) of positive or nonspecific sera from the ELISA, remained unchanged and in accordance with OIE recommendations. We conclude that the network approach offers a powerful alternative for diagnostic test validation when the number of samples from infected animals is scarce. Results reported in our study facilitated certification through the Subcommittee of Animal Health Laboratory Standards and accreditation through the National Association of Testing Authorities (Approved tests, https://goo.gl/MC1e9A). Ongoing monitoring of the HeVsG iELISA is carried out through the LEADDR PT program and follows recommendations as described previously.3
Since 2014, a HeV vaccine has been used in Australian horses. The HeVsG iELISA does not distinguish between antibodies resulting from natural infection with HeV or vaccination. Careful interpretation of results is necessary in view of vaccination and/or outbreak investigation history. The soluble G protein has been used in different ELISA formats and Luminex-based multiplexed microsphere assays and showed promising results to detect HeV antibodies, including differentiation of antibody responses caused by vaccination in horses.9
Acknowledgments
We thank colleagues from network state laboratories for testing of sera from non-infected horses in their respective regions and for active participation during ILC and PT rounds, provision of results, and constructive discussion for improvement and standardization of the test protocol at regular LEADDR network meetings. We also thank the AAHL Proficiency Testing Team for selection and preparation of serum panels and evaluation and reporting of results.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: We acknowledge the generous financial support by the Australian Department of Agriculture and Water Resources, to carry out the network approach.
References
- 1. Bossart KN, et al. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble g glycoprotein of Hendra virus. J Virol 2005;79:6690–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bossart KN, et al. Neutralization assays for differential henipavirus serology using Bio-Plex Protein Array Systems. J Virol Methods 2007;142:29–40. [DOI] [PubMed] [Google Scholar]
- 3. Crowther JR, et al. Aspects of kit validation for tests used for the diagnosis and surveillance of livestock diseases: producer and end-user responsibilities. Rev Sci Tech Off Int Epiz 2006;25:913–935. [PubMed] [Google Scholar]
- 4. Daniels P, et al. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes Infect 2001;3:289–295. [DOI] [PubMed] [Google Scholar]
- 5. Feldman KS, et al. Design and evaluation of consensus PCR assays for henipaviruses. J Virol Methods 2009;161:52–57. [DOI] [PubMed] [Google Scholar]
- 6. Greiner M, Gardner IA. Epidemiological issues in the validation of veterinary diagnostic tests. Prev Vet Med 2000;45:3–22. [DOI] [PubMed] [Google Scholar]
- 7. Greiner M, et al. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med 2000;45:23–41. [DOI] [PubMed] [Google Scholar]
- 8. Halpin K, et al. Henipavirus Ecology Research Group. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg 2011;85:946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McNabb L, et al. Henipavirus microsphere immuno-assays for detection of antibodies against Hendra virus. J Virol Methods 2014;200:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith IL, et al. Development of a fluorogenic RT-PCR assay (TaqMan) for the detection of Hendra virus. J Virol Methods 2001;98:33–40. [DOI] [PubMed] [Google Scholar]
- 11. World Organization for Animal Health (OIE). Principles and methods of validation of diagnostic assays for infectious diseases, Chapter 1.1.6 In: OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris, France: OIE, 2017. [Google Scholar]
- 12. World Organization for Animal Health (OIE). Nipah and Hendra virus diseases, Chapter 2.1.14 In: OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris, France: OIE, 2017. [Google Scholar]


