Abstract
Histophilus somni is an opportunistic pathogen responsible for respiratory and systemic diseases of cattle and sheep. Rapid and accurate detection of H. somni is essential to distinguish H. somni from other potential pathogens for proper control and treatment of infections. Nanomaterial optical fiber biosensors (NOFS) recognize analyte interactions, such as DNA hybridization, with high specificity and sensitivity, and were applied to detect H. somni DNA in culture and clinical samples. An ionic self-assembled multilayer (ISAM) film was fabricated on a long-period grating optical fiber, and a biotinylated, nucleotide probe complementary to the H. somni 16S rDNA gene was coupled to the ISAM film. Exposure of the ISAM::probe to ⩾100 killed cells of H. somni strain 2336 without DNA amplification resulted in attenuation of light transmission of ⩾9.4%. Exposure of the complexed fiber to Escherichia coli or non–H. somni species of Pasteurellaceae reduced light transmission by ⩽3.4%. Exposure of the ISAM::probe to blood, bronchoalveolar fluid, or spleen from mice or calves infected with H. somni resulted in ⩾24.3% transmission attenuation. The assay correctly detected all 6 strains of H. somni tested from culture, or tissues from 3 separate mice and calves tested in duplicate. Six heterologous strains (representing 6 genera) reacted at below the cutoff value of 4.87% attenuation of light transmission. NOFS detected at least 100 H. somni cells without DNA amplification within 45 min with high specificity. Although different fibers could vary in signal sensitivity, this did not affect the sensitivity or specificity of the assay.
Keywords: Histophilus somni, optical fiber biosensor assay
Introduction
Bovine respiratory disease (BRD) is a major health problem confronting cattle producers, and is the primary cause of morbidity, mortality, and economic losses to the cattle industry in North America, as well as decreased weight gain and performance.20,23 Histophilus somni is an opportunistic pathogen that is part of the normal mucosal flora of cattle and sheep, but also one of the predominant bacterial agents responsible for BRD.2,8,14 In fact, H. somni is the single most important cause of morbidity and mortality in stocker and feedlot cattle in North America, and is responsible for major economic losses to the cattle industry.5,13,19,37 Disease caused by H. somni is largely underdiagnosed, particularly myocarditis, because small fatal lesions in the heart can be overlooked at autopsy.22 Furthermore, serologic assays may yield false-positive results because of prior infection or colonization, and because of cross-reactive antibodies.6,36 Moreover, isolation of this bacterium from organs, particularly the lungs, may be difficult when other pathogenic organisms are concurrently present given the slow growth and highly fastidious nature of H. somni.15,21 Therefore, precise identification of H. somni infection is challenging, and a reliable test is needed to accurately detect H. somni in clinical samples.
Biosensor systems integrate biological components (such as enzymes, antibodies, other proteins, or nucleic acids) with a physicochemical transducer (optical, electrochemical, thermometric, piezoelectric) to yield a measurable signal. Optical fibers have become an important part of sensor technology.26 Light weight, low cost, and limited interference from outside signals (e.g., electrical noise) are among the main advantages of optical fiber biosensors (Sabri et al. Toward optical sensors: review and applications. J Physics: Conference Series, 43:012064; Jan 2013; Jakarta, Indonesia). Such fiber optic sensors are analytical devices that serve as transduction elements, and transmit light on the principle of total internal reflection. The aim of fiber optic sensor technology is to generate a signal proportional to the concentration of a chemical or biochemical to which the biological element reacts. Optical fiber grating devices, in which periodic variation is induced in the refractive index of the optical fiber core, operate by inducing a large decrease in the transmittance of light through the fiber at a specific wavelength. This wavelength can be modified by temperature, pressure, or binding events.17
Ionic self-assembled multilayer (ISAM) films (also commonly referred to as layer-by-layer films) are a unique class of materials that allow detailed structural and thickness control at the nanometer level, combined with uncomplicated manufacturing and low cost.9,10 Nanoscale self-assembled coatings on optical fibers greatly enhance the recognition of antigen–antibody binding or DNA hybridization through direct transmission measurements in a rugged, portable, inexpensive format.3,33 Therefore, fiber optic biosensors may be useful in culture-free, rapid detection tests. We report herein the successful application of nanomaterial optical fiber biosensors (NOFS) for the detection of H. somni DNA in culture and in infected animal tissues without the need for PCR amplification.
Materials and methods
Bacterial strains, growth conditions, and oligonucleotides used
All bacterial strains (Table 1) were stored in sterile skim milk at −80°C, and cultured onto Columbia agar with 5% sheep blood at 37°C and 6% CO2 for 24 h. For most experiments, the bacteria were grown in brain–heart infusion (BHI) broth to mid-log phase (~109 colony-forming units [CFU]/mL), determined spectrophotometrically, and confirmed by viable plate count. For H. somni, Haemophilus parasuis, and Haemophilus influenzae, the bacteria were grown in BHI broth supplemented with 0.1% Trizma base (Millipore Sigma, Burlington, MA), 1% horse serum, and 0.01% thiamine monophosphate supplemented with either 10 μg/mL of nicotinamide adenine dinucleotide (NAD; H. parasuis) or 1 μg/mL of NAD and 1 μg/mL of hemin (H. influenzae). The cells were harvested by centrifugation, and washed with and resuspended in phosphate-buffered saline (PBS; pH 7.0). The bacteria were killed and lysed to release genomic DNA (gDNA) by boiling for 10 min, as described.25 When necessary, gDNA was isolated from freshly grown cells (DNeasy blood and tissue kit, Qiagen, Valencia, CA).
Table 1.
Attenuation of light transmission through the optical fiber when the ISAM::probe duplex was exposed to bacterial strains, animal tissues, or animal fluids.
| Bacterial strains or animal tissues used* | No. of CFU present | % attenuation of light transmission (mean ± SD) |
|---|---|---|
| Histophilus somni 2336 | 100 | 9.4 ± 0.3 |
| 400 | 13.7 ± 0.4 | |
| 1,000 | 16.8 ± 0.1 | |
| 2,000 | 27.7 ± 1.5 | |
| 12,500 | 34.7 ± 1.4 | |
| H. somni 19 | 10,000 | 19.0 ± 2.1 |
| H. somni 58 | 10,000 | 24.3 ± 1.5 |
| H. somni 738 | 10,000 | 28.6 ± 2.2 |
| H. somni 797 | 10,000 | 22. 6 ± 0.9 |
| H. somni 1297 | 10,000 | 31.7 ± 3.7 |
| Actinobacillus pleuropneumoniae J45 | 10,000 | −1.2 ± 0.7 |
| Escherichia coli DH5α | 50,000 | 1.6 ± 1.9 |
| Haemophilus influenzae Eag | 10,000 | 3.4 ± 1.2 |
| Haemophilus parasuis Nagasaki | 10,000 | 0.4 ± 0.2 |
| Mannheimia haemolytica M42548 | 10,000 | 2.1 ± 1.4 |
| Pasteurella multocida X73 | 10,000 | 2.9 ± 1.9 |
| Fluids or tissues from animals infected with H. somni† | ||
| Mouse blood | ND‡ | 33.0 ± 1.1 |
| Calf BAL fluid | ND‡ | 24.3 ± 2.8 |
| Calf spleen | ND‡ | 24.7 ± 1.8 |
| Fluids or tissues from animals infused with PBS† | ||
| Mouse blood | ND‡ | 2.4 ± 0.4 |
| Calf BAL fluid | ND‡ | 2.2 ± 1.2 |
| Calf spleen | ND‡ | 3.8 ± 1.1 |
BAL = bronchoalveolar lavage; CFU = colony-forming unit; ISAM = ionic self-assembled multilayer; ND = none detected; PBS = phosphate-buffered saline.
Bacteria were grown in broth media (described in Materials and methods) to mid-log phase, washed, and suspended in PBS. The ISAM::probe duplex was exposed to indicated bacterial counts of each strain (in 0.5-mL volumes of PBS) to determine the attenuation of light transmission.
Blood was collected from mice, and BAL fluid and spleen tissues were collected from calves. Blood or BAL fluid was diluted at 1:10 in PBS, and 0.5-mL volumes of resulting suspensions were exposed to ISAM::probe duplex. Spleen tissues were thoroughly rubbed with cotton swabs, the swabs were subsequently immersed and swirled in 2-mL volumes of PBS, and 0.5-mL volumes of the resulting suspensions were exposed to ISAM::probe duplex.
When dilutions of fluid or tissue suspensions were plated on agar media, bacterial colonies were not recovered.
Oligonucleotide probes to the 16S ribosomal DNA (rDNA) gene (Table 2) were designed based on the published genome sequence for H. somni strain 2336,28 and were produced by Integrated DNA Technologies, Collinsville, IL. Biotin was synthesized into the 5’-end of the probe to determine if the addition of streptavidin to add an additional layer would enhance sensitivity of the assay. Digoxigenin was introduced into the 3’-end of the probe to enable colorimetric detection by ELISA (Table 2).
Table 2.
Oligonucleotide probes and primers to the 16S ribosomal DNA gene of Histophilus somni.
| Probe name | Probe sequence (5’–3’) | Comment |
|---|---|---|
| HS-Probe-BIOTIN | GTAAAGTTCTTTCGGTGATGAGGAAGGCGATTAGTTTAAGAGATTAATTGATTGACGATAATCACAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCG CGGT | Probe was biotinylated at the 5’-end |
| HS-Probe-DIG | GCGTTAATCGGAATGACTGGGCGTAAAGGGCACGCAGGTGGTGACTTAAGTGAGGTGTGA | Probe was labeled with digoxigenin at the 3’-end |
| HS-Primer-453F | GAAGGCGATTAGTTTAAGAG | Angen et al.1 |
| HS-Primer-860R | TTCGGGCACCAAGTRTTCA | Angen et al.1 |
PCR
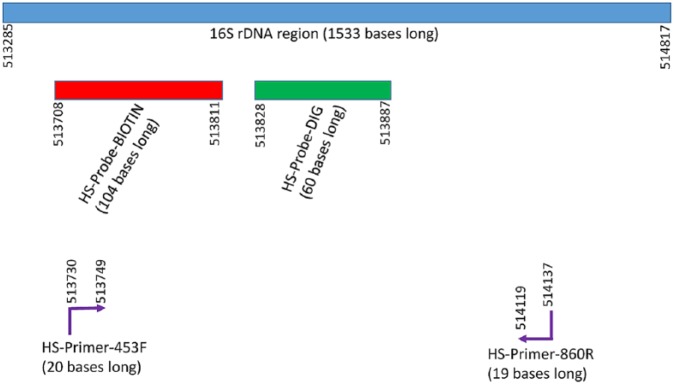
A PCR assay targeting the 16S rDNA gene of H. somni has been shown to specifically amplify DNA from H. somni.1 Therefore, this assay, which included specific primers (Fig. 1), annealing temperature, and magnesium concentration, was used to amplify the 16S rDNA region as the target for bacterial identification. GenBank accession of strain 2336 genome is CP000947.1, and the locus tag of the 1,533-bases long 16S rDNA sequence is HSM_R0014. PCR was used to confirm the specificity of the amplified region for identification of H. somni, and to generate amplicons for use in the NOFS and magnetic bead ELISA (MB-ELISA).
Figure 1.
Locations of the oligonucleotide primers and probes within the 16S ribosomal DNA region of the genome of Histophilus somni strain 2336. The numbers represent the sequential nucleotide locations of the H. somni strain 2336 genome (GenBank accession CP000947.1). The locus tag of the 1,533-bases long 16S rDNA sequence is HSM_R0014. Figures are not to the exact proportions. The website for access to gene and rDNA information is https://www.ncbi.nlm.nih.gov/nuccore/CP000947.1?report=GenBank.
The PCR was carried out as described previously.1 Briefly, the PCR assay (in a 25-μL volume) included 10 pmol each of the primers HS-Primer-453F and HS-Primer-860R (Table 2), 1 mM MgCl2, 200 μM dNTPs, 1× concentration of One Taq standard reaction buffer (New England Biolabs, Ipswich, MA), 1.25 units of One Taq DNA polymerase (New England Biolabs), and template DNA. Reaction conditions were initial denaturation at 95°C for 5 min, followed by 30 cycles at 95°C for 1 min, 57°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 10 min.
MB-ELISA
MB-ELISA was used to validate that the oligonucleotide probe to the 16S rDNA region (Table 2, Fig. 1) would bind to all strains of H. somni DNA tested and not to DNA from other bacterial species. MagnaLink streptavidin magnetic beads (Solulink, San Diego, CA) were used as the solid-phase support; the protocol was a modification of the procedure recommended by the manufacturer. Briefly, 60 pmol of the probe (HS-Probe-BIOTIN; Table 2) that was covalently coupled to biotin (Integrated DNA Technologies), was suspended in 750 μL of nucleic acid binding and wash buffer (50 mM Tris–HCl, 150 mM NaCl, 0.05% Tween 20, pH 8.0) and incubated with the streptavidin magnetic beads in 1.5-mL microcentrifuge tubes. The PCR products, or gDNA isolated from strain 2336, were incubated with the beads coupled to the biotinylated probe in 3× saline–sodium citrate hybridization buffer (0.45 M NaCl and 45 mM trisodium citrate) and 0.05% Tween 20. A 60-pmol aliquot of a digoxigenin-labeled probe (HS-Probe-DIG; Table 2) in 750 μL of hybridization buffer was then incubated with the bead::probe::DNA triplex. The binding of the digoxigenin-labeled probe onto the triplex was determined (Roche DIG detection starter kit, Millipore Sigma).
Fabrication of the ISAM film
Technical details of fabrication of the ISAM films and the NOFS operation have been described.3,34 The ISAM method involves the alternate dipping of a charged substrate (optical fiber) into an aqueous solution of a polycation and then an aqueous solution of a polyanion at room temperature (Supplementary Fig. 1). The optical fiber with a turnaround point long-period grating (TAP-LPG) was immersed in an aqueous 10 mM polyallylamine hydrochloride (pH 7.0) solution for 5 min, followed by rinsing 3 times in ultrapure water.3 The fiber was then immersed in aqueous 10 mM poly-1-[p-(3’-carboxy-4’-hydroxyphenylazo)benzenesulfonamido]-1,2-ethanediyl (PCBS; pH 7.0) solution for 5 min, and rinsed again. The final layer was always the negatively charged (because of terminal carboxyl groups) PCBS. These 2 steps were repeated until the desired number of bilayers was achieved, which was 4 for this assay.
Coupling the probe to the ISAM film
The ISAM film was exposed to a ~0.6-mL solution of 0.17 M freshly prepared solution of N-(3-dimethylaminodipropyl)-N’-ethylcarbodiimide (EDC), 0.17 M N-hydroxysulfosuccinimide (NHSS), and 60 pmol of the biotinylated oligonucleotide probe in PBS (pH 7.0). The mixture was incubated at room temperature for 30 min. EDC, in the presence of the NHSS cross-linker, couples an NHSS stable intermediate to carboxyl groups (on the film) forming an ester that enables efficient conjugation to amine groups (on the biotin) at physiologic pH (Thermo Scientific. Easy molecular bonding crosslinking technology; Reactivity chemistries, applications and structure references. In: Thermo Scientific, Crosslinking Technical Handbook, 2018:3–6). Biotin has the additional advantage of binding tightly and spontaneously to streptavidin (see below).
Conjugation of streptavidin to the ISAM film
In an alternative method to couple the probe onto the ISAM film using a streptavidin intermediate, 4 bilayers were deposited onto the optical fiber, leaving PCBS with negatively charged carboxyl groups exposed. Forty μL of streptavidin (1 mg/mL in PBS, pH 7.0) was mixed with 0.6 mL of cross-linker solution (0.17 M EDC and 0.17 M NHSS in PBS, pH 7.0). The mixture was added to the fiber and incubated for 8 h with mixing every 15 min. The fiber was rinsed, and the H. somni 16S rDNA biotinylated probe was added to the streptavidin-coated fiber for spontaneous coupling to streptavidin, as described above. The ISAM::streptavidin::probe triplex was then incubated with serial dilutions of Escherichia coli strain DH5α or H. somni strain 2336. The optical fiber used for this study was different from the one used in the prior section and had lower signal sensitivity. This sensitivity is determined primarily by the maximum attenuation strength of the fiber, which is determined by the fabrication conditions of the fiber. The signal sensitivity does not correlate with sensitivity of the assay, or the capability to identify true-positive samples.
NOFS assay
The ISAM::probe duplex was incubated with the sample (gDNA; amplicon from PCR using gDNA; amplicon from PCR using dilutions of lysed, killed cells; or dilutions of tissue or fluid extracts from infected animals) to enable hybridization between the probe and DNA in the sample, as described previously.3 The TAP-LPG in the optical fiber induces a broad attenuation band 1,450–1,600 nm, as shown previously.3 The strength of this attenuation increases proportionately as additional material binds to the surface of the fiber, providing the fundamental sensing mechanism of the assay. Because of the uniform growth of the attenuation across the wavelength range, it is possible to characterize the response of the assay simply by recording the change in the transmitted light at 1,550 nm, which is the peak of the attenuation band. Thus, in the experiments described herein, light in the range of 1,400–1,700 nm was transmitted through the ISAM fiber to an optical analyzer, and the difference in the attenuation in light transmission at 1,550 nm before and after exposure to the sample in PBS buffer was reported. This difference occurred as a result of increased coupling of light out of the core of the optical fiber as a result of the sample binding to the DNA probe.
Animals and animal specimens
The bovine samples used in our study were obtained during earlier challenge experiments.11 Three calves were challenged by the intranasal or transtracheal routes with H. somni strain 2336, and 3 calves were inoculated with saline alone. Bronchoalveolar lavage (BAL) fluid or spleen sections from these calves were stored frozen at −20°C.11 Six male BALB/c mice weighing 26–32 g (Charles River Laboratories, Wilmington, MS), were housed under controlled conditions of temperature, humidity, and lighting to acclimate for 7 d. The mice were divided into 2 groups: 3 mice were given intraperitoneal (IP) injections of 1 × 107 cells of H. somni strain 2336 in 100 µL of saline, and 3 mice were given IP inoculations of saline only. At 7 d post-injection, the mice were euthanized by exposure to excess carbon dioxide. BAL fluid was collected as described,16 and blood and spleens were collected. All murine samples were tested fresh shortly after postmortem. All bovine and murine samples tested by NOFS were also cultured for H. somni. All procedures were approved by the Institutional Animal Care and Use Committee (protocol 13-188 CVM).
Detection of H. somni DNA from blood and tissues of infected animals
Blood and/or BAL fluids were diluted 1:10 in 2 mL of PBS. Spleen tissues were rubbed thoroughly with cotton swabs, and subsequently the swabs were immersed in 2 mL of PBS. All samples were then boiled for 10 min to release bacterial DNA into the suspension. Serial dilutions of the cell suspensions were cultured onto Columbia blood agar to determine if viable bacteria were present.
Statistical analyses
The means and standard deviations (SDs) were calculated from the results of assays repeated at least 3 times. Analysis of variance was calculated using an online calculator (http://www.danielsoper.com/statcalc3/calc.aspx?id=43), and used to compare the transmission attenuation among different culture dilutions. The Student t-test was calculated using an online calculator (http://www.socscistatistics.com/tests/studentttest/Default2.aspx), and used to compare the transmission attenuation exposures of the probe between infected versus control animal extracts. Similar t-tests were performed to compare the transmission attenuations that resulted from exposure of the probe to 2 different PCR products. The mean difference in transmission attenuation between groups was considered significant if p ⩽ 0.05.
Results
Specificity of the DNA probe for H. somni
A PCR using oligonucleotide primers to the 16S rDNA region (Table 2) amplified a 400-bp amplicon when 200 ng of gDNA from H. somni strain 2336 was used in the PCR assay. A similar size amplicon was also produced when at least 7 × 103 killed cells of strain 2336 were used in this PCR (data not shown). However, no DNA products were amplified when similar numbers of killed cells or quantity of gDNA from other bacterial species (Table 1) were used (data not shown), indicating that the primers were specific to this region of H. somni DNA.
Validation of the DNA probe used for hybridization with target DNA
As predicted, subsequent addition of a second complementary digoxigenin probe and color developer to the above assay resulted in a colorimetric change, indicating that successful hybridization of the probe onto the target PCR DNA occurred (data not shown). However, when H. somni strain 2336 gDNA (up to 4,000 ng) was used in the absence of prior PCR, there was no reactivity with the probe. This qualitative assay was performed to confirm that specific binding (hybridization) of the selected oligonucleotide probe to the respective target sequence of the bacterium occurred.
NOFS assay to detect H. somni in culture or tissues from infected animals
Incubation of the ISAM::probe duplex with PCR products from 7 × 103 or 7 × 104 H. somni strain 2336 cells (equivalent to ~29.4 pg or 29.4 ng of DNA, respectively)18 resulted in 10.6% and 58.7% attenuation of light transmission, respectively (Fig. 2). There was no detectable attenuation of light transmission from control samples lacking DNA. The NOFS assay was then tested for detection of specific DNA from serial dilutions of H. somni strain 2336 cells without PCR amplification. Incubation of the ISAM::probe duplex with 50,000 E. coli cells resulted in <1.6% attenuation of transmission, whereas incubation of the same duplex with 100, 400, 1,000, 2,000, or 12,500 lysed cells of H. somni strain 2336 produced >9.4% attenuation of light transmission (p < 0.0001 for all results; Table 1; Fig. 3). Increased attenuation of transmission correlated with an increase in the number of cells incubated with the ISAM::probe duplex. For example, there was >35% light attenuation when 12,500 H. somni strain 2336 cells were added to the probe and 16.8% attenuation when 1,000 cells were added. Similar results for the NOFS assay were obtained with all strains of H. somni tested (Table 1).
Figure 2.
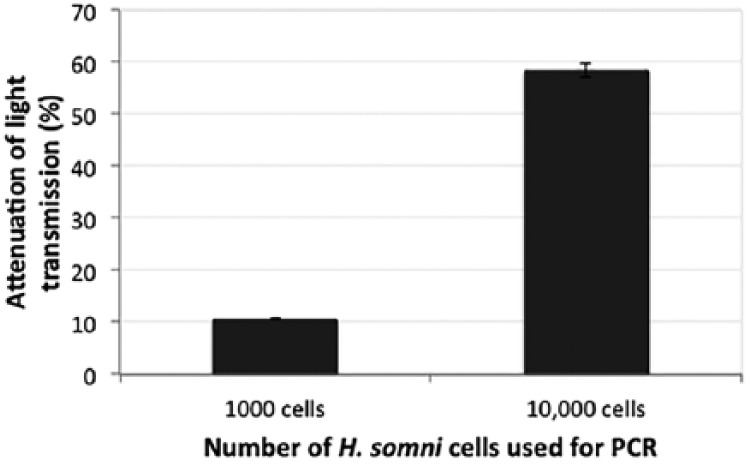
Attenuation of light transmission with the ionic self-assembled multilayer (ISAM)::probe duplex incubated with PCR amplicons. The signal attenuations (percent reduction of signal) resulting from exposure of the ISAM::probe to PCR products from 7 × 103 or 7 × 104 cells of Histophilus somni strain 2336 were significantly different (p = 0.000028). Error bars represent mean ± 1 SD.
Figure 3.
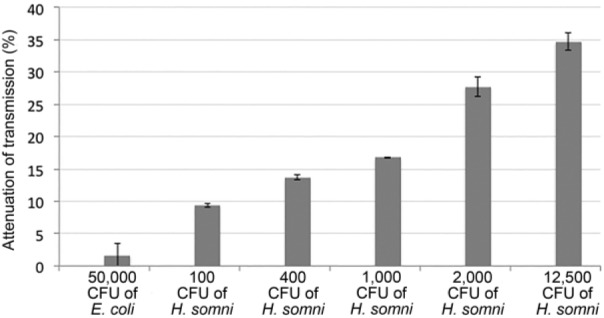
Attenuation of light transmission with the ionic self-assembled multilayer (ISAM)::probe duplex incubated with lysed cells of Histophilus somni or Escherichia coli. The signal attenuations (percent reduction of signal) resulting from exposure of the ISAM::probe to concentrations of H. somni strain 2336 from 100 CFU to 12,500 CFU were significantly different between each dilution, and with 50,000 CFU of E. coli (p < 0.0001). Error bars represent mean ± 1 SD.
Incubation of the ISAM::probe duplex with 10,000 cells each of other members of the Pasteurellaceae family (Actinobacillus pleuropneumoniae, H. influenzae, H. parasuis, Mannheimia haemolytica, and Pasteurella multocida) resulted in ⩽3.4% attenuation of transmission (Table 1). Therefore, as few as 100 cells of H. somni strain 2336 in a sample could be detected using the NOFS assay without the need for prior PCR, with a high degree of specificity for H. somni.
The NOFS assay was also used to determine if H. somni could be detected directly from clinical specimens from infected mice or calves. Blood from mice previously given IP inoculations of H. somni strain 2336, and BAL fluid or spleen sections from calves that had been challenged by intranasal or transtracheal inoculation, were tested.11 These specimens were tested shortly after challenge, and all were culture positive for H. somni. Attenuation of transmission between infected versus control tissues were 33.0% and 2.4% (p < 0.00001) for mouse blood, 24.3% and 2.2% (p = 0.000164) for calf BAL fluid, and 24.7% and 3.8% (p = 0.000102) for calf spleen tissue (Table 1; Fig. 4). However, live bacteria were not isolated from bovine BAL fluids or spleens by viable plate count, likely as a result of the samples being stored at only –20°C and freeze–thawed several times, resulting in loss of bacterial viability.
Figure 4.
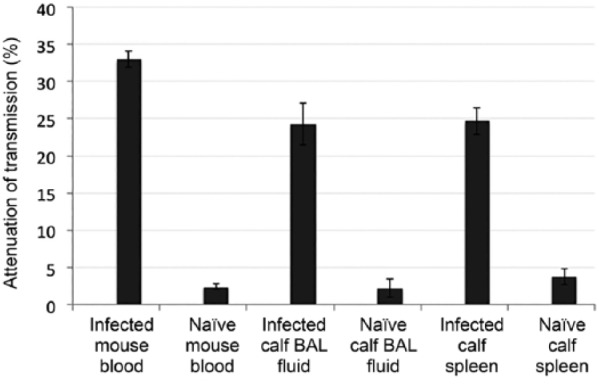
Attenuation of light transmission with the ionic self-assembled multilayer (ISAM)::probe duplex incubated with clinical samples from infected mice and calves. The signal attenuations (percent reduction of signal) resulting from exposure of the ISAM::probe to blood from infected mice, and bronchoalveolar lavage (BAL) fluid or spleen tissue samples from infected calves versus control animals were significantly different for mouse blood (p < 0.00001), calf BAL fluid (p = 0.000164), and calf spleen tissue (p = 0.000102). Error bars represent mean ± 1 SD.
Addition of a streptavidin linker to the NOFS assay
The use of streptavidin as a linker between the fiber and hybridizing DNA was tested to determine if the limit of detection (LOD) of H. somni could be further enhanced. Because of damage to the optical fiber, the optical fiber used in this study was different from the one used in the prior section and had lower signal sensitivity. For this second fiber, the maximum attenuation strength was 10 dB compared to 27 dB for the first fiber. Thus, the total magnitudes of light attenuation in this section were smaller than those reported above. Although the magnitude of the attenuation for the latter fiber was lower, there was no change in the LOD of the 2 fibers. Control samples contained all components except streptavidin. Incubation with 500–12,500 cells of E. coli using the NOFS with or without streptavidin resulted in negative attenuation values of 0.6% and 0.9%, respectively (Fig. 5). The negative signals corresponded to an increase in light transmission given random noise fluctuations. The attenuation of transmission of H. somni strain 2336 when tested by NOFS with and without streptavidin was 5.7% and 4.5% (p = 0.0599) for 500 cells, 7.1% and 5.8% (p = 0.0016) for 1,250 cells, 12.6% and 9.5% (p = 0.0009) for 5,000 cells, and 14.5% and 10.9% (p = 0.0024) for 12,500 cells. Therefore, treatment of the ISAM films with streptavidin enhanced the LOD of the NOFS assay, but was not necessary for effective detection.
Figure 5.
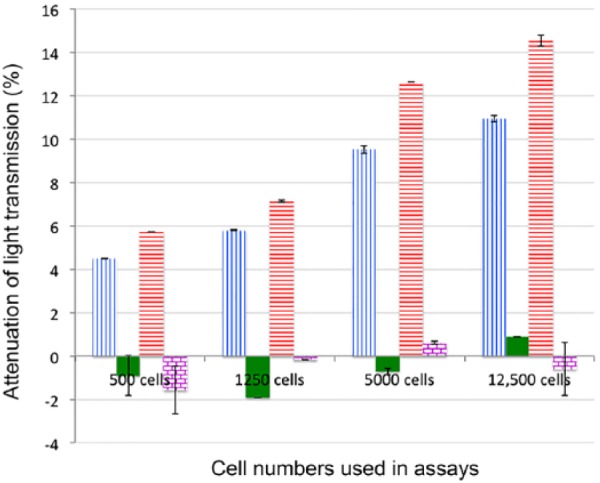
Effect of streptavidin bound to ionic self-assembled multilayer (ISAM) films on attenuation of light transmission. ISAM films that were conjugated with or without streptavidin were incubated with Histophilus somni strain 2336 or Escherichia coli strain DH5α. Blue vertical-line columns indicate H. somni strain 2336 extracts incubated with ISAM lacking streptavidin; solid green columns indicate E. coli strain DH5α extracts incubated with ISAM lacking streptavidin; red horizontal-line columns indicate H. somni strain 2336 extracts incubated with ISAM containing streptavidin; columns with purple grids indicate E. coli strain DH5α extracts incubated with ISAM containing streptavidin. The signal attenuations (percent reduction of signal) resulting from exposure of the streptavidin-coated ISAM::probe versus non-coated ISAM::probe to H. somni strain 2336 were significantly different when ≥500 cells were tested (p = 0.0599 for 500 cells, p = 0.00164 for 1,250 cells, p = 0.000936 for 5,000 cells, and p = 0.00239 for 12,500 cells). Error bars represent mean ± 1 SD.
The NOFS assay for testing the sample, and negative and positive controls, could be completed in 45 min. Based on the transmission attenuation percentages generated by cultures of all bacterial strains, from H. somni–positive animal tissues and fluids, and from controls, a cutoff value of light attenuation percentage of 4.87%, without incorporation of streptavidin, was established. This value was determined by multiplying the mean of the light attenuation of the true-negative isolates (1.53%) plus 2 times the SD (1.67).29 At this cutoff level, all samples tested were properly identified as positive for H. somni and negative for other bacterial species. Thus, the NOFS assay described has the potential to detect H. somni in clinical specimens rapidly and with a high degree of sensitivity and specificity. The inclusion of streptavidin has the potential to further enhance the LOD of the assay.
Discussion
Real-time PCR (rtPCR) assays have been developed for same-day identification of infectious disease agents.12,27,31 However, rtPCR technology is expensive and generally only available to well-established, large laboratories that have the experienced staff required to maintain and operate the equipment. A PCR test has previously been described to detect H. somni following culture,1 and directly from tissues.30 However, identification of the PCR products was carried out by gel electrophoresis, which is presumptive, and should be confirmed for specificity of the product by sequencing or hybridization to a specific probe. Sequencing of the PCR products confirms bacterial identity, but is time consuming and expensive. Therefore, more rapid, user-friendly assays are needed for smaller laboratories or clinics. Biosensors for detection of amplified PCR products have been developed using electrodes and impedance spectroscopy,7,35 and piezoelectric gold electrodes.32 However, such biosensors may be cost prohibitive and require trained staff to operate and maintain them. Therefore, they may not be suitable for small laboratories or clinics. Optical transduction methods such as surface plasmon resonance (SPR) are rapid and sensitive approaches that have been developed for detection of bacterial agents (Yang et al. Using loop-mediated isothermal DNA amplification [LAMP] and spectral surface plasmon resonance (SPR) to detect methicillin-resistant S. aureus [MRSA]. Intl Conf Biomed Engineer Biotechnol; May 2012; Macao, China). However, SPR assays require the use of light-emitting diodes and spectroscopy to generate excited light and receive a signal, making SPR sensors expensive and their operation requiring personnel with expertise in optics.24 A wide variety of biosensors have been described for detection of methicillin-resistant Staphylococcus aureus within 1–2 h, and which are applicable to most other bacteria.4 Although some reports describe the amount of DNA that can be detected under controlled conditions by biosensors, most assays cannot detect <104 CFU/mL after DNA amplification.4 In contrast, in our study, the NOFS assay had a detection limit of ~102 CFU/mL, confirming that the NOFS assay is a highly sensitive assay.
Unlike other biosensors that have been reported, the NOFS described herein accommodates deposition of a variety of materials into multiple layers, each just a nanometer thick, with TAP-LPGs covalently bound to a biotinylated oligonucleotide probe that is specific to the bacterium. Binding of complementary (target) H. somni DNA to the oligonucleotide probe altered the thickness and refractive index of the attached thin film, which in turn modifies the transmission characteristics of the fiber and produces an observable output indicating the presence of the target DNA. The assay does not require personnel with specialized training, and the components are relatively inexpensive.
BLAST analysis of the H. somni strain 2336 genome was used to design a probe with 100% identity at the nucleotide level with the 16S rDNA gene of H. somni. The MB-ELISA confirmed that the biotinylated oligonucleotide probe could be detected on a streptavidin-linked solid phase. However, only PCR-amplified DNA could be detected by the MB-ELISA, but not purified gDNA or DNA from lysed cells. In contrast, the NOFS assay was capable of detecting as few as 100 cells of H. somni strain 2336 with 100% sensitivity and specificity with or without prior PCR amplification, demonstrating that fewer bacterial cells could be detected by the NOFS assay than by MB-ELISA. Of interest was that PCR amplification resulted in greater attenuation of transmission with higher concentrations of cells (e.g., >10,000) than lower concentrations (e.g., 1,000). Regardless, all of the tested H. somni strains strongly attenuated light transmission (>19%) through the fiber. However, similar concentrations of other bacterial species, including other members of Pasteurellaceae, caused little or no attenuation of transmission (<4%). NOFS also detected the presence of H. somni strain 2336 DNA in mouse blood, or in calf BAL fluid or spleen from animals previously challenged with H. somni strain 2336. Although H. somni could be cultured from the fresh murine blood and tissue samples, cultures of the bovine samples for viable H. somni were negative, likely because the tissues had been frozen at −20°C for several years, and occasionally thawed. Therefore, NOFS may be able to detect H. somni from samples in which H. somni cannot be recovered by culture because of loss of viability or overgrowth by contaminants. Furthermore, the NOFS assay can be completed in a shorter period of time than PCR or rtPCR assay.
The addition of streptavidin as a linker between the ISAM film and the probe significantly enhanced the signal at all cell concentrations tested that contained >500 bacterial cells. The fiber used in this latter assay was slightly different from the one used in prior assays, and the percent attenuation exhibited by this fiber was always slightly less than the percentages obtained by the previous fiber, but the relative results were always the same. Although the fibers can be resurfaced and reused, they do have a half-life and will eventually break. Each fiber can vary in regard to the maximum attenuation strength of the fiber, which is determined by the fabrication process of the fiber. However, although different fibers may generate somewhat different absolute numbers, following standardization of mass production of large quantities of fibers, the sensitivity and specificity of the assay should remain the same.
Supplemental Material
Supplemental material, DS1_JVDI_10.1177_1040638718803665 for Identification of Histophilus somni by a nanomaterial optical fiber biosensor assay by Aloka B. Bandara, Ziwei Zuo, Kelly McCutcheon, Siddharth Ramachandran, James R. Heflin and Thomas J. Inzana in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank Ben Thomas for excellent technical assistance, and Indra Sandal and Shaadi Elswaifi for providing the calf samples used in the assays.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Financial support for the research described was provided, in part, by the U.S. Department of Agriculture–National Institute of Food and Agriculture (USDA-NIFA) award 2013-67015-21314 to TJ Inzana, and Agricultural Experiment Station funds through USDA-NIFA NC 1192 Multi-State Research Project (An integrated approach to bovine respiratory disease).
References
- 1. Angen O,et al. Development of a PCR test for identification of Haemophilus somnus in pure and mixed cultures. Vet Microbiol 1998;63:39–48. [DOI] [PubMed] [Google Scholar]
- 2. Arcangioli MA, et al. The role of Mycoplasma bovis in bovine respiratory disease outbreaks in veal calf feedlots. Vet J 2008;177:89–93. [DOI] [PubMed] [Google Scholar]
- 3. Bandara AB, et al. Detection of methicillin-resistant staphylococci by biosensor assay consisting of nanoscale films on optical fiber long-period gratings. Biosens Bioelectron 2015;70:433–740. [DOI] [PubMed] [Google Scholar]
- 4. Ceylan Koydemir H, et al. MEMS biosensors for detection of methicillin resistant Staphylococcus aureus. Biosens Bioelectron 2011;29:1–12. [DOI] [PubMed] [Google Scholar]
- 5. Corbeil LB. Histophilus somni host-parasite relationships. Anim Health Res Rev 2007;8:151–160. [DOI] [PubMed] [Google Scholar]
- 6. Corbeil LB, et al. Haemophilus somnus: bovine reproductive and respiratory disease. Can Vet J 1986;26:90–93. [PMC free article] [PubMed] [Google Scholar]
- 7. Corrigan DK, et al. Impedimetric detection of single-stranded PCR products derived from methicillin resistant Staphylococcus aureus (MRSA) isolates. Biosens Bioelectron 2012;34:178–184. [DOI] [PubMed] [Google Scholar]
- 8. Czuprynski CJ, et al. Complexities of the pathogenesis of Mannheimia haemolytica and Haemophilus somnus infections: challenges and potential opportunities for prevention? Anim Health Res Rev 2004;5:277–282. [DOI] [PubMed] [Google Scholar]
- 9. Decher G. Fuzzy nanoassemblies. Science 1997;277:1232–1237. [Google Scholar]
- 10. Decher GH, et al. Buildup of ultrathin multilayer films by a self-assembly process: consecutively alternating adsorption of anionic and cationic polyelectrolytes on charged surfaces. Thin Solid Films 1992;210/211:831–835. [Google Scholar]
- 11. Elswaifi SF, et al. The role of lipooligosaccharide phosphorylcholine in colonization and pathogenesis of Histophilus somni in cattle. Vet Res 2012;43:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fellahi S, et al. Comparison of SYBR green I real-time RT-PCR with conventional agarose gel-based RT-PCR for the diagnosis of infectious bronchitis virus infection in chickens in Morocco. BMC Res Notes 2016;9:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gagea MI, et al. Diseases and pathogens associated with mortality in Ontario beef feedlots. J Vet Diagn Invest 2006;18:18–28. [DOI] [PubMed] [Google Scholar]
- 14. Griffin D,et al. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract 2010;26:381–394. [DOI] [PubMed] [Google Scholar]
- 15. Haines DM, et al. Immunohistochemical study of Hemophilus somnus, Mycoplasma bovis, Mannheimia hemolytica, and bovine viral diarrhea virus in death losses due to myocarditis in feedlot cattle. Can Vet J 2004;45:231–234. [PMC free article] [PubMed] [Google Scholar]
- 16. Han H,et al. Thymic stromal lymphopoietin amplifies the differentiation of alternatively activated macrophages. J Immunol 2013;190:904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kersey AD, et al. Fiber grating sensors. J Lightwave Tech 1997;15:1442–1463. [Google Scholar]
- 18. Kubitschek HE, Freedman ML. Chromosome replication and the division cycle of Escherichia coli B/r. J Bacteriol 1971;107:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loneragan GH, et al. Trends in mortality ratios among cattle in US feedlots. J Am Vet Med Assoc 2001;219:1122–1127. [DOI] [PubMed] [Google Scholar]
- 20. Martin SW, et al. The association of titers to Haemophilus somnus, and other putative pathogens, with the occurrence of bovine respiratory disease and weight gain in feedlot calves. Can J Vet Res 1998;62:262–267. [PMC free article] [PubMed] [Google Scholar]
- 21. Moisan PG, Fitzgerald SD. Haemophilus somnus myocarditis in feedlot cattle. Agri-Practice 1995;16:21–24. [Google Scholar]
- 22. O’Toole D,et al. Diagnostic exercise: myocarditis due to Histophilus somni in feedlot and backgrounded cattle. Vet Pathol 2009;46:1015–1017. [DOI] [PubMed] [Google Scholar]
- 23. O’Toole D, Sondgeroth KS. Histophilosis as a natural disease. Curr Top Microbiol Immunol 2016;396:15–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Piliarik M, Homola J. Surface plasmon resonance (SPR) sensors: approaching their limits? Opt Express 2009;17:16505–16517. [DOI] [PubMed] [Google Scholar]
- 25. Queipo-Ortuno MI, et al. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol 2008;15:293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramsden JJ. Optical biosensors. J Mol Recognit 1997;10:109–120. [DOI] [PubMed] [Google Scholar]
- 27. Schmalz G,et al. Detection of five potentially periodontal pathogenic bacteria in peri-implant disease: a comparison of PCR and real-time PCR. Diagn Microbiol Infect Dis 2016;85:289–294. [DOI] [PubMed] [Google Scholar]
- 28. Siddaramappa S,et al. Horizontal gene transfer in Histophilus somni and its role in the evolution of pathogenic strain 2336, as determined by comparative genomic analyses. BMC Genomics 2011;12:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh G. Determination of cutoff score for a diagnostic test. Internet J Lab Med 2006;2:1–4. [Google Scholar]
- 30. Tegtmeier C,et al. Comparison of bacterial cultivation, PCR, in situ hybridization and immunohistochemistry as tools for diagnosis of Haemophilus somnus pneumonia in cattle. Vet Microbiol 2000;76:385–394. [DOI] [PubMed] [Google Scholar]
- 31. Thorn M,et al. Active cytomegalovirus infection diagnosed by real-time PCR in patients with inflammatory bowel disease: a prospective, controlled observational study. Scand J Gastroenterol 2016;51:1075–1080. [DOI] [PubMed] [Google Scholar]
- 32. Tombelli S, et al. A DNA-based piezoelectric biosensor: strategies for coupling nucleic acids to piezoelectric devices. Talanta 2006;68:806–812. [DOI] [PubMed] [Google Scholar]
- 33. Wang Z, et al. Highly sensitive optical response of optical fiber long period gratings to nanometer thick ionic self-assembled multilayers. Appl Phys Lett 2005;86:223104–223101. [Google Scholar]
- 34. Wang Z,et al. Biosensors employing ionic self-assembled multilayers adsorbed on long-period fiber gratings. Sensors Actuators B 2009;139:618–623. [Google Scholar]
- 35. Wang Z,et al. Label-free, electrochemical detection of methicillin-resistant Staphylococcus aureus DNA with reduced graphene oxide-modified electrodes. Biosens Bioelectron 2011;26:3881–3886. [DOI] [PubMed] [Google Scholar]
- 36. Widders PR, et al. Experimental abortion and the systemic immune response to “Haemophilus somnus” in cattle. Infect Immun 1986;54:555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wollums AR. The bronchopneumonias (respiratory disease complex of cattle, sheep, and goats) In: Smith BP, ed. Large Animal Internal Medicine. 5th ed. St. Louis, MO: Elsevier, 2015:584–617. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS1_JVDI_10.1177_1040638718803665 for Identification of Histophilus somni by a nanomaterial optical fiber biosensor assay by Aloka B. Bandara, Ziwei Zuo, Kelly McCutcheon, Siddharth Ramachandran, James R. Heflin and Thomas J. Inzana in Journal of Veterinary Diagnostic Investigation