Abstract
Accurate segmentation of the prostate and organs at risk (e.g., bladder and rectum) in CT images is a crucial step for radiation therapy in the treatment of prostate cancer. However, it is a very challenging task due to unclear boundaries, large intra- and inter-patient shape variability, and uncertain existence of bowel gases and fiducial markers. In this paper, we propose a novel automatic segmentation framework using fully convolutional networks with boundary sensitive representation to address this challenging problem. Our novel segmentation framework contains three modules. First, an organ localization model is designed to focus on the candidate segmentation region of each organ for better performance. Then, a boundary sensitive representation model based on multi-task learning is proposed to represent the semantic boundary information in a more robust and accurate manner. Finally, a multi-label cross-entropy loss function combining boundary sensitive representation is introduced to train a fully convolutional network for the organ segmentation. The proposed method is evaluated on a large and diverse planning CT dataset with 313 images from 313 prostate cancer patients. Experimental results show that the performance of our proposed method outperforms the baseline fully convolutional networks, as well as other state-of-the-art methods in CT male pelvic organ segmentation.
Keywords: Image Segmentation, Fully Convolutional Network, Boundary Sensitive, Male Pelvic Organ, Prostate Cancer, CT
1. Introduction
Prostate cancer is the second most common cancer and one of the leading causes of cancer death in American men. The American Cancer Society estimates that there will be about 164,690 new cases of prostate cancer and 29,430 deaths from prostate cancer in the United States in 20182. Nowadays, external beam radiation therapy (EBRT) is one of the most commonly used and effective treatments for prostate cancer. During a standard protocol for EBRT planning, the prostate volume and the neighboring organs at risk, such as the bladder and rectum, are manually delineated in the planning CT images for killing cancer cells while preserving normal cells. However, manual annotation is a highly time-consuming and laborious task, even for experienced radiation oncologists. Moreover, manual annotation often suffers large intra- and inter-observer variability which then affects treatment planning. Hence, in order to minimize the workload of oncologists and improve the accuracy of the annotation process, an automatic and reliable method of segmenting male pelvic organs in CT images is highly desired in clinical settings.
Accurate automatic segmentation of male pelvic organs (i.e. prostate, bladder, and rectum) in CT images, however, is a very challenging task for the following reasons. First, due to the low contrast of CT images, the boundaries between male pelvic organs and surrounding structures are usually unclear, especially when two nearby organs are adjacent to each other. As shown in Fig. 1, the boundaries are hard to distinguish even after careful contrast adjustment. Second, as they are soft tissues, the shapes of these organs have high variability and can change significantly across patients, as observed in the slices of three patients in Fig. 1(a), (b), and (c). Third, the existence of bowel gases makes the appearance of the rectum uncertain (as shown in Fig. 1(b) and (c)). Finally, the presence of fiducial markers used to guide dose delivery brings interference to the image intensity of the surrounding organs and causes information loss, as shown in Fig. 1(d).
Figure 1.
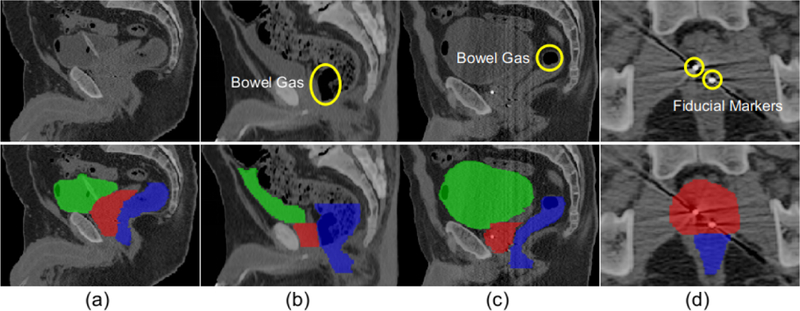
Typical CT images and their pelvic organ segmentations. The four columns indicate images from four patients. The first row shows the original CT slices and the second row shows the same corresponding slices overlaid with their respective segmentations. Yellow circles in (b) and (c) indicate different amounts of bowel gas in the rectum, while yellow circles in (d) indicate two fiducial markers in the prostate. Red denotes the prostate, green denotes the bladder, and blue denotes the rectum. The first three columns are shown in the sagittal view and the last column is shown in the axial view.
To address the aforementioned challenges, many computer-aided pelvic organ segmentation methods have been proposed over the past few years, such as Zhan and Shen (2003); Huang et al. (2006); Haas et al. (2008); Feng et al. (2010); Chen et al. (2011); Liao et al. (2013); Gao et al. (2016); Acosta et al. (2017). Theoretically, these methods mainly involve two key technologies: (1) feature extraction, in which some hand-crafted features based on expert knowledge, such as Gabor filters (Zhan and Shen (2003)), intensity histograms (Chen et al. (2011)), and Haar features (Gao et al. (2016)) are extracted for CT image analysis; and (2) model design, in which the state-of-the-art methods adopt various models based on different prior information, such as boundary regression model (Huang et al. (2006)) and pixel-wise classification model (Liao et al. (2013)). Recently, convolutional networks (Simonyan and Zisserman (2014)) which can achieve feature learning and model training in an end-to-end framework, have seen great success in semantic segmentation. Further, after the proposal of fully convolutional networks (Long et al. (2015)), images with arbitrary sizes can now be segmented with more efficient inference and learning. For medical image segmentation, one of the most successful fully convolutional networks is U-net (Ronneberger et al. (2015)), which is designed with a U-shaped architecture and has won many segmentation competitions, such as the ISBI challenges for segmentation of neuronal structures in electron microscopic stacks (Arganda-Carreras et al. (2015)) and for computer-automated detection of caries in bitewing radiography (Wang et al. (2016)).
However, the direct use of these fully convolutional networks to segment the male pelvic organs in CT images cannot achieve good results for the following reasons. First, these networks have no effective mechanism to address the challenges of unclear boundaries and large shape variations in CT male pelvic organ segmentation. Second, these networks aim to label each voxel that belongs to a target organ, and thus do not make full use of the meaningful boundary information. Finally, the CT images scanned from the whole pelvic area introduce complex backgrounds for each organ segmentation and make these networks hard to optimize.
To this end, we propose an automatic segmentation framework using fully convolutional networks with boundary sensitive representation to accomplish the challenging CT male pelvic organ segmentation. Specifically, an organ localization model is first designed to extract proposals which can fully cover organs and contain less background compared to original images. Then, a boundary sensitive representation involving both low-level spatial cue and high-level semantic cue is used to assign soft labels to voxels near the boundary. Finally, a fully convolutional network under a multi-label cross-entropy loss function is trained to reinforce the network with semantic boundary information.
In summary, our main contributions are listed below:
To make better use of boundary information to guide segmentation, we propose a novel boundary representation model deduced from a multi-task learning framework, which assigns soft labels to voxels near the boundary based on both low-level spatial cue and high-level semantic cue, and then produces a more robust and accurate representation of the boundary information.
To address the challenges of unclear boundaries and large shape variations, we design a multi-label cross-entropy loss function to train the segmentation network, where each voxel can not only contribute to the foreground and background, but also adaptively contribute to the boundary with different probabilities.
To reduce the complexity of optimization, we introduce a localization model to focus on the candidate segmentation region of each organ, which can contribute significantly to even better performance.
We perform comprehensive experiments on a challenging dataset with 313 planning CT images from 313 prostate cancer patients to demonstrate the effectiveness of our method. The experimental results show that our method can significantly improve performance compared to the baseline fully convolutional networks and also outperforms other state-of-the-art segmentation methods.
2. Related Works
The proposed algorithm is a deep learning-based method for segmenting male pelvic organs in CT images. Therefore, in this section, we will review the literature related to CT male pelvic organ segmentation and deep learning for medical image segmentation.
2.1. CT Male Pelvic Organ Segmentation
The segmentation of male pelvic organs in CT images has been investigated for a long time and the existing methods can be roughly categorized into three classes: registration-based methods, deformable-model-based methods, and machine-learning-based methods.
The registration-based methods (Davis et al. (2005); Lu et al. (2011); Chen et al. (2009)) first optimize a series of transformation matrices from the current treatment image to the planning and previous treatment images, and then their respective segmentation images are transformed to the current treatment image and further combined to obtain the final segmentation result for the current treatment image. Davis et al. (2005) combined large deformation image registration with a bowel gas segmentation and deflation algorithm to locate the prostate. Lu et al. (2011) formulated the segmentation and registration steps using a Bayesian framework and further constrained them together using an iterative conditional model strategy to segment the prostate, rectum, and bladder. Chen et al. (2009) adopted meshless point sets modeling and 3D nonrigid registration to align the planning image to the treatment image. Even though the registration-based methods have achieved attractive performance, their segmentation accuracy demonstrates less robustness in the case of inconsistent appearance changes due to reasons such as different amounts of urine in the bladder or bowel gas in the rectum.
The deformable-model-based methods (Costa et al. (2007); Yokota et al. (2009); Chen et al. (2011)) also depend on the planning and previous treatment images. They first initialize a deformable model using the learned organ shapes from planning and previous treatment images and then optimize the deformable model based on the image-specific knowledge. Costa et al. (2007) designed a coupled 3D deformable model for the joint segmentation of the prostate and bladder. Yokota et al. (2009) developed a hierarchical multi-object statistical shape model to represent the diseased hip joint structure and applied it to segment the femur and pelvis from 3D CT images. Chen et al. (2011) incorporated anatomical constraints from surrounding bones and a learned appearance model to segment the prostate and rectum. The deformable-model-based methods are strongly limited by the deformable model initialization, so the final segmentation result is sensitive to large organ motions and shape variations.
The machine-learning-based methods (Lu et al. (2012); Zhen et al. (2016); Gao et al. (2016); Ma et al. (2017)) have recently attracted much interest. They usually formulate the segmentation problem as a standard classification or regression problem, where classifiers or regressors trained from training images are used to predict the likelihood map and then obtain the segmentation result. Lu et al. (2012) incorporated information theory into the learning-based approach for pelvic organ segmentation. Ma et al. (2017) trained a population model based on the data from a group of patients and a patient-specific model based on the data of the individual patient for prostate segmentation. Instead of learning a single segmentation task, some approaches adopt multi-task learning to improve the performance by exploiting commonalities and differences across tasks. For example, Zhen et al. (2016) modeled the estimation of each coordinate of the points on shape contours as a regression task and then jointly learned coordinate correlations to capture holistic shape information. Gao et al. (2016) proposed a multi-task random forest to learn the displacement regressor jointly with the organ classifier to segment CT male pelvic organs. The machine-learning-based methods are more flexible and efficient than the registration-based and deformable-model-based methods since they do not require prior information. However, the performance of these methods is always compromised by the limited expressive power of the hand-crafted features, especially for the very challenging segmentation of male pelvic organs in CT images.
2.2. Deep Learning for Medical Image Segmentation
Instead of extracting features manually, deep learning incorporates feature engineering into a learning step and automatically learns effective feature hierarchies from a set of data. Deep learning has been widely used for medical image segmentation (Lai (2015)), with respective reviews given in Shen et al. (2017) and Litjens et al. (2017). Carneiro et al. (2012) decoupled the rigid and nonrigid classifiers based on deep belief networks for automatic left ventricle segmentation. Xu et al. (2014) used deep learning for the automatic extraction of feature representation to accomplish classification and segmentation tasks in medical image analysis and the experimental results showed that the automatic feature learning outperforms manual identification. Havaei et al. (2017) presented a convolutional neural network architecture by exploiting both local features as well as global contextual features for brain tumor segmentation. In order to incorporate low-level and high-level features, a special fully convolutional network, namely U-net (Ronneberger et al. (2015)), was proposed and has been successfully applied to many different medical segmentation tasks.
Deep learning is also driving advances for male pelvic organ segmentation in CT images. For example, Men et al. (2017) proposed a multi-scale convolutional architecture based on the deep dilated convolutional neural network to segment the bladder. Gordon et al. (2017) combined deep learning and level-sets for the segmentation of the inner and outer bladder wall. Shi et al. (2017) constructed an easy-to-hard transfer sequence for CT prostate segmentation by leveraging the manual annotation as guidance. Although these works showed some effectiveness in male pelvic organ segmentation, they were designed for segmenting a specific pelvic organ which cannot guarantee their effectiveness for segmenting other pelvic organs. Moreover, the network architectures adopted in these works did not take into account the challenging characteristics of CT pelvic organ segmentation, thus harming their performance.
3. Method
In this section, we will give a detailed description of the proposed framework for the segmentation of the prostate, bladder, and rectum in CT images. The whole architecture of the proposed framework is shown in Fig. 2, which mainly consists of three modules: organ localization used to focus on the candidate segmentation region for each organ for better performance (as shown in Fig. 2(a)); boundary sensitive representation aimed at representing the semantic boundary information with robust and accurate soft labels (as shown in Fig. 2(b)); multi-label segmentation introducing a multi-label cross-entropy loss function to reinforce the network with semantic boundary information for segmentation (as shown in Fig. 2(c)). Specifically, boundary sensitive representation and multi-label segmentation are performed on the organ proposals extracted based on organ localization, instead of the original images. Moreover, we adopt a U-net (Ronneberger et al. (2015)) like architecture as the backbone due to its excellent performance in medical image segmentation.
Figure 2.
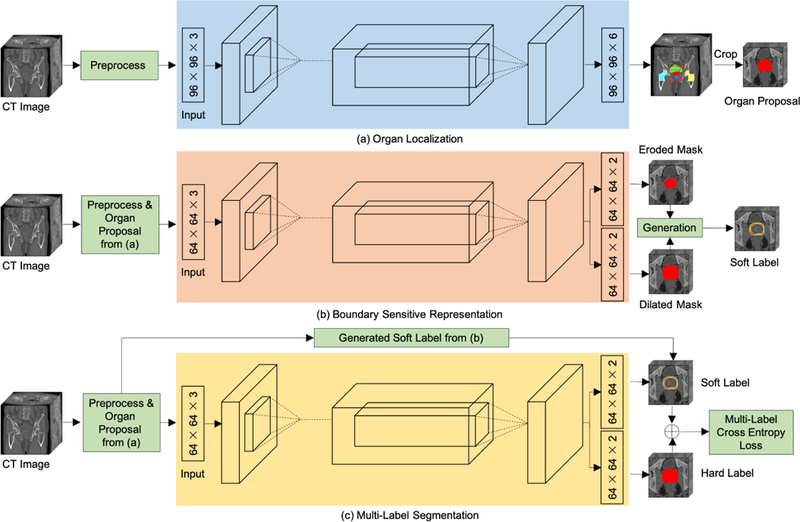
An illustration of the proposed framework for prostate, bladder, and rectum segmentations in CT images (with the prostate segmentation as an example). (a) Organ proposals of raw CT images are first generated by an organ localization network to focus on the candidate regions. (b) Then, a boundary sensitive representation network with multi-task learning is trained to generate soft boundary labels of the corresponding organ proposals. (c) Finally, a multi-task network that uses soft labels and hard labels as supervision is learned to perform voxel-wise segmentation in organ proposals.
3.1. Organ Localization
In segmenting each organ separately, the original CT image scanned from the whole pelvic area covers a large background relative to the small organ. This irrelevant background information not only increases the complexity of the feature space optimization, but also greatly reduces computational efficiency. Therefore, before fine segmentation of each organ, an organ localization model is first trained to extract a more accurate region of each organ from the original image. Here, in order to save training cost, the organ localization model is designed to perform multiple organ segmentations jointly rather than segment each organ separately. Besides the three target organs (prostate, bladder, and rectum), the right and left femoral heads are also jointly segmented due to their saliency and stability that can help to locate the target organs. As shown in Fig. 2(a), a U-net (Ronneberger et al. (2015)) like architecture is adopted for this multi-class (background, prostate, bladder, rectum, right and left femoral heads) segmentation problem. Overlapping patches across three sequential slices in the axial plane are used as input and the middle slices of the corresponding label patches are used as the expected prediction. For the segmentation of unseen images, we sample patches with a large stride and average the results as the final prediction for the whole images.
Based on the segmentation results, the centroid of each organ can be determined and then by taking the centroid as the center of a cube, the organ proposal can be extracted from the original image. In our implementation, the cropped proposal sizes for the prostate, bladder, and rectum are 128 × 128 × 128, 160 × 160 × 128, and 128 × 128 × 160 respectively. While the average sizes (with standard deviation) of the prostate, bladder, and rectum in our dataset are 56 ×49 × 47 (9 × 9 × 8), 76 × 88 × 57 (12 × 15 × 17), and 49 × 64 × 90 (11 × 11 × 14) in voxels, with the voxel resolution of 1mm × 1mm × 1mm. The cropped sizes are large enough to cover each organ and contain sufficient background context information for segmentation. The remaining operations are all performed on the organ proposals, instead of the original images.
3.2. Boundary Sensitive Representation with Soft Label
Theoretically, organ boundaries indicate the transition from the foreground to background or vice versa and can then be used to address the segmentation problem. Some methods under the guidance of boundary information have been proposed for medical image segmentation, such as Shao et al. (2015); He et al. (2017); Wang et al. (2018). However, they all rely on spatial distances to represent the boundary information while neglecting the semantic differences, leading to the inaccurate representation of organ information. For example, it is unreasonable to equally treat one easy voxel (correctly and confidently segmented) and one hard voxel with the same distance to the boundary. Therefore, in order to avoid introducing semantic gap and also maximize guidance ability, we focus on hard voxels near the boundary to represent the boundary information. Because these voxels are close to the boundary and have lower semantic discriminability, we call them boundary sensitive voxels. To achieve robust and accurate representation, we assign a soft label to each voxel based on those detected boundary sensitive voxels by a multi-task framework as follows.
First, two different types of morphological masks, i.e., eroded and dilated masks, are generated by applying morphological erosion and dilation operations on the ground truth segmentation. Next, a multi-task fully convolutional network as shown in Fig. 2(b) is trained under supervision of the generated eroded and dilated masks. If voxels are the hard samples near the boundary, they tend to be induced by the morphological masks rather than the ground truth segmentation, namely, correct segmentation in terms of the morphological masks and incorrect segmentation in terms of the ground truth segmentation. Then, the boundary sensitive voxels are detected based on the differences between the ground truth segmentation and the predicted eroded and dilated masks. Based on this, we finally assign each voxel a soft label as follows, to represent the boundary information.
| (1) |
where
| (2) |
vi denotes the i-th voxel, Cj denotes the j-th voxel on the contour C, B is the union set of boundary sensitive voxels and contour voxels, ϵis a small value to prevent the soft labels of boundary sensitive voxels from being zero, and δ is a predefined parameter (δ = 3 here).
An illustration of the soft label generation is shown in Fig. 3. Using this method to generate soft labels, we can see that the hard voxels near the boundary contribute to the boundary in a weighted manner based on how close they are to the contour, while easy voxels are less focused. Therefore, the boundary can be represented in a more accurate and robust way without leading to semantic confusion.
Figure 3.
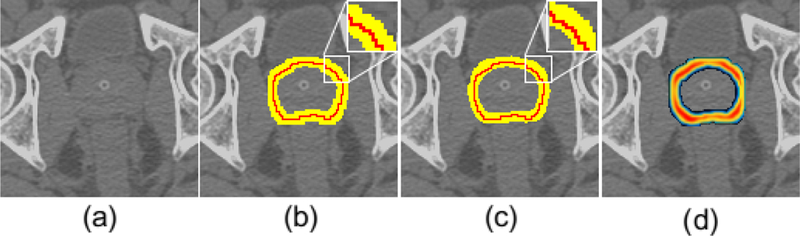
An illustration of the soft label generation (prostate is used as an example). (a) represents the original image; (b) shows the differential voxels (yellow) between the ground truth segmentation and corresponding morphological masks on both sides of the contour (red); (c) shows the mined boundary sensitive voxels (yellow), which are different from the differential voxels in (b); (d) shows the generated soft label map.
3.3. Multi-Label Segmentation for Male Pelvic Organs
By incorporating low-level and high-level features, the conventional U-net (Ronneberger et al. (2015)) can be trained from few images and produces pixel-wise segmentation results. However, for CT pelvic organ segmentation, it cannot deal with the region near the boundary very well due to unclear boundaries and large shape variations. Therefore, we introduce the generated soft labels into the segmentation process to train a more powerful network. To this end, each voxel is labeled with multi labels, i.e., one hard label indicating whether or not the voxel belongs to the foreground, and one soft label indicating the sensitive probability to the boundary. Based on this, we introduce a multi-label cross-entropy loss function to optimize the segmentation network:
| (3) |
where f (vi) ∈ {0, 1} denotes whether vi belongs to the foreground class, l(vi) ∈ [0, 1] is the generated soft label, and are the softmax output class probabilities corresponding to the hard label and soft label, respectively, and λ is a balance parameter (λ = 1 here). For new unseen images, only the predicted likelihood map of hard labels is used to perform the final segmentation.
We process different organs with separately trained models, so one voxel may be segmented as different organs. To deal with this overlapping segmentation, we adopt a max function to determine the final labels of voxels if predicted as different organs using different models:
| (4) |
where is the final predicted label of the voxel vi, and is the predicted organ probability using the segmentation network for organ c.
3.4. Basic Network Architecture
For all three models mentioned above, we implement a slightly modified version of the U-net (Ronneberger et al. (2015)) architecture. Our implementation differs from U-net (Ronneberger et al. (2015)) by setting the number of feature channels at the first convolutional layer to 32, instead of 64, and using Batch Normalization (BN) after each convolutional layer except the last 1 × 1 convolutional layer. Moreover, we do not include cropping due to the use of zero-padding.
4. Experiments and Results
4.1. Dataset and Preprocessing
We verify the performance of the proposed method on 313 planning CT images acquired from 313 prostate cancer patients. These images were collected from the North Carolina Cancer Hospital but were scanned using various types of CT equipment, causing the original in-plane resolution of these images to range from 0.932 to 1.365 mm and the inter-slice thickness to range from 1 to 3 mm. To deal with this, we resample all the images to the same resolution of 1 mm × 1 mm × 1 mm. While the average size of original images is 512×512×203, the average image size after resampling becomes 583 × 583 × 354. Two senior oncologists were invited to delineate the five pelvic organs (prostate, bladder, rectum, and two femoral heads), and the averaged results after their cooperative correction are used as the ground truth.
The fiducial markers and bone tissues cause the image intensity values to be in a large range that is far beyond the scope of the target segmentation organs. Therefore, we adopt a truncation function to reserve the intensity values from 500 to 1500 and then normalize the values into [0, 1]. Before inputting to the network, we subtract the mean value computed on the whole training set from each voxel. We perform five-fold cross-validation to conduct a fair comparison with the state-of-the-art methods. Specifically, the whole dataset is first randomly shuffled and then split into five folds. In each round, four folds are used for training and validation (with a ratio of 7:1) and the remaining fold is used for testing. Five repeated rounds are performed until each of the five folds is traversed as the testing set.
4.2. Implementation Details
The proposed model is based on the open-source deep learning framework Caffe (Jia et al. (2014)) for implementation and Nesterovs Accelerated Gradient method (NAG) for optimization. The batch size is set to 20, and momentum to 0.9. The learning rate is initially set to 10−3 and then decreased using an inv policy with gamma of 0.0001 and power of 0.53. The weights of all layers are initialized with Xavier initialization. At the training phase, we randomly crop patches from the whole image to train the organ localization network and use the cropped organ proposals to train the multi-task and multi-label segmentation networks. The corresponding patch sizes are 96 × 96 × 3 and 64 × 64 × 3, respectively. At the testing phase, we divide the image and organ proposal into overlapping patches with stride sizes of (24, 24, 1) and (16, 16, 1), respectively.
4.3. Evaluation Criteria
To evaluate the segmentation performance of different methods, we adopt the following commonly used metrics in medical image segmentation (Guo et al. (2016); Taha and Hanbury (2015)) as evaluation criteria:
- Dice Similarity Coefficient (DSC):
(5) - Positive Predictive Value (PPV):
(6) - Sensitivity (SEN):
(7) - Average Symmetric Surface Distance (ASD):
(8)
where Vgt and Vseg denote the voxel sets of manually labeled ground truth and automatically segmented organs respectively, and Sgt and Sseg are the corresponding surface voxel sets of Vgt and Vseg, d(p, Sgt) is the minimum Euclidean distance of voxel p ∈ Sseg to all voxels in Sgt. For DSC, PPV, and SEN, the larger the value, the better the performance, while for ASD, the smaller the value the better performance. If not specifically noted, the reported results are all based on the cropped organ proposals, instead of the original images.
In addition, we define two other metrics to evaluate the effectiveness of organ localization, namely the average ratio (AvgR) and the target-to-target ratio (TTR) as defined below:
| (9) |
and
| (10) |
where X and Y denote the set of the cropped organ proposals and the original images, respectively, gi is the ground truth label of the i-th voxel, and 1A(.) is the indicator function. A smaller AvgR means more useless background voxels are dropped and TTR = 1 means that all organ voxels are covered in the organ proposals, compared with the original whole images.
4.4. Impact of Morphological Kernel Size
To effectively capture boundary information in pelvic organ segmentation, we define soft labels under the guidance of erosion and dilation masks obtained by applying morphological erosion and dilation operations on the ground truth and introduce them into the segmentation task. Specifically, the kernel size K for the generation of morphological masks is a key parameter that affects the efficiency of soft labels. Intuitively, a larger size K causes more voxels around the boundary to be assigned as non-zero values which may cover more redundant information, while a smaller size K causes fewer voxels around the boundary to be assigned as non-zero values which may maintain less information for accurate boundary representation. Therefore, in order to investigate the optimal K for better performance, we use different K to generate the corresponding erosion and dilation masks, respectively, and then we train a corresponding multi-label prostate segmentation network based on each choice of K. The dataset used here is the extracted prostate proposals and split in the way mentioned in Sec. 4.1. Fig. 4 shows the boxplot of DSC values of five-fold cross-validation of the prostate segmentation with respect to different K values on the testing dataset. We can observe that the optimal K for prostate segmentation is 7, and both smaller values with insufficient information and larger values with more interference lead to inferior performance. Therefore, in our implementation, we set K = 7.
Figure 4.
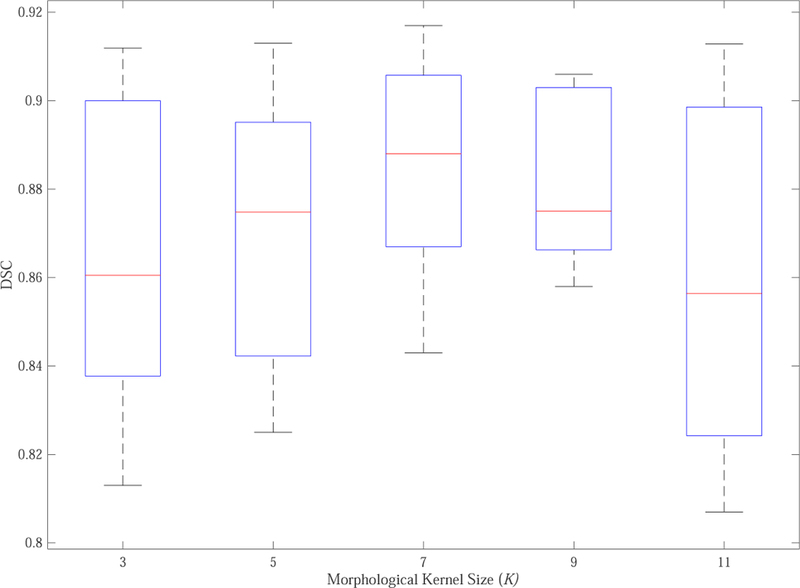
Boxplot of DSC values of five-fold cross-validation of the prostate segmentation with respect to different morphological kernel size (K).
4.5. Impact of Parameters δ and λ
There are two other parameters that impact the segmentation performance, namely, δ in Eq. 2 and λ in Eq. 3. To evaluate the sensitivity of segmentation performance to them, we change δ and λ separately and report the segmentation performance for the prostate based on DSC values in Fig. 5. From Fig. 5(a), we can see that the smaller or larger δ values will lead to too big or too small differences in soft labels of adjacent voxels, which cannot form an optimal representation of boundary information. When λ = 0, our multi-label segmentation model degenerates into the conventional U-net. When the value of λ gradually increases from 0 to 1, the performance of the model is improving as shown in Fig. 5(b). However, if the value of λ continues to increase, the performance will drop even worse than the conventional U-net (λ = 2). This is mainly because the boundary information dominates the segmentation which is not as robust as the whole organ information. Therefore, we set δ = 3 and λ = 1 in our implementation.
Figure 5.
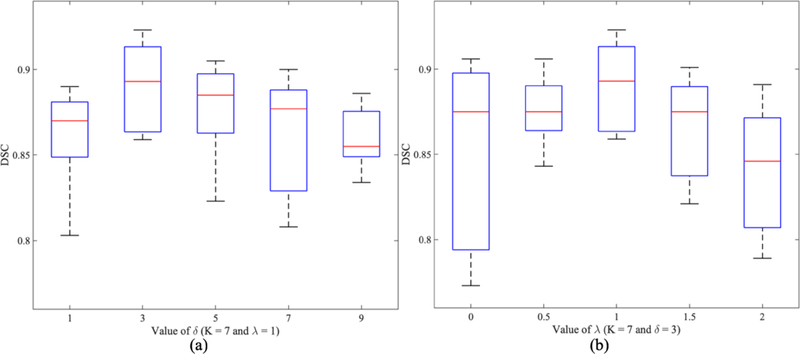
Boxplot of DSC values of our proposed method for the segmentation of prostate with respect to different values of δ (a) and λ (b).
4.6. Comparison with Fully Convolutional Networks
We achieve the segmentation of three pelvic organs (prostate, bladder, and rectum) in organ proposals based on the proposed multi-label segmentation network. In order to verify its effectiveness, we adopt several of the most successful fully convolutional networks using the same organ proposal data for the segmentation of three organs. The first one is FCNs (Long et al. (2015)), which is the first work to train convolutional networks in an end-to-end way for pixel-wise prediction and has shown outstanding performance in many fields. For comparison, we use the FCN-8s (Long et al. (2015)) model trained on the PAS-CAL VOC 2012 dataset (Everingham et al.) to finetune to the 313 CT image dataset. The second one is U-net (Ronneberger et al. (2015)), which is the winner of many segmentation challenges and also the basic architecture of our model. We adopt the same configuration as ours for comparison. The last one is modified from the dilated convolution network (Yu and Koltun), in which we replace the convolutional layers in the contracting path of U-net with dilated convolutions and call this architecture “Dilated U-net”.
Table 1 makes a quantitative comparison of mean DSC and ASD with standard deviation of FCN-8s (Long et al. (2015)), U-net (Ronneberger et al. (2015)), Dilated U-net, and our multi-label segmentation network. Moreover, we perform paired t-test between the results of each comparison method and our method to get the corresponding p values. It can be seen that our method achieves the best performance for segmenting the prostate, bladder, and rectum and outperforms other state-of-the-art methods with statistical significance. The segmentation performance of FCN-8s (Long et al. (2015)) shows that the conventional very deep fully convolutional network can obtain acceptable but unremarkable results for this challenging segmentation task. U-net (Ronneberger et al. (2015)), the very successful architecture for medical image segmentation achieves better performance than FCN-8s (Long et al. (2015)) in DSC, but it still cannot deal with the unclear boundary very well, as can be seen from the ASD values. Utilizing dilated convolutions to expand the receptive field without losing resolution, Dilated U-net further improves performance in terms of both DSC and ASD. Even so, our network outperforms comparison networks by a large margin. In particular, for the prostate and rectum, the performance improvement is especially meaningful due to their more indiscriminative boundary. These comparison results demonstrate that our multi-label loss imposes better supervision for the challenging CT pelvic organ segmentation, and the soft label used not only provides better representation for the boundary information but also contributes to guiding the organ segmentation.
Table 1:
Quantitative comparison of the segmentation results of three male pelvic organs by different networks. (The best results are indicated in bold, with asterisks implying statistical significance (p < 0.05)).
| Metric | FCN-8s | U-net | Dilated U-net | Ours | |
|---|---|---|---|---|---|
| Prostate | DSC p value |
0.83±0.08 4.3E-8∗ |
0.85±0.07 1.1E-5∗ |
0.86±0.05 1.8E-6∗ |
0.89±0.03 N/A |
| ASD p value |
2.12±1.02 4.3E-10∗ |
2.01±1.14 2.9E-10∗ |
1.78±0.81 4.6E-8∗ |
1.32±0.76 N/A |
|
| Bladder | DSC p value |
0.89±0.13 2.0E-4∗ |
0.91±0.04 1.9E-3∗ |
0.91±0.06 2.4E-4∗ |
0.94±0.03 N/A |
| ASD p value |
2.12±2.41 7.0E-11∗ |
4.30±2.07 2.8E-13∗ |
1.53±1.25 1.6E-8∗ |
1.15±0.65 N/A |
|
| Rectum | DSC p value |
0.84±0.06 1.4E-5∗ |
0.86±0.05 9.8E-5∗ |
0.87±0.06 1.6E-4∗ |
0.89±0.04 N/A |
| ASD p value |
2.02±0.94 7.2E-4∗ |
2.14±1.86 3.9E-5∗ |
1.71±1.45 2.6E-4∗ |
1.53±0.91 N/A |
|
Some representative segmentation results of different networks are shown in Fig. 6, which delineates the predictions overlaid on the raw images using different colors for the three organs in the axial plane. For better observation, these images have been preprocessed using the method we mentioned. In Fig. 6, we can see that the boundaries are hard to distinguish, and the shapes have large variations across patients. Nevertheless, our method still achieves more robust and accurate results compared with other methods, especially when dealing with complex scenes.
Figure 6.
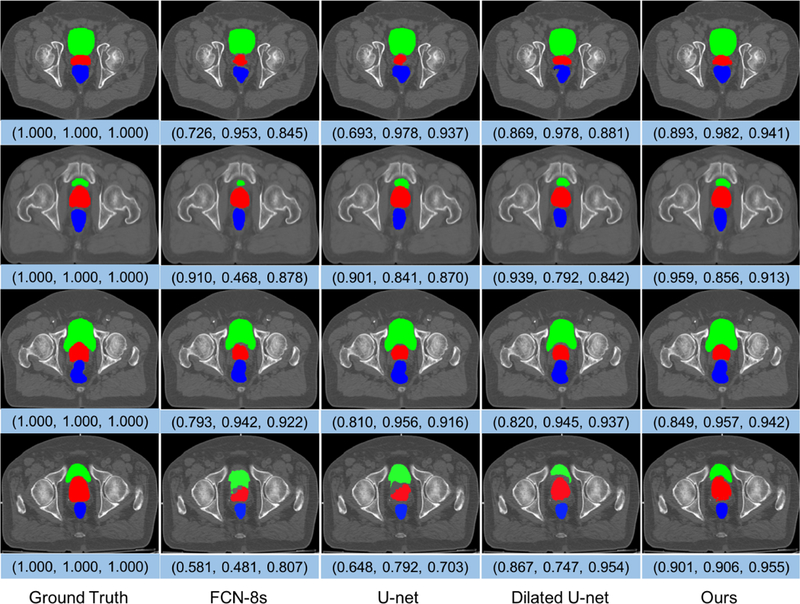
Representative cases of the segmentation results of the prostate (red), bladder (green), and rectum (blue) obtained by FCN-8s, U-net, Dilated U-net, and our multi-label segmentation network. These images are from different subjects and shown in the axial view. The triple array at the bottom of each image denotes the DSC values of the prostate, bladder, and rectum, respectively.
We also show several typical comparisons between the ground truth segmentation and automatic segmentation of each individual organ in Fig. 7, and select a particular region in each image to zoom in for detailed observation. It is obvious that FCN-8s (Long et al. (2015)), U-net (Ronneberger et al. (2015)), and Dilated U-net is weak in dealing with unclear boundaries, while the prediction results obtained by our method are more consistent with the ground truth segmentation. This is also a good proof of the effectiveness of our boundary representation method.
Figure 7.
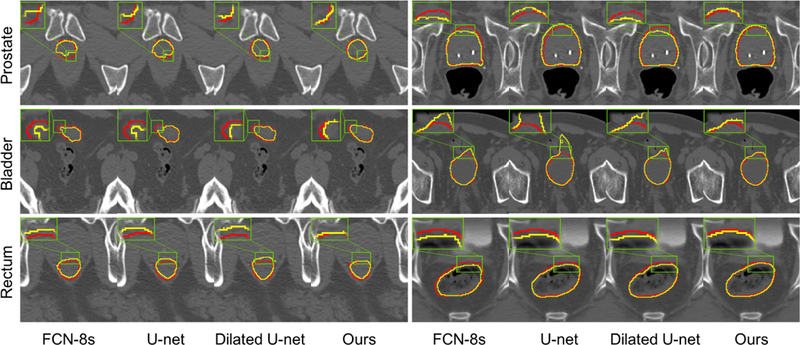
Comparison between segmentations produced by different networks and the ground truth segmentation. Each row shows two representative cases for each organ. Red curves indicate the manual delineations, and yellow curves indicate the automatic segmentations.
4.7. Effectiveness of Organ Localization
Instead of segmenting the original CT image covering the whole pelvic area, an organ localization model is first applied to allow us to focus on the candidate segmentation region for each organ. In order to verify its effectiveness in improving performance, we compare the DSC values of U-net (Ronneberger et al. (2015)) in the segmentation of organs in original CT images and cropped organ proposals. Moreover, we also report the AvgR and TTR values to indicate how many useless background voxels have been dropped and whether the whole organ volume has been preserved after organ localization. The results are reported in Table 2. We can see that organ localization can effectively remove some background voxels that have negative impacts on segmentation performance (in terms of AvgR and DSC) while preserving all target organ volumes (in terms of TTR).
Table 2:
DSC values of U-net using original whole images and organ proposals, respectively, and also AvgR and TTR values after organ localization, for the three male pelvic organs.
| Prostate | Bladder | Rectum | ||
|---|---|---|---|---|
| DSC | Image | 0.73±0.10 | 0.84±0.08 | 0.72±0.13 |
| Proposal | 0.85±0.07 | 0.91±0.04 | 0.86±0.05 | |
| AvgR | 1.65% | 2.58% | 2.06% | |
| TTR | 1.0 | 1.0 | 1.0 | |
4.8. Effectiveness of Boundary Sensitive Representation
In our implementation, we design a new model to represent the boundary information and then guide the organ segmentation. To evaluate its importance and effectiveness, we define two other boundary soft label definition methods for comparison. The first one is named as Unlearning-Soft-Label (USL), which is based on spatial distances between voxels and boundaries to define soft labels. The other one is named as Single-Task-Soft-Label (STSL), which uses single-task learning to segment the boundary masks (dilating the organ contour line with the same kernel size as generating morphological masks) directly and performs soft labeling on the predicted boundary voxels. Table 3 reports the segmentation performance based on three different soft label definitions. In addition to DSC and ASD, we also perform paired t-test between the performance of each comparison method and our method to get the corresponding p values. From these results, we can see that USL outperforms conventional U-net in terms of both DSC and ASD, which demonstrates the guidance capability of boundary information for segmentation. Compared with our method, STSL is not very effective although it can slightly improve the performance of the conventional U-net. From p values, it is observed that our method can improve the segmentation performance more effectively, compared with other methods with statistically significant improvement.
Table 3:
Verification of the effectiveness of our boundary sensitive representation model based on DSC and ASD (mm). (The best results are indicated in bold, with asterisks implying statistical significance (p < 0.05)).
| Metric | U-net | USL | STSL | Ours | |
|---|---|---|---|---|---|
| Prostate | DSC p value |
0.85±0.07 2.8E-6∗ |
0.86±0.05 2.1E-4∗ |
0.87±0.06 1.1E-4∗ |
0.89±0.03 N/A |
| ASD p value |
2.01±1.14 1.4E-11∗ |
1.65±1.29 1.5E-10∗ |
1.53±1.41 5.0E-9∗ |
1.32±0.76 N/A |
|
| Bladder | DSC p value |
0.91±0.04 4.0E-3∗ |
0.93±0.06 9.6E-4∗ |
0.94±0.04 7.2E-2 |
0.94±0.03 N/A |
| ASD p value |
4.30±2.07 8.3E-11∗ |
1.86±0.59 1.6E-6∗ |
1.22±0.57 1.3E-1 |
1.15±0.65 N/A |
|
| Rectum | DSC p value |
0.86±0.05 1.0E-5∗ |
0.88±0.07 4.8E-3∗ |
0.87±0.05 8.4E-4∗ |
0.89±0.04 N/A |
| ASD p value |
2.14±1.86 6.9E-6∗ |
1.75±1.24 5.5E-6∗ |
1.80±1.19 1.9E-5∗ |
1.53±0.91 N/A |
|
Moreover, we output the soft label probability maps of multi-label segmentation networks under three representation methods. One representative example for each organ is shown in Fig. 8. Due to considering only the low-level spatial cue and ignoring the high-level semantic cue, USL is introduced with a lot of noise interference, weakening the value of the boundary information. Therefore, in Fig. 8, the predicted boundaries of USL are easily interfered which results in unreasonable guidance for segmentation. For STSL, taking voxels around boundaries as a new class is not a good choice while many hard-segmented voxels are still not well recognized. In contrast, our method detects the boundary sensitive voxels to represent the boundary information, improving the accuracy of the representation and also the robustness of the prediction.
Figure 8.
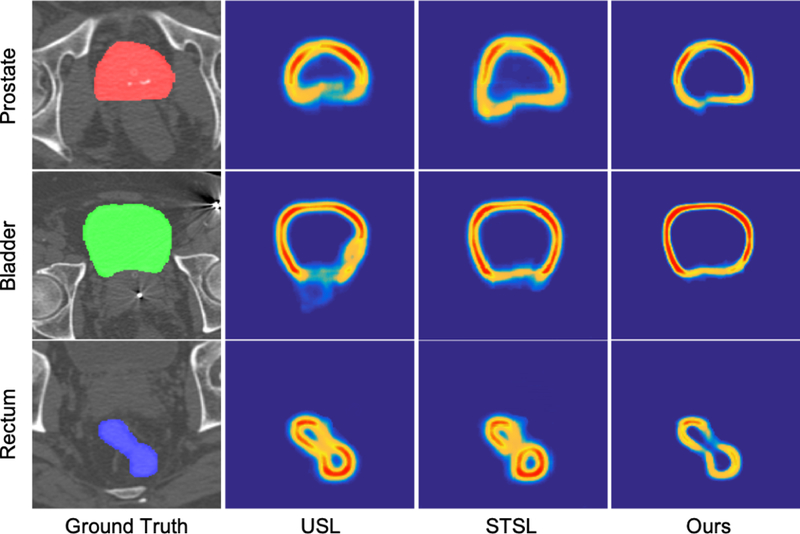
Visualization of soft label probability maps under different boundary representation methods.
4.9. Comparison with Other State-of-the-art Methods
We compare our method with other existing methods for CT male pelvic organ segmentation using the same dataset based on their reported performance, and their brief introductions are summarized as follows:
Locally-constrained Boundary Regression (LcBR, Shao et al. (2015)) exploits the regression forest for boundary detection and designs an auto-context model to improve the segmentation results in an iterative way.
Joint Regression and Classification (JRC, Gao et al. (2015)) guides deformable models by jointly learning image regressor and classifier.
Iterative JRC (IJRC, Gao et al. (2016)) is the extended version of JRC (Gao et al. (2015)), which introduces an auto-context model to iteratively enforce structural information during voxel-wise prediction.
Tables 4 and 5 report a quantitative comparison between our method and the three previous works on the same dataset using the mean DSC and ASD with standard deviations. For prostate, the three comparison methods achieve similar performance using DSC, with our method having a 2% improvement. Meanwhile, it is remarkable that our method greatly improves the performance for ASD, which measures the segmentation results based on the average distance from all voxels on the boundary of the automatic segmentation to the boundary of the ground truth. This improvement is mainly due to our complementary multi-label loss, which not only produces the segmentation between the foreground and background, but also tries to delineate the accurate boundary. The bladder with wall structure is relatively easy to segment compared to the prostate and rectum. All methods can obtain satisfactory DSC values, but the ASD results show that our method is more capable of accurately identifying the boundary. For the rectum, our method has the best DSC and the second best ASD, behind only IJRC (Gao et al. (2016)).
Table 4:
Comparison with previous works on the same dataset using DSC. (The best results are indicated in bold)
| Method | Prostate | Bladder | Rectum |
|---|---|---|---|
| JRC | 0.86±0.05 | 0.91±0.10 | 0.79±0.20 |
| LcBR | 0.88±0.02 | 0.86±0.08 | 0.84±0.05 |
| IJRC | 0.87±0.04 | 0.92±0.05 | 0.88±0.05 |
| Ours | 0.89±0.03 | 0.94±0.03 | 0.89±0.04 |
Table 5:
Comparison with previous works on the same dataset using ASD (mm). (The best results are indicated in bold)
| Method | Prostate | Bladder | Rectum |
|---|---|---|---|
| JRC | 1.85±0.74 | 1.71±3.74 | 2.13±2.97 |
| LcBR | 1.86±0.21 | 2.22±1.01 | 2.21±0.50 |
| IJRC | 1.77±0.66 | 1.37±0.82 | 1.38±0.75 |
| Ours | 1.32±0.76 | 1.15±0.65 | 1.53±0.91 |
Further, we also illustrate our results along with some state-of-the-art methods (Freedman et al. (2005) (Freedman); Costa et al. (2007) (Costa); Chen et al. (2011) (Chen); Lay et al. (2013) (Lay); Mart´ınez et al. (2014) (Martinez); Men et al. (2017) (Men); Shi et al. (2017) (Shi)) using different datasets. Since these approaches segment different subjects of the three pelvic organs using different metrics, we separate the results into Table 6 (DSC and ASD) and Table 7 (PPV and SEN). Please note that the reported results are only for reference.
Table 6:
Illustration of the results of other works using different datasets based on DSC and ASD (mm).
| Metric | Lay | Martinez | Men | Shi | Ours | |
|---|---|---|---|---|---|---|
| Prostate | DSC | N/A | 0.87 | N/A | 0.89 | 0.89 |
| ASD | 3.57 | N/A | N/A | 1.64 | 1.32 | |
| Bladder | DSC | N/A | 0.89 | 0.93 | N/A | 0.94 |
| ASD | 3.08 | N/A | N/A | N/A | 1.15 | |
| Rectum | DSC | N/A | 0.82 | N/A | N/A | 0.89 |
| ASD | 3.97 | N/A | N/A | N/A | 1.53 | |
Table 7:
Illustration of the results of other works using different datasets based on PPV and SEN. The values before and after the slash denote the median and mean values respectively.
| Metric | Freedman | Costa | Chen | Ours | |
|---|---|---|---|---|---|
| Prostate | PPV | 0.85/− | −/0.85 | 0.87/− | 0.93/0.93 |
| SEN | 0.83/− | −/0.81 | 0.84/− | 0.88/0.85 | |
| Bladder | PPV | N/A | −/0.80 | N/A | 0.95/0.94 |
| SEN | N/A | −/0.75 | N/A | 0.96/0.93 | |
| Rectum | PPV | 0.85/− | N/A | 0.76/− | 0.93/0.92 |
| SEN | 0.74/− | N/A | 0.71/− | 0.88/0.86 | |
5. Conclusion and Discussion
In this paper, we have presented an automatic algorithm for the segmentation of male pelvic organs in CT images. To address the challenges of unclear boundaries and large shape variations, we propose a boundary sensitive representation method to characterize the boundary and capture the shape with less noise. Then, we introduce the boundary information into the supervision and define a multi-label cross-entropy loss function to reinforce the network with more discriminative capability. Extensive experiments on a large and diverse planning CT dataset show that our proposed method outperforms the baseline fully convolutional networks, and also achieves better segmentation accuracy than other state-of-the-art methods for CT male pelvic organ segmentation.
In future work, we will further improve the segmentation performance from the following three aspects. 1) To make the generated soft labels more suitable to the dataset, we can investigate a more effective end-to-end fashion, instead of the current multi-step one. 2) The generated soft labels not only can guide the network optimization, but also can be further explored. For example, we can combine the output hard labels and the soft labels together to refine the segmentation results. 3) Since using the whole boundary to represent the organ shape may bring lots of redundant information, we can only use soft labels around some meaningful locations, such as the anterior-most, posterior-most, left, and right points, which may provide a more effective manner to represent the organ shape.
Highlights.
To make better use of boundary information to guide segmentation, we propose a novel boundary representation model deduced from a multi-task learning framework, which assigns soft labels to voxels near the boundary based on both low-level spatial cue and high-level semantic cue, and then produces a more robust and accurate representation of the boundary information.
To address the challenges of unclear boundaries and large shape variations, we design a multi-label cross-entropy loss function to train the segmentation network, where each voxel can not only contribute to the foreground and background, but also adaptively contribute to the boundary with different probabilities.
To reduce the complexity of optimization, we introduce a localization model to focus on the candidate segmentation region of each organ, which can contribute significantly to even better performance.
We perform comprehensive experiments on a challenging dataset with 313 planning CT images from 313 prostate cancer patients to demonstrate the effectiveness of our method. The experimental results show that our method can significantly improve performance compared to the baseline fully convolutional networks and also outperforms other state-of-the-art segmentation methods.
6. Acknowledgements
This work was supported in part by NIH grant (CA206100).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The inv policy updates the learning rate as multiplication of initial learning rate and (1 + gamma ∗ iter)−power
References
- Acosta O, Mylona E, Le Dain M, Voisin C, Lizee T, Rigaud B, Lafond C, Gnep K, de Crevoisier R, 2017. Multi-atlas-based segmentation of prostatic urethra from planning ct imaging to quantify dose distribution in prostate cancer radiotherapy. Radiotherapy and Oncology 125, 492–499. [DOI] [PubMed] [Google Scholar]
- Arganda-Carreras I, Turaga SC, Berger DR, Cireşan D, Giusti A, Gambardella LM, Schmidhuber J, Laptev D, Dwivedi S, Buhmann JM, et al. , 2015. Crowdsourcing the creation of image segmentation algorithms for connectomics. Frontiers in neuroanatomy 9, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro G, Nascimento JC, Freitas A, 2012. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Transactions on Image Processing 21, 968–982. [DOI] [PubMed] [Google Scholar]
- Chen S, Lovelock DM, Radke RJ, 2011. Segmenting the prostate and rectum in ct imagery using anatomical constraints. Medical image analysis 15, 1–11. [DOI] [PubMed] [Google Scholar]
- Chen T, Kim S, Zhou J, Metaxas D, Rajagopal G, Yue N, 2009. 3d meshless prostate segmentation and registration in image guided radiotherapy, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 43–50. [DOI] [PubMed] [Google Scholar]
- Costa MJ, Delingette H, Novellas S, Ayache N, 2007. Automatic segmentation of bladder and prostate using coupled 3d deformable models, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 252–260. [DOI] [PubMed] [Google Scholar]
- Davis BC, Foskey M, Rosenman J, Goyal L, Chang S, Joshi S, 2005. Automatic segmentation of intra-treatment ct images for adaptive radiation therapy of the prostate, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 442–450. [DOI] [PubMed] [Google Scholar]
- Everingham M, Van Gool L, Williams CKI, Winn J, Zisserman A,. The PASCAL Visual Object Classes Challenge 2012. (VOC2012) Results http://www.pascal-network.org/challenges/VOC/voc2012/workshop/index.html.
- Feng Q, Foskey M, Chen W, Shen D, 2010. Segmenting ct prostate images using population and patient-specific statistics for radiotherapy. Medical physics 37, 4121–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman D, Radke RJ, Zhang T, Jeong Y, Lovelock DM, Chen GT, 2005. Model-based segmentation of medical imagery by matching distributions. IEEE transactions on medical imaging 24, 281–292. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lian J, Shen D, 2015. Joint learning of image regressor and classifier for deformable segmentation of ct pelvic organs, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 114–122. [Google Scholar]
- Gao Y, Shao Y, Lian J, Wang AZ, Chen RC, Shen D, 2016. Accurate segmentation of ct male pelvic organs via regression-based deformable models and multi-task random forests. IEEE transactions on medical imaging 35, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M, Hadjiiski L, Cha K, Chan HP, Samala R, Cohan RH, Caoili EM, 2017. Segmentation of inner and outer bladder wall using deep-learning convolutional neural network in ct urography, in: Medical Imaging 2017: Computer-Aided Diagnosis, International Society for Optics and Photonics p. 1013402. [Google Scholar]
- Guo Y, Gao Y, Shen D, 2016. Deformable mr prostate segmentation via deep feature learning and sparse patch matching. IEEE Transactions on Medical Imaging 35, 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B, Coradi T, Scholz M, Kunz P, Huber M, Oppitz U, Andre L, Lengkeek V, Huyskens D, Van Esch A, et al. , 2008. Automatic segmentation of thoracic and pelvic ct images for radiotherapy planning using implicit anatomic knowledge and organ-specific segmentation strategies. Physics in Medicine & Biology 53, 1751. [DOI] [PubMed] [Google Scholar]
- Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H, 2017. Brain tumor segmentation with deep neural networks. Medical image analysis 35, 18–31. [DOI] [PubMed] [Google Scholar]
- He X, Lum A, Sharma M, Brahm G, Mercado A, Li S, 2017. Automated segmentation and area estimation of neural foramina with boundary regression model. Pattern Recognition 63, 625–641. [Google Scholar]
- Huang TC, Zhang G, Guerrero T, Starkschall G, Lin KP, Forster K, 2006. Semi-automated ct segmentation using optic flow and fourier interpolation techniques. Computer methods and programs in biomedicine 84, 124–134. [DOI] [PubMed] [Google Scholar]
- Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T, 2014. Caffe: Convolutional architecture for fast feature embedding, in: Proceedings of the 22nd ACM international conference on Multimedia, ACM; pp. 675–678. [Google Scholar]
- Lai M, 2015. Deep learning for medical image segmentation. arXiv preprint arXiv:1505.02000.
- Lay N, Birkbeck N, Zhang J, Zhou SK, 2013. Rapid multi-organ segmentation using context integration and discriminative models, in: International Conference on Information Processing in Medical Imaging, Springer; pp. 450–462. [DOI] [PubMed] [Google Scholar]
- Liao S, Gao Y, Lian J, Shen D, 2013. Sparse patch-based label propagation for accurate prostate localization in ct images. IEEE transactions on medical imaging 32, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI, 2017. A survey on deep learning in medical image analysis. Medical image analysis 42, 60–88. [DOI] [PubMed] [Google Scholar]
- Long J, Shelhamer E, Darrell T, 2015. Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Lu C, Chelikani S, Papademetris X, Knisely JP, Milosevic MF, Chen Z, Jaffray DA, Staib LH, Duncan JS, 2011. An integrated approach to segmentation and nonrigid registration for application in image-guided pelvic radiotherapy. Medical Image Analysis 15, 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zheng Y, Birkbeck N, Zhang J, Kohlberger T, Tietjen C, Boettger T, Duncan JS, Zhou SK, 2012. Precise segmentation of multiple organs in ct volumes using learning-based approach and information theory, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 462–469. [DOI] [PubMed] [Google Scholar]
- Ma L, Guo R, Zhang G, Schuster DM, Fei B, 2017. A combined learning algorithm for prostate segmentation on 3d ct images. Medical physics 44, 5768–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mart´ınez F, Romero E, Dréan G, Simon A, Haigron P, De Crevoisier R, Acosta O, 2014. Segmentation of pelvic structures for planning ct using a geometrical shape model tuned by a multi-scale edge detector. Physics in Medicine & Biology 59, 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men K, Dai J, Li Y, 2017. Automatic segmentation of the clinical target volume and organs at risk in the planning ct for rectal cancer using deep dilated convolutional neural networks. Medical physics 44, 6377–6389. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer; pp. 234–241. [Google Scholar]
- Shao Y, Gao Y, Wang Q, Yang X, Shen D, 2015. Locally-constrained boundary regression for segmentation of prostate and rectum in the planning ct images. Medical image analysis 26, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Wu G, Suk HI, 2017. Deep learning in medical image analysis. Annual review of biomedical engineering 19, 221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yang W, Gao Y, Shen D, 2017. Does manual delineation only provide the side information in ct prostate segmentation?, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
- Taha AA, Hanbury A, 2015. Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC medical imaging 15, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Huang CT, Lee JH, Li CH, Chang SW, Siao MJ, Lai TM, Ibragimov B, Vrtovec T, Ronneberger O, et al. , 2016. A benchmark for comparison of dental radiography analysis algorithms. Medical image analysis 31, 63–76. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wei L, Wang L, Gao Y, Chen W, Shen D, 2018. Hierarchical vertex regression-based segmentation of head and neck ct images for radiotherapy planning. IEEE Transactions on Image Processing 27, 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mo T, Feng Q, Zhong P, Lai M, Eric I, Chang C, 2014. Deep learning of feature representation with multiple instance learning for medical image analysis, in: Acoustics, Speech and Signal Processing (ICASSP), 2014 IEEE International Conference on, IEEE; pp. 1626–1630. [Google Scholar]
- Yokota F, Okada T, Takao M, Sugano N, Tada Y, Sato Y, 2009. Automated segmentation of the femur and pelvis from 3d ct data of diseased hip using hierarchical statistical shape model of joint structure, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 811–818. [DOI] [PubMed] [Google Scholar]
- Yu F, Koltun V,. Multi-scale context aggregation by dilated convolutions, in: ICLR. [Google Scholar]
- Zhan Y, Shen D, 2003. Automated segmentation of 3d us prostate images using statistical texture-based matching method, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 688–696. [Google Scholar]
- Zhen X, Yin Y, Bhaduri M, Nachum IB, Laidley D, Li S, 2016. Multitask shape regression for medical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 210–218. [Google Scholar]


