Abstract
We conducted a cross-sectional study to detect trypanosome infections of horses and donkeys in the Riyadh Province of Saudi Arabia. DNA was extracted from blood samples collected from 368 horses and 142 donkeys, and subjected to universal first ribosomal internal transcribed spacer region (ITS1)-PCR followed by Trypanosoma evansi species–specific RoTat1.2-PCR. The universal ITS1-PCR revealed T. evansi infection in horses (n = 12; 3.3%) and donkeys (n = 4; 2.8%). There was no significant effect of sex or age on the prevalence of trypanosomiasis in horses or donkeys. Application of the RoTat1.2-PCR revealed that the RoTat1.2 VSG gene was absent from the positive ITS1-PCR samples of 3 horses and 1 donkey. This discrepancy could be explained by the circulation of T. evansi type B in Saudi Arabia; however, this suspicion requires confirmation.
Keywords: Equine, ITS1, PCR, RoTat1.2, Saudi Arabia, trypanosomiasis
Trypanosomiasis, or surra, caused by Trypanosoma evansi, is a serious disease that affects camels and horses in tropical and subtropical countries and often leads to reduced productivity and economic losses.6 The parasite is found in both intra- and extravascular fluids of multiple hosts and is transmitted mechanically by several genera of hematophagous flies such as Tabanus, Chrysops, Atylotus, Haematopota, and Stomoxys.11 In camels and horses, trypanosomiasis is found in acute and chronic forms, and clinical signs include intermittent fever, lacrimation, conjunctival petechiae, anemia, edema, enlarged lymph nodes, abortion, decreased fertility, and loss of body weight, which can result in death.11
In Saudi Arabia, the horse population is estimated to be >33,000 (http://www.fao.org/faostat/en/#data/QA, using filters: Saudi Arabia, Stocks, Horses, 2016), and more than 500 horses are imported annually from different countries such as the United Arab Emirates, the United States, and Europe. Horses are used for various purposes, including husbandry activities, transportation, racing, showing, and breeding. Diagnosis of equine trypanosomiasis relies on conventional parasitologic examinations and serologic assays.2,3 DNA-based technologies including PCR have been widely used in the diagnosis of trypanosomiasis infection in camels, horses, cattle, and pets, given its sensitivity and specificity in detecting all stages of parasitic infection.4,12,14,15,17 There is little information available on the prevalence or epidemiology of equine trypanosomiasis in Saudi Arabia. Thus, we aimed to estimate the prevalence of Trypanosoma spp. in horses and donkeys in the Riyadh Province of Saudi Arabia.
Our study was reviewed and approved by the Ethics Committee of the Department of Biological Science, Shaqra University (Riyadh, Saudi Arabia). This cross-sectional investigation was conducted from May 2015 to August 2017 in Riyadh Province, which is in the central part of Saudi Arabia between 24º38ºN and 46º43º E (Fig. 1). Riyadh Province has very hot summers, when the temperature approaches 50ºC. The average high temperature in July is 45ºC. Winters are cold but seldom dropping below 0ºC, with windy nights. The overall climate is arid, receiving very little annual rainfall (21.4 mm), with a relative humidity of 10–47% throughout the year. Riyadh Province is also known to have many dust storms (http://sdwebx.worldbank.org/climateportal/index.cfm?page=country_historical_climate&ThisCCode=SAU#).
Figure 1.

Map of Riyadh Province, Saudi Arabia.
Blood samples (n = 510) were collected from 368 horses and 142 donkeys. All of the animals were apparently healthy at the time of blood collection. Blood samples were collected from each animal (5–10 mL) from the jugular vein into blood collection tubes (BD EDTA Vacutainer tube, Gribbles Pathology, VIC, Australia) and transported to the parasitology laboratory at the Department of Biological Sciences, Faculty of Science and Humanities, Shaqra University, for DNA extraction.
Total genomic DNA was isolated (DNeasy blood and tissue kit, Qiagen, Hilden, Germany), eluted in 100 μL of elution buffer according to the manufacturer’s instructions, and stored at −80°C prior to being sent to the molecular laboratory at the School of Biological and Marine Sciences, University of Plymouth (Plymouth, UK) for PCR analysis.
A 2-step PCR protocol was used to detect Trypanosoma spp. in DNA samples. In the first step, first ribosomal internal transcribed spacer (ITS1)-PCR was performed to amplify a 250–700-bp fragment from the ITSI region using the ITS1 CF forward primer (5’-CCGGAAGTTCACCGATATTG-3’) and the ITS1 BR reverse primer (5’-TGCTGCGTTCTTCAACGAA-3’), which differentiates trypanosome species by their respective band size, with 250, 400, 480, and 700 bp indicating T. vivax, T. simiae, T. brucei subspecies, and T. congolense savannah, respectively.12 Then, all positive 480-bp ITS1-PCR samples were further subjected to T. evansi species–specific RoTat1.2-PCR using a primer set that amplifies 151 bp of the T. evansi RoTat1.2 VSG gene (TeRoTat920F: 5’-CTGAAGAGGTTGGAAATGGAGAAG-3’; TeRoTat1070R: 5’-GTTTCGGTGGTTCTGTTGTTGTTA-3’).10 The 2 PCRs were performed in a final reaction volume of 50 µL, containing 25 µL of 2× DNA polymerase master mix (Dream Taq, Thermo Scientific, Nalgene, UK), 0.4 µM (1 µL) of each primer, and 4 µL of DNA template; the reaction was brought to the final volume of 50 μL with 19 μL of PCR-grade water (Invitrogen, Paisley, UK). Positive and negative controls were included in each reaction. The thermocycling conditions consisted of an initial 2-min incubation at 95°C, followed by 40 cycles of denaturation at 95°C for 30 s, primer annealing at 58°C for 30 s and extension at 72°C for 1 min, followed by a final extension step at 72°C for 5 min, after which the samples were held at 4°C. The PCR product was electrophoresed on 1.5% agarose gel containing 10 μL/mL SYBER Safe (Thermo Scientific) in Tris acetate–EDTA buffer at 100 V for 45 min and photographed under ultraviolet transilluminators (ImageQuant Laz4000, GE Healthcare Life Science, Hammersmith, UK). The size of each product was estimated by comparison with a GeneRuler 100-bp DNA ladder marker (Thermo Scientific).
To determine the trypanosome species, positive samples of T. evansi were sent to Macrogen Europe (The Netherlands) for sequencing of the ITS1 region, and the results were compared with sequences available in GenBank using BLAST (http://blast.ncbi.nlm.nih.gov/). Phylogenetic analyses were constructed based on comparing the identified sequence in our study with the related sequences from GenBank by the neighbor-joining method with the distance algorithms available in MEGA v.7 (https://www.megasoftware.net/). Bootstrap values were determined with 1,000 replicates of the datasets. The sequence was deposited in GenBank under accession MH087231 (see Supplementary Figure 1).
Statistical analyses were performed (SPSS v.17.0, IBM, New York, NY). Animals were divided into 2 age groups: animals ⩽5-y-old (young) and animals >5-y-old (mature). The associations between the prevalence of Trypanosoma and risk factors such as sex and age were determined using the chi-square test; p ⩽ 0.05 was considered significant.
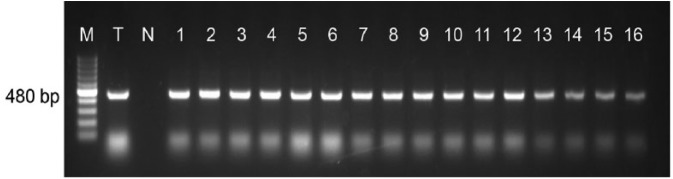
ITS1-PCR revealed that the prevalence of T. evansi infection was 3.3% in horses and 2.8% in donkeys (Table 1; Fig. 2). However, RoTat1.2-specific PCR revealed that 4 samples that tested positive by ITS1-PCR were negative for the RoTat1.2 gene (Fig. 3). Phylogenetic data and BLAST results of ITS1 showed that T. evansi is closely related to T. evansi isolate STIB 780, accession KU552341, from Kenyan camels (Fig. 4). Both horses and donkeys in Riyadh Province were infected with a single trypanosome species (T. evansi), and no mixed-species infection was found. There was no significant effect of sex or age on the prevalence of T. evansi in horses or donkeys (Table 1).
Table 1.
Chi-square analysis of the association of sex and age risk factors with Trypanosoma spp. infection in horses and donkeys in the Riyadh Region of Saudi Arabia.
| Risk factor | Horses |
Donkeys |
||||||
|---|---|---|---|---|---|---|---|---|
| n | Positive | χ2 | p value | n | Positive | χ2 | p value | |
| Sex | ||||||||
| Male | 127 | 3 (2.4) | 0.481 | 0.496 | 55 | 1 (1.8) | 0.567 | 0.327 |
| Female | 241 | 9 (3.7) | 87 | 3 (3.4) | ||||
| Total | 368 | 12 (3.3) | 142 | 4 (2.8) | ||||
| Age | ||||||||
| ⩽5 y | 217 | 4 (1.8) | 3.368 | 0.066 | 49 | 0 (0.0) | 2.168 | 0.141 |
| >5 y | 151 | 8 (5.3) | 93 | 4 (4.3) | ||||
| Total | 368 | 12 (3.3) | 142 | 4 (2.8) | ||||
p > 0.05 = nonsignificant. Numbers in parentheses are percentages.
Figure 2.
Results of the 1.8% agarose gel electrophoresis showing amplified DNA of the 480-bp band that is specific for Trypanosoma brucei subspecies (T. evansi) using ITS1-PCR. Lane M = 100-bp molecular size marker (GeneRuler); lane T = Trypanosoma spp. positive control DNA; lane N = negative PCR control (water); lanes 1–12 = template DNA isolated from horse blood samples; lanes 13–16 = template DNA isolated from donkey blood samples.
Figure 3.
Results of 1.8% agarose gel electrophoresis separating the amplicons of the RoTat1.2 VSG gene of Trypanosoma evansi. Lane M = 100-bp molecular size marker (GeneRuler); lane T.e = T. evansi positive control DNA; lane N = negative PCR control (water); lanes 1–12 = template DNA isolated from horse blood samples; lanes 13–16 = template DNA isolated from donkey blood samples. All samples were weakly positive except samples 3, 6, 9, and 16, which were negative. The 151-bp band is specific for T. evansi using the TeRoTat920F/TeRoTat1070R primer set.
Figure 4.

Phylogenetic relationships based on the partial nucleotide sequences (4983-bp) of ITS1 ribosomal DNA of Trypanosoma (Riyadh equine) with other variants of T. evansi in GenBank. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model. Evolutionary analyses were conducted in MEGA v.7. Numbers at nodes are bootstrap values derived from 1,000 replicates.
We detected only T. evansi and not T. vivax using PCR with the generic primers ITS1 CF and ITS1 BR, which allow for the detection and differentiation of all pathogenic trypanosomes, as has been shown in previous studies.12,15,16 In contrast, the T. evansi type A species–specific RoTat1.2-PCR confirmed the presence of parasite DNA in 12 ITS1-PCR–positive samples and its absence in 4 ITS1-PCR–positive samples. The latter finding could be attributed to the presence of non–RoTat1.2 T. evansi type B outside of Africa, although this should be confirmed by further investigation.17
Overall, T. evansi was more prevalent in horses (n = 12; 3.3%) than in donkeys (n = 4; 2.8%) in our study, but the prevalence in horses was lower than identified in other studies. In Jordan, the prevalence of T. evansi was reported to be 9.6% (n = 83) in horses and 0.0% (n = 40) in donkeys.1 In Sudan, trypanosomes were more prevalent in horses 12.7% (n = 393) than in donkeys (3.4%, n = 116).15 However, in Gambia, the prevalence of trypanosome infection was 93.4% (n = 183) in horses and 82.7% (n = 58) in donkeys.8 In Israel, the seroprevalence of T. evansi was 4.6% (n = 614) in horses.5 In Pakistan, the prevalence of T. evansi was 0.5% (n = 375) in horses with the Woo test, 1.3% with both ITS1 CF/BR PCR and RoTat1.2 PCR, and 14.4% with a card agglutination test for trypanosomiasis (CATT)/Trypanosoma evansi.17 However, in India, PCR testing indicated that 6.8% (n = 44) of donkeys were infected with T. evansi.13
Differences in results from previous studies may be attributed to management and environmental differences of the studies as well as sample sizes and methodology (i.e., microscopic, serologic, and PCR techniques). The lower prevalence of infection in donkeys than horses could be related to the feeding preference of vectors for horses over donkeys or may be related to the behavior of donkeys, which prevent flies from feeding by their head movements.8
The nonsignificant difference between the prevalences of infection according to sex or age group is in agreement with previous studies, which demonstrated that sex and age groups are equally exposed to and affected by trypanosomiasis.7,17 However, other studies indicated that male and adult donkeys were more often affected by trypanosomiasis than female and young donkeys.9
Supplemental Material
Supplemental material, DS1_JVDI_10.1177_1040638718798688 for Molecular detection of equine trypanosomiasis in the Riyadh Province of Saudi Arabia by Abdullah D. Alanazi, Robert Puschendorf, Bashir Salim, Mohamed S. Alyousif, Ibrahim O. Alanazi and Hajri R. Al-shehri in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank the staff members of the Biological Science Department, Faculty of Science and Humanities, Shaqra University, and the staff members of the molecular laboratory at the School of Biological Science, University of Plymouth, UK for their technical support.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was supported by the Deanship of Scientific Research at Shaqra University.
References
- 1. Abo-Shehada MN, et al. Prevalence of Surra among camels and horses in Jordan. Prev Vet Med 1999;38:289–293. [DOI] [PubMed] [Google Scholar]
- 2. Al-Khalifa MS, et al. Blood parasites of livestock in certain Regions in Saudi Arabia. Saudi J Biol Sci 2009;16:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alanazi AD, et al. Seroprevalence study on Theileria equi and Babesia caballi antibodies in horses from central province of Saudi Arabia. J Parasitol 2012;98:1015–1017. [DOI] [PubMed] [Google Scholar]
- 4. Barghash SM, et al. Prevalence of Trypanosoma evansi in Maghrabi camels (Camelus dromedarius) in northern-west coast, Egypt using molecular and parasitological method. Acta Parasitol Glob 2014;5:125–132. [Google Scholar]
- 5. Berlin A, et al. Prevalence of Trypanosoma evansi in horses in Israel evaluated by serology and reverse dot blot. Res Vet Sci 2012;93:1225–1230. [DOI] [PubMed] [Google Scholar]
- 6. Desquesnes M, et al. Trypanosoma evansi and surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Res Int 2013;2013:194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eyob A, et al. A cross-sectional study of equine trypanosomosis and its vectors in Wolayta zone, southern Ethiopia. J Vet Med Anim Health 2011;3:21–26. [Google Scholar]
- 8. Faye D, et al. Prevalence and incidence of trypanosomosis in horses and donkeys in the Gambia. Vet Parasitol 2001;101:101–114. [DOI] [PubMed] [Google Scholar]
- 9. Hussain M, et al. Molecular detection and seasonal prevalence of Trypanosoma brucei and its effect on hematobiochemical parameters in donkeys from Dera Ghazi Khan District in Southern Punjab, Pakistan. Pak J Zool 2016;48:1781–1786. [PubMed] [Google Scholar]
- 10. Konnai S, et al. Development and application of a quantitative real-time PCR for the diagnosis of surra in water buffaloes. Infect Genet Evol 2009;9:449–452. [DOI] [PubMed] [Google Scholar]
- 11. Luckins AG, et al. Non-tsetse-transmitted animal trypanosomiasis. In: Maudlin I et al., eds. The Trypanosomiases. Oxfordshire, UK: CABI, 2004:269–281. [Google Scholar]
- 12. Njiru ZK, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 2005;95:186–192. [DOI] [PubMed] [Google Scholar]
- 13. Ravindran R, et al. Trypanosoma evansi in camels, donkeys and dogs in India: comparison of PCR and light microscopy for detection—short communication. Veterinarski Arhiv 2008;78:89–94. [Google Scholar]
- 14. Rjeibi MR, et al. First report of surra (Trypanosoma evansi infection) in a Tunisian dog. Parasite 2015;22:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salim B, et al. Molecular detection of equine trypanosomes in the Sudan. Vet Parasitol 2014;200:246–250. [DOI] [PubMed] [Google Scholar]
- 16. Salim B, et al. Molecular epidemiology of camel trypanosomiasis based on ITS1 rDNA and RoTat 1.2 VSG gene in the Sudan. Parasit Vectors 2011;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tehseen S, et al. Field investigation of Trypanosoma evansi and comparative analysis of diagnostic tests in horses from Bahawalpur, Pakistan. Turk J Vet Anim Sci 2017;41:288–293. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS1_JVDI_10.1177_1040638718798688 for Molecular detection of equine trypanosomiasis in the Riyadh Province of Saudi Arabia by Abdullah D. Alanazi, Robert Puschendorf, Bashir Salim, Mohamed S. Alyousif, Ibrahim O. Alanazi and Hajri R. Al-shehri in Journal of Veterinary Diagnostic Investigation