Abstract
Porcine circovirus–associated diseases (PCVADs), caused by porcine circovirus 2 (PCV-2), have a significant economic impact on the swine industry worldwide. In Africa, there is little information, to date, regarding the occurrence of PCV-2, and it has not been reported in Mozambique’s swine population. We randomly collected mesenteric lymph nodes (n = 111) from slaughtered pigs from 9 districts in southern Mozambique. PCV-2 DNA was detected in 54% (62 of 111) of the samples and 78% (23 of 31) of the farms. PCV-2 antigen was detected by immunohistochemistry in lymph nodes (6 of 62; 10%) that were positive for PCV-2 by PCR. Histopathologic changes observed in these lymph nodes were lymphoid depletion, multifocal nodal necrosis, and infiltrates of histiocytes and multinucleate giant cells. One positive sample from each district was selected in order to obtain sequences covering the ORF2 region. Five sequences clustered with PCV-2d, of which 3 sequences from Maputo, Namaacha, and Moamba were grouped with PCV-2d-2; 2 sequences from Manhiça and Matola were grouped as PCV-2d-1; and 4 sequences from Boane, Matutuíne, Chibuto, and Xai-Xai were closely related to PCV-2b-1A/B genotypes. Our study indicates that a diversity of PCV-2 viruses is circulating in the Mozambican swine population.
Keywords: Immunohistochemistry, Matola City, PCR, pigs, porcine circovirus 2.
Species Porcine circovirus 2 (PCV-2; genus Circovirus, family Circoviridae) is a small nonenveloped virus containing a single-stranded circular DNA genome.1 The genome of PCV-2 contains 1,766–1,768 nt with at least 4 open reading frames (ORFs). ORF1 encodes 2 proteins associated with replication, designated Rep and Rep′. ORF2 encodes the capsid protein,18 ORF3 encodes a protein that is thought to play a role in apoptosis,15 and ORF4 encodes a newly discovered protein that aids in the suppression of caspase activity and in the regulation of CD4+ and CD8+ T-lymphocytes.10
PCV-2 is subdivided into 4 genotypes (a–d); PCV-2d was previously named PCV-2b mutant.24 Since 2000, PCV-2b has caused outbreaks in many parts of the world1 and has become the most prevalent genotype within domestic pig populations worldwide.14,20 PCV-2a is subdivided into 5 clusters (2A–2E), and PCV-2b into 3 clusters (1A–1C).18 PCV-2d can be further divided into PCV-2d-1 and PCV-2d-2.24 Natural inter- and intra-genotype recombinations have been reported between PCV-2 strains.2,18
PCV-2 is the primary causative agent of porcine circovirus–associated disease (PCVAD), which includes several syndromes: postweaning multisystemic wasting syndrome (PMWS), porcine respiratory disease complex, porcine dermatitis and nephropathy syndrome, reproductive disease, and enteritis.22 PMWS is the most common and economically important clinical syndrome associated with PCV-2 infection, and affects pigs 25–120 d of age, with the greatest number of cases occurring at 60–80 d of age.3 Diagnosis of PCVAD is based on clinical signs, histologic lesions, and detection of PCV-2 antigens or DNA within characteristic lesions.3,22 In both subclinical and clinical PCVAD, PCV-2 antigens and nucleic acids are detected primarily in lymphoid tissues.8,22
To date, there is little information concerning the presence of PCV-2 in Africa; data are limited to South Africa5 and Uganda.17 PCV-2 has not yet been studied in the swine population in Mozambique; thus, our aim was to determine the occurrence of PCVAD microscopic lesions as well as PCV-2 antigens and DNA in mesenteric lymph nodes from slaughter pigs. In addition, we characterized phylogenetically the PCV-2 strains circulating in pigs in Mozambique.
Our study was conducted from December 2014 to February 2015 (1st period) and December 2015 to February 2016 (2nd period), in a slaughterhouse in Matola City, Maputo Province, Mozambique (25o 55’ 26’’ S, 32o 27’ 57’’ E). This slaughterhouse was selected because it is the largest pig abattoir in southern Mozambique, and receives animals from several districts in the region.
Mesenteric lymph nodes (n = 111) were randomly collected from slaughtered pigs. To avoid cross-contamination between herds, pigs from the same owner and the same district were slaughtered in sequence. The knives and gloves used for sampling were changed or decontaminated between sampling different groups of pigs. Duplicate samples from each mesenteric lymph node were collected from each pig; one sample was stored at −20°C until DNA extraction, and the other sample was fixed in 10% buffered formalin for 24–48 h for routine histologic procedures. The origin (district and owner or farm) of the slaughtered pigs provided by the slaughterhouse was recorded. We also recorded the pig’s live weight, but the age of the pigs was not available.
Total DNA was extracted from 25 mg of tissue homogenates (QIAamp DNA mini kit, Qiagen, Santa Clarita, CA) according to the manufacturer’s recommendations. To avoid cross-contamination, samples were processed individually. DNA was eluted in 200 µL of elution buffer, quantified (NanoDrop 2000/2000c spectrophotometer, Thermo Fisher Scientific, Wilmington, NC), and stored at −20°C.
Total DNA from mesenteric lymph nodes was used in PCR with pairs of primers described previously.13 PCR mix conditions were the following: 2.5 µL of 10× buffer, 1.5 mM of MgCl2, 200 mM of dNTPs, 10 pmol of each primer, 1 U DNA polymerase enzyme (Ludwig Biotecnologia, Porto Alegre, Brazil), 1 µL of DNA sample, and water up to 25 μL. PCR was performed with an initial cycle of 95°C for 2 min, 35 cycles at 95°C for 30 s, 50°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 5 min, which amplified a 263-bp product. PCR products were electrophoresed in agarose gel using 1 µL of Blue Green dye (LGC Biotecnologia, São Paulo, Brazil) according to the manufacturer’s protocol.
Mesenteric lymph node samples were fixed in 10% neutral buffered formalin, dehydrated, embedded in paraffin wax, and sectioned at 3–5 µm. The sections were stained with hematoxylin and eosin. Only PCV-2 PCR–positive samples were examined under an optical microscope and subsequently analyzed with immunohistochemistry (IHC).
IHC was performed on all mesenteric lymph node samples that were positive by PCV-2 PCR. Antigen retrieval was obtained with 0.5% XIV protease (Sigma Chemical, Poole, UK) at room temperature for 15 min. Endogenous peroxidase activity in tissue sections was inhibited by incubation in 10% hydrogen peroxide for 15 min; nonspecific reactions were blocked with 5% skim milk for 15 min. Sections were incubated with an anti–PCV-2 polyclonal antibody (Iowa State University, Ames, IA) at 1:1,000 dilution overnight at room temperature. Signal was amplified and visualized (MACH 4 universal HRP polymer detection, Biocare Medical, Concord, CA) with the chromogen 3,3’-diaminobenzidine (Dako North America, Carpinteria, CA), respectively. The sections were counterstained with Harris hematoxylin for 30 s. As positive control, a PCV-2–positive pig intestine sample was used.25 Negative controls were established by omission of primary antibody. Immunostaining intensity was classified as mild (+), moderate (++), and severe (+++) according to the number of PCV-2–positive cells detected.9
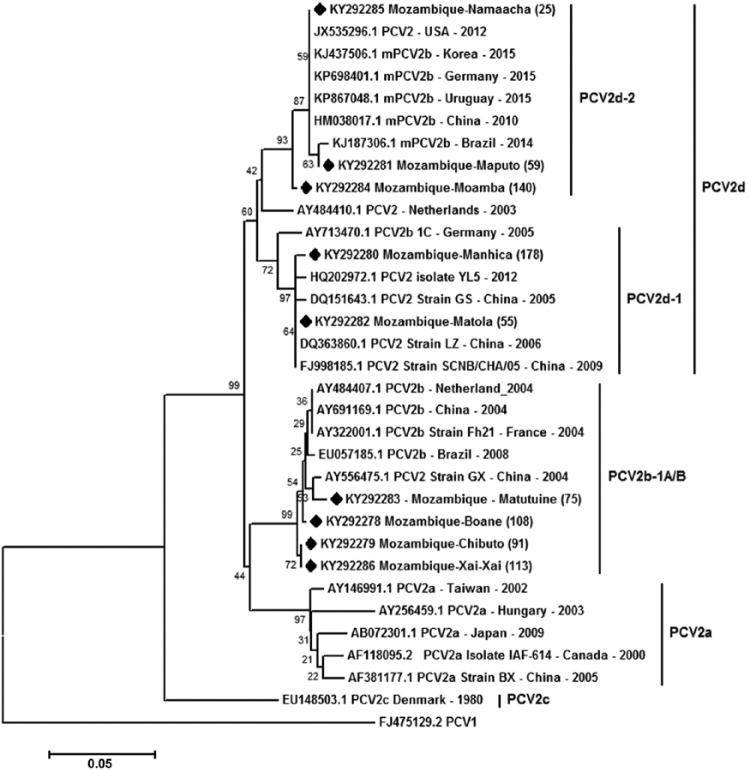
One positive sample from each of 9 districts was selected in order to obtain sequences covering the ORF2 region, using primers and PCR conditions described previously.4 The amplification products (30–45 ng) were purified (NucleoSpin II kit, Macherey-Nagel, Düren, Germany), labeled with 3.2 pmol of each primer and 2 µL of BigDye Terminator v3.1 cycle sequencing RR-100 (Applied Biosystems, Foster City, CA), and sequenced (ABI Prism 3100 genetic analyzer, Applied Biosystems) armed with 50-cm capillaries and POP6 polymer (Macherey-Nagel). The sequences were edited using BioEdit v.7.0.7 Alignment was performed using ClustalW (http://www.clustal.org/omega/) and MEGA5.23 Our dataset comprised the 9 sequences that represented 9 districts of Mozambique as well as representative PCV-2 sequences retrieved from GenBank (accessions are available in Fig. 3). Phylogenetic analysis was based on amino acid sequences using the neighbor-joining method–p-distance16 with 1,000 bootstraps, using MEGA5.23
Figure 3.
Phylogenetic tree constructed using the neighbor-joining method21 based on a dataset of 213 amino acid positions of the porcine circovirus 2 (PCV-2) capsid region (ORF2). All 35 sequences are represented by their GenBank accessions. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method16 and are in units representing the number of amino acid differences per site. The analysis involved 33 amino acid sequences. All positions containing gaps and missing data were eliminated. The optimal tree with the sum of branch length = 0.5963 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches.6 The 9 sequences from our study are marked with a black diamond. Evolutionary analysis was conducted in MEGA5.23 A PCV-1 sequence (FJ475129.2) was used as an outgroup.
A total of 457 pigs were slaughtered (average: 22 pigs/day) with a body weight of 5–120 kg (mean: 64 kg). The sampled pigs included exotic breeds (mainly Large White and Landrace), an indigenous breed (black pigs), and mixed breeds from 9 districts of southern Mozambique, namely: Matola, Matutuíne, Namaacha, Boane, Moamba, Manhiça (Maputo Province), Xai-Xai and Chibuto (Gaza Province), and Maputo (Maputo City Province; Fig. 1). Clinical signs were not evaluated, and gross anatomic lesions were not detected in mesenteric lymph nodes.
Figure 1.
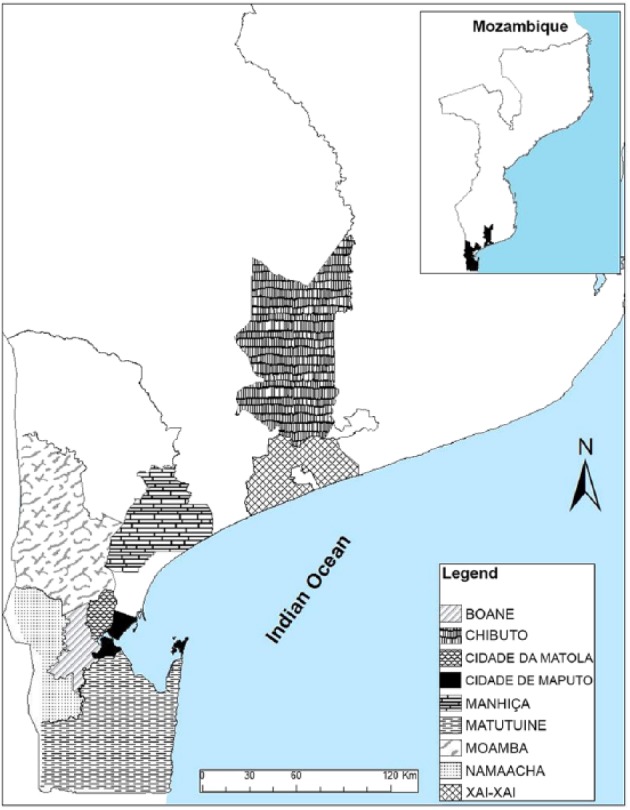
Map of Mozambique demonstrating the 9 districts of origin of the pigs sampled.
PCV-2 DNA was detected in 54% (62 of 111) of the samples and 78% (21 of 31) of the farms from all 9 districts (Table 1).
Table 1.
Number and percentage of PCV-2–positive farms and pigs from each Mozambique district.
| District | No. |
Positive farms |
Positive pigs |
|||
|---|---|---|---|---|---|---|
| Farms | Pigs | No. | % | No. | % | |
| Maputo | 2 | 4 | 2 | 100 | 4 | 100 |
| Matola | 6 | 14 | 5 | 83 | 12 | 86 |
| Matutuíne | 3 | 22 | 2 | 67 | 14 | 64 |
| Namaacha | 4 | 26 | 4 | 100 | 17 | 65 |
| Boane | 7 | 17 | 5 | 71 | 7 | 41 |
| Moamba | 4 | 9 | 2 | 50 | 2 | 22 |
| Manhiça | 1 | 4 | 1 | 100 | 1 | 25 |
| Xai-Xai | 1 | 2 | 1 | 100 | 1 | 50 |
| Chibuto | 3 | 13 | 1 | 33 | 4 | 31 |
| Total | 31 | 111 | 23 | 78 | 62 | 54 |
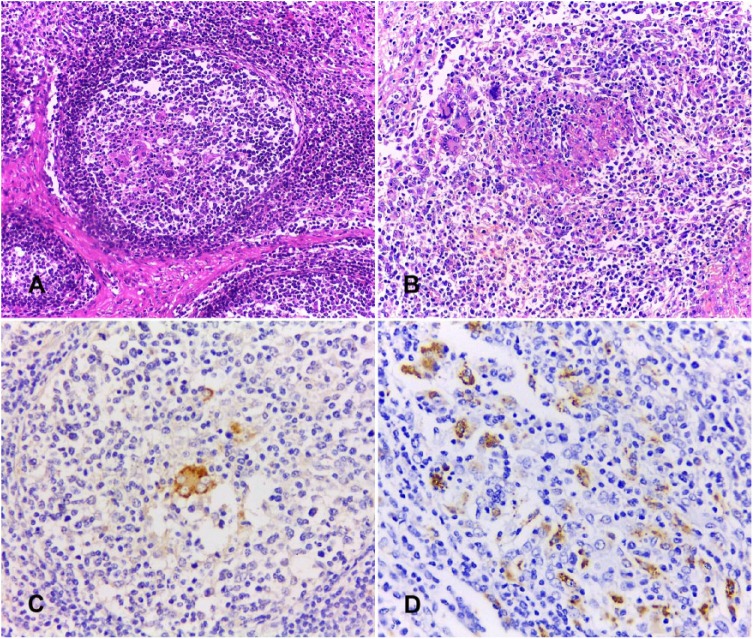
Histologic lesions were present in 64% (40 of 62) of the samples that were positive by PCV-2 PCR. The main lesions observed were eosinophil infiltration (17 of 62), lymphoid depletion (16 of 62), histiocyte infiltration (9 of 62), lymphoid hyperplasia (9 of 62), multinucleate giant cells (4 of 62), and necrotic areas (1 of 62). Lymphoid depletion, histiocyte infiltration, and multinucleate giant cells were frequently observed in the center of the lymphoid follicles (Fig. 2A). One sample with severe microscopic lesions exhibited multifocal-to-coalescing areas of necrosis associated with areas of diffuse histiocyte infiltration and foci of giant cell infiltration (Fig. 2B). Intracytoplasmic inclusion bodies were not evident in any of the lymph nodes with PCV-2–associated lesions.
Figure 2.
Histology and anti–porcine circovirus 2 (PCV-2) immunohistochemistry (IHC) of mesenteric lymph nodes from slaughtered pigs. A. Depleted lymphoid follicle with histiocyte and multinucleate giant cell infiltrate. H&E. B. Lymphoid depletion, focal necrosis, and histiocyte and multinucleate giant cell infiltrate. H&E. C. Immunostaining for PCV-2 in a multinucleate giant cell in the center of a lymphoid follicle. IHC, DAB chromogen, hematoxylin counterstain. D. Diffuse PCV-2 antigens in histiocyte cytoplasm. IHC, DAB chromogen, hematoxylin counterstain.
PCV-2 antigens were detected in 6 of 62 (10%) of the PCR-positive samples. Immunostaining was mild in 3 of 6 samples, moderate in 2 of 6, and severe in 1 of 6. Positive staining was seen within the cytoplasm of multinucleate giant cells (Fig. 2C) and histiocytes (Fig. 2D), located primarily in the depleted lymphoid follicles. PCV-2 antigens were detected in all lymph nodes that exhibited multinucleate giant cell infiltrates (4 of 6).
Phylogenetic analysis was performed using one sequence from each district. The sequences from our study covered 213 amino acids of the capsid region (ORF2). Five sequences from Mozambique clustered with PCV-2d, but 3 sequences were grouped in the PCV-2d-2 clade (KY292281, KY292285, KY292284) from the Maputo, Namaacha, and Moamba Districts, respectively. In addition, 2 sequences were grouped in the PCV-2d-1 clade (KY292280, KY292282) from the Manhiça and Matola Districts, respectively. The other 4 sequences (KY292283, KY292278, KY292279, KY292286), from Matutuíne, Boane, Chibuto, and Xai-Xai Districts, respectively, grouped with PCV-2b-1A/B sequences (Fig. 3).
PCV-2 DNA was detected in 78% of farms from 9 districts of southern Mozambique, suggesting that the virus is widely distributed throughout the swine population in this region of the country. Histologic lesions observed in mesenteric lymph nodes that were positive for PCV-2 by IHC were consistent with those described previously.8,12 The diffuse PCV-2 antigen immunostaining in one lymph node with a predominantly necrotic lesion has been described in cases of necrotizing lymphadenitis associated with PCV-2 infection.12 The mild or absent histologic lesions and low levels of PCV-2 antigens detected by immunostaining observed in our study are consistent with subclinical infection with PCV-2.22 On the other hand, marked lymphoid depletion and granulomatous infiltrates associated with moderate-to-severe immunostaining was observed in 3 samples, indicating that clinical PCVAD may occur in the pig population in Mozambique as well. However, the eosinophil infiltrate and lymphoid hyperplasia detected in our study could be associated with infection with pathogens other than PCV-2.
PCV-2 sequences from 4 districts clustered in PCV-2b-1A/B clades with sequences from The Netherlands, China, France, and Brazil (Fig. 3). PCV-2b-1A/B has also been identified in Thailand11 and Uruguay.20 In China, the PCV-2b-1A/B cluster was reported as the major subtype from 2001 to 2009.14
A global molecular genetic analysis of PCV-2 sequences demonstrated increased emergence of the PCV-2d genotype in swine.24 In our study, the PCV-2d sequences were found in PCV-2d-1 and 2d-2 clades. PCV-2d-1 sequences indicated a close relationship with sequences from China where PCV-2d was found to be an emerging and predominant PCV-2 subtype,2 whereas PCV-2d-2 grouped with sequences from different countries and even from different continents. Our results corroborate the increased presence of PCV-2d worldwide24; this is probably a global genotype shift of PCV-2b, as occurred with PCV-2a to PCV-2b >10 y ago.19
Given that the PCV-2 genotypes detected in our study have also been detected in many countries in Asia, Europe, and America, this close phylogenetic relationship of viruses may have resulted from swine trade between countries. The pig population in Mozambique includes European exotic breeds such as Large White and Landrace. A PCV-2–positive farm from Namaacha imports pigs from South Africa, which reported PMWS in the early 2000s caused by an outbreak of PCV-2a that was introduced into South Africa via pig semen imported from the United States.5 In 2013, a European-like PCV-2b was detected in the lymph nodes of slaughtered pigs in Uganda,17 but the sequences are not available in GenBank to be evaluated. Our study demonstrates that PCV-2b-1A/B and PCV-2d genotypes are circulating in herds in Mozambique and confirms the emergence of PCV-2d in pigs, as found in the last 7 y in other countries. Studies with a larger and more representative number of sequenced samples are necessary to estimate the true prevalence of PCV-2 genotypes of various Mozambican pig populations.
Acknowledgments
We thank the abattoir staff for their support during sampling, and the staff of the Division of Pathology, Veterinary Faculty, Eduardo Mondlane University for their work on the preparation of histologic samples.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: CJ Laisse’s PhD scholarship was co-funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and The World Academy of Science (TWAS): CNPq-TWAS Fellowships Programme 2012. This research was also supported by the National Research Fund (FNI) of Mozambiqu.
ORCID iDs: Cláudio J. Laisse  https://orcid.org/0000-0002-9257-9229
https://orcid.org/0000-0002-9257-9229
Matheus V. Bianchi  https://orcid.org/0000-0002-3951-0038
https://orcid.org/0000-0002-3951-0038
References
- 1. Allan GM, Ellis JA. Porcine circovirus: a review. J Vet Diagn Invest 2000;12:3–14. [DOI] [PubMed] [Google Scholar]
- 2. Cai L, et al. Identification of an emerging recombinant cluster in porcine circovirus type 2. Virus Res 2012;165:95–102. [DOI] [PubMed] [Google Scholar]
- 3. Chae C. Postweaning multisystemic wasting syndrome: a review of aetiology, diagnosis and pathology. Vet J 2004;168:41–49. [DOI] [PubMed] [Google Scholar]
- 4. Dezen D, et al. Multiply-primed rolling-circle amplification (MPRCA) of PCV2 genomes: applications on detection, sequencing and virus isolation. Res Vet Sci 2010;88:436–440. [DOI] [PubMed] [Google Scholar]
- 5. Drew TW, et al. Genetic similarities between porcine circovirus type 2 isolated from the first reported case of PMWS in South Africa and North American isolates. Vet Rec 2004;155:149–151. [DOI] [PubMed] [Google Scholar]
- 6. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39:783–791. [DOI] [PubMed] [Google Scholar]
- 7. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Series 1999;41:95–98. [Google Scholar]
- 8. Hansen MS, et al. Occurrence and tissue distribution of porcine circovirus type 2 identified by immunohistochemistry in Danish finishing pigs at slaughter. J Comp Pathol 2010;142:109–121. [DOI] [PubMed] [Google Scholar]
- 9. Haruna J, et al. The role of immunostimulation in the development of postweaning multisystemic wasting syndrome in pigs under field conditions. Can J Vet Res 2006;70:269–276. [PMC free article] [PubMed] [Google Scholar]
- 10. He J, et al. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol 2013;87:1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jantafong T, et al. Genetic characterization of porcine circovirus type 2 in piglets from PMWS-affected and -negative farms in Thailand. Virol J 2011;8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J, Chae C. Necrotising lymphadenitis associated with porcine circovirus type 2 in pigs. Vet Rec 2005;156:177–178. [DOI] [PubMed] [Google Scholar]
- 13. Larochelle R, et al. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods 1999;80:69–75. [DOI] [PubMed] [Google Scholar]
- 14. Li W, et al. Genetic analysis of porcine circovirus type 2 (PCV2) strains isolated between 2001 and 2009: genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes 2010;40:244–251. [DOI] [PubMed] [Google Scholar]
- 15. Liu J, et al. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol 2005;79:8262–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press, 2000. [Google Scholar]
- 17. Ojok L, et al. Detection and characterisation of porcine circovirus 2 from Ugandan pigs. Indian J Vet Pathol 2013;37:77–80. [Google Scholar]
- 18. Olvera A, et al. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 2007;357:175–185. [DOI] [PubMed] [Google Scholar]
- 19. Patterson AR, Opriessnig T. Epidemiology and horizontal transmission of porcine circovirus type 2 (PCV2). Anim Health Res Rev 2010;11:217–234. [DOI] [PubMed] [Google Scholar]
- 20. Ramos N, et al. Molecular analysis of porcine circovirus type 2 strains from Uruguay: evidence for natural occurring recombination. Infect Genet Evol 2013;19:23–31. [DOI] [PubMed] [Google Scholar]
- 21. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406–425. [DOI] [PubMed] [Google Scholar]
- 22. Segalés J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res 2012;164:10–19. [DOI] [PubMed] [Google Scholar]
- 23. Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiao CT, et al. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol 2015;96:1830–1841. [DOI] [PubMed] [Google Scholar]
- 25. Zlotowski P, et al. Intestinal lesions in pigs affected with postweaning multisystemic wasting syndrome. Pesq Vet Bras 2008;28:313–318. [DOI] [PubMed] [Google Scholar]