Abstract
Macrophages hold great potential in cancer drug delivery because they can sense chemotactic cues and home to tumors with high efficiency. However, it remains a challenge to load large amounts of therapeutics into macrophages without compromising cell functions. Here we report a silica-based drug nanocapsule approach to solve this issue. Our nanocapsule consists of a drug-silica complex filling and a solid silica sheath, and it is designed to minimally release drug molecules in the early hours of cell entry. While taken up by macrophages at high rates, the nanocapsules minimally affect cell migration in the first 6–12 h, buying time for macrophages to home to tumors and release drugs in situ. In particular, we show that doxorubicin (Dox) as a representative drug can be loaded into macrophages up to 16.6 pg/cell using this approach. When tested in a U87MG xenograft model, intravenously (i.v.) injected Dox-laden macrophages show comparable tumor accumulation as untreated macrophages. Therapy leads to efficient tumor growth suppression, while causing little systematic toxicity. Our study suggests a new cell platform for selective drug delivery, which can be readily extended to the treatment of other types of diseases.
Keywords: macrophage, nanoparticle, doxorubicin, glioblastoma, cancer, cell-mediated drug delivery
Introduction
Exploiting immune cells for drug delivery is an emerging area of research.[1–3] Many types of leukocytes, including macrophages, neutrophils, and dendritic cells, can sense chemokine and cytokine cues and home to inflamed tissues. Macrophages or their predecessor monocytes in particular, can respond to cancer-related cytokines (e.g., CSF-1, VEGF, PDGF, TNF, IL-1, IL-5, etc.) and chemokines (e.g., CCL-5, 7, 8, 12, etc.),[4–6] and navigate to the diseased sites, passing multiple biological barriers along the way. This holds true for central tumor areas, which are often avascular and inaccessible to conventional therapeutics. These unique properties make macrophages a potentially appealing vehicle for cancer drug delivery.
Despite the promises, it remains a challenge to load large quantities of drugs into macrophages. Conventionally, the most common cell loading strategy is to conjugate drug molecules or tether drug-loaded nanoparticles onto the cell plasma membrane.[7] This so-called “backpack” approach has been exploited by others and us to load therapeutics onto stem cells,[8–11] leukocytes,[12–15] red blood cells,[16] and T cells[17] with varying success. However, plasma membrane is essential for cell functions and plasticity, and nanoparticle loading may adversely affect cell signal transduction, adhesion, and migration. According to Irvine et al.,[17,18] nanoparticles can occupy up to 5% of plasma membrane without significantly affecting cell functions. This translates to a drug loading rate of less than 1.0 μg per million cells (Supporting Information). Considering that in a normal cell transfer procedure 1–10 million cells are injected, the amount of drugs that can be delivered using this approach is very limited; not to mention that macrophages are phagocytes, and membrane-bound nanoparticles are often quickly engulfed by cells rather than residing on the surface.
An alternative strategy is to load drugs into the cell cytosol. This approach is considered challenging or not feasible because most chemotherapeutics are highly toxic to macrophages. Incubating macrophages with high concentrations of drugs induces immediate cell death, whilst sub-lethal dose incubation causes insufficient drug loading. There have been some successes of exploiting macrophage plasma membrane as a camouflage to improve nanoparticles’ pharmacokinetics.[19,20] A few groups have attempted to load liposome or polymer nanoparticles into live macrophages,[21–23] but the difficulties to achieve high drug loading while maintaining cell functions have limited the related developments.
Herein we report a unique nanocapsule technology to solve the macrophage drug loading issue. We and others observe that it takes 6–12 h for intravenously (i.v.) injected macrophages to migrate to inflamed tissues.[6,24] We reason that if drug-loaded nanoparticles do not release the payloads in the early hours of cell entry, the adverse impacts can be held in check in spite of a high apparent drug content. This would buy time for macrophages to traffic to tumors, and release therapeutics in situ to induce efficient and selective cancer cell killing (Scheme 1). For the purpose, it is desired that nanoparticles have a two-phase drug release profile, with minimal drug liberation in the first 6–12 h and controlled release afterwards. This is challenging because nanoparticles after internalization are trapped in the phagolysosomes of macrophages, which are rich in hydrolytic enzymes and reactive oxygen species (ROS), and can quickly digest conventional drug carriers.[25,26] To solve the issue, we created a drug-silica nanocapsule platform, consisting of a drug-silica nanocomplex core, and a solid silica sheath. The silica coating is more resistant to degradation and oxidation than alternative materials such as polymers or liposomes; by fine-tuning the coating thickness, stalled drug released would be achieved and the degree of extended release adjusted. The drug-silica nanocomplex is more susceptible to degradation than the shell as drug molecules create de facto defects in the silica matrix,[27] leading to two-phased drug release. Meanwhile, because drug molecules are electrostatically bound with silica, burst drug release, which is commonly seen with conventional drug carriers, should be avoided. All these properties should allow for high drug loading into macrophages while minimally affecting cell migration. We tested this hypothesis with doxorubicin (Dox) as a representative chemotherapeutic drug, and we evaluated the efficiency of drug delivery first in vitro and then in vivo in U87MG tumor bearing mice.
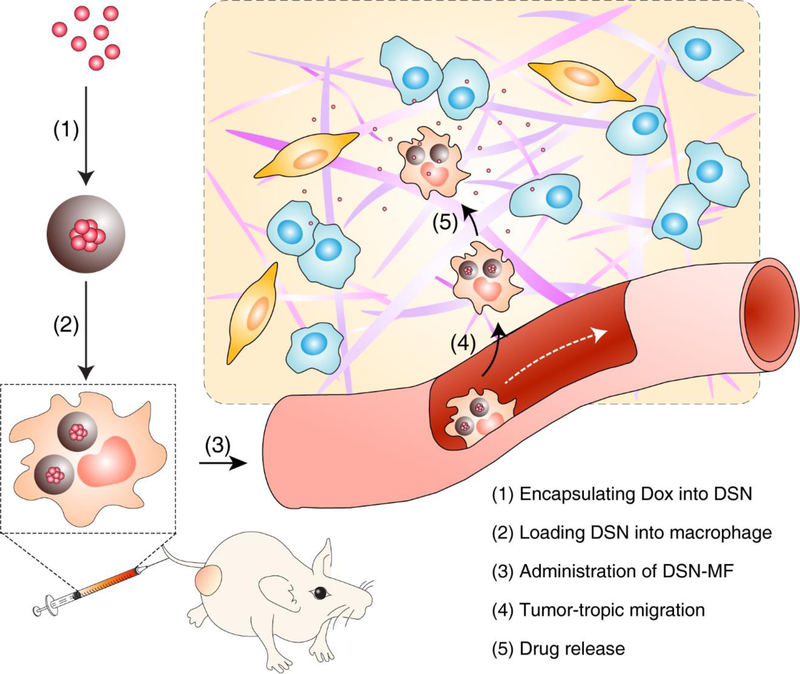
Scheme 1. Nanocapsule-laden macrophages for drug delivery to tumors.
(1) Antineoplastic drug, in this particular case Dox, was first loaded into a carefully tailored nanocapsule called drug-silica nanocomplex (DSN); (2) DSN nanoparticles were engulfed by macrophages ex vivo; (3) DSN-laden macrophages (DSN-MF) were i.v. injected to a tumor bearing mouse; (4) chemotactic migration of DSN-MF to tumors; (5) DSN-MF releases Dox inside tumor to selectively kill cancer cells.
Results
DSN synthesis and physical characterizations
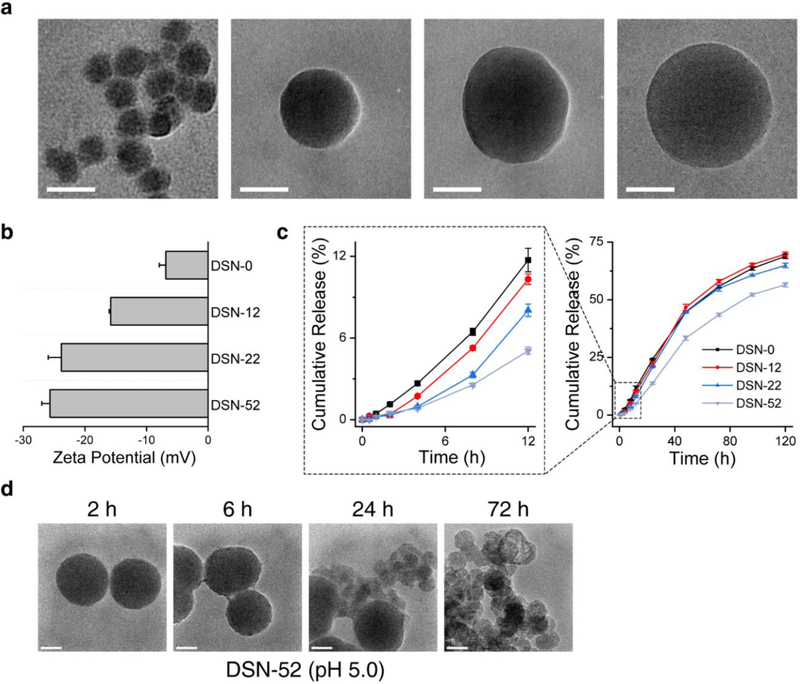
We first synthesized Dox-silica nanocomplexes (referred to as DSN-0) by co-condensation of Dox and tetraethyl orthosilicate (TEOS, Dox: TEOS molar ratio of 1:13) in ethanol. Dox is positively charged at this condition and electrostatically bound with TEOS, whilst being intercalated into the growing silica matrix.[27] The resulting DSN-0 nanoparticles were 28.4 ± 3.4 nm in diameter (Figure 1a). They were stably dispersed in PBS (Figure S1b, S1c), with a slightly negatively charged surface (−6.9 ± 1.0 mV, Figure 1b). Based on UV-Vis spectroscopic analysis, it was estimated that the Dox accounted for 16.7 wt% of the DSN-0 weight (Table S2).
Figure 1.
Physical characterizations of DSN nanoparticles. a) TEM images and b) zeta potential of DSN-0, DSN-12, DSN-22, and DSN-52 nanoparticles. c) Drug release profiles of DSN-0, DSN-12, DSN-22, and DSN-52 nanoparticles, measured at pH 5.0. d) TEM images of DSN-52 nanoparticles after incubating in a pH 5.0 solution for different times. Scale bars, 50 nm.
Despite the strong electrostatic interaction, up to 11.7% of Dox was released within 12 h at pH 5.0 (close to lysosome pH;[28] Figure 1c). To minimize drug release in the early hours, we imparted a silica capsule onto the surface of DSN-0 through the Stöber method. By varying the TEOS precursor amounts, we were able to prepare DSN nanocapsules with silica coating thicknesses of 12, 22, and 52 nm (Figure 1a), and the resulting nanoparticles were referred to as DSN-12, DSN-22, and DSN-52, respectively. We found that a thicker silica coating was associated with more negative surface charge (Figure 1b) and more extended drug release (Figure 1c, S1a). Specifically, DSN-12 and DSN-22 released 10.3% and 8.0% of their Dox contents at 12 h (pH 5.0), and for DSN-52, this number was reduced to 5.1%. We also investigated DSN-52’s morphology changes over time in both neutral and acidic solutions. At pH 7.4, DSN-52 remained intact for over 72 h (Figure S1d). At pH 5.0, on the other hand, we found no obvious morphology changes in the first 6 h, but signs of nanoparticle degradation at 24 h (Figure 1d). The gradual erosion of the silica coating then exposes the DSN core. Unlike solid silica, where adjacent silicon atom is covalently linked by an oxygen bridge, the silica matrix of DSN is more susceptible to hydrolysis due to the Dox dopants. In samples taken at later time points of incubation, we spotted many hollow capsules, suggesting the degradation of the DSN cores (Figure 1d). This correlates with faster drug release recorded after 12 h. For comparison, we also assessed drug release with Dox-loaded liposomes (i.e., Doxove) and mesoporous silica nanoparticles at the same condition. In both cases, we found significant early-hour burst release, with 26.2% (Doxove, Figure S2a) and 58.6% (mesoporous silica nanoparticle, Figure S2b) of their Dox contents liberated at 12 h, respectively.
Imparting silica coating diluted the drug content in the nanoparticles. Specifically, the Dox loading was 11.2, 8.9, and 5.1 wt%, respectively, for DSN-12, DSN-22, and DSN-52, compared to 16.7 wt% for DSN-0 (Table S2, S3). While it is possible to further increase the silica coating thickness and stall the drug release process, it is speculated that a too diluted drug content in the particles (e.g. less than 5%) may adversely affect Dox loading into macrophages. Due to this consideration, we selected the DSN-52 formulation for subsequent cell and animal studies.
Loading DSN-52 nanocapsules into macrophages
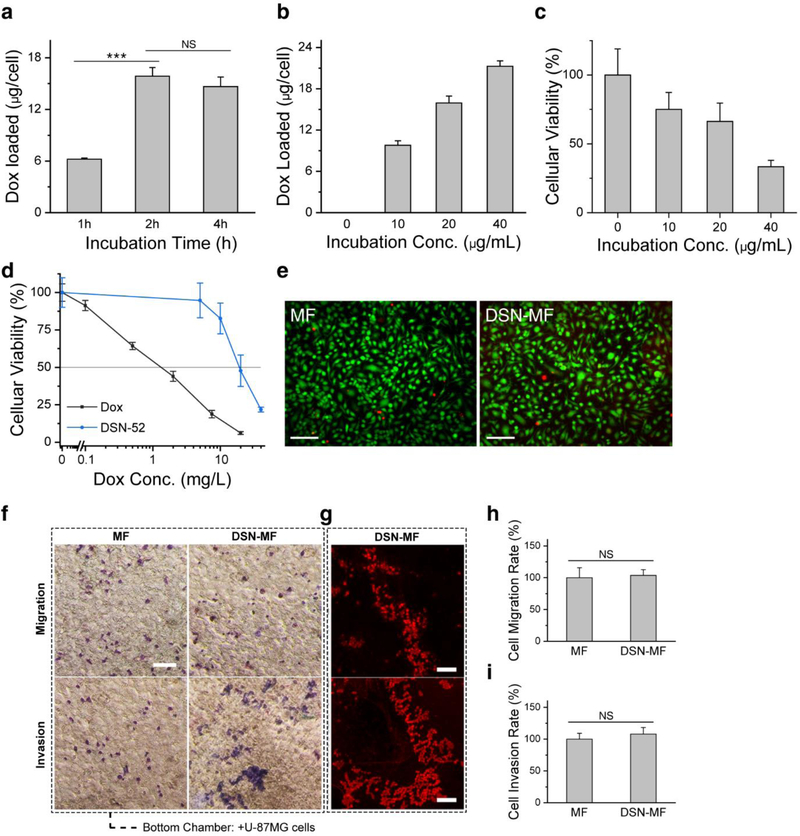
We incubated DSN-52 with RAW264.7 cells, a murine macrophage cell line, and stopped the incubation at different times to analyze nanocapsule uptake by measuring the amount of cellular Dox on a per cell basis. We found that 2 h incubation led to efficient Dox uptake, while extending incubation further minimally increased the cellular Dox contents (Figure 2a). The uptake is attributed to macrophage phagocytosis of nanoparticles, which was observed by others with nanoparticles of comparable sizes.[22] The uptake was concentration dependent. When incubated with DSN-52 at 10, 20, and 40 μg Dox mL−1, the cell Dox content at 2 h was 9.8, 15.9, and 21.3 pg cell−1, respectively (Figure 2b). As a comparison, Doxove showed a Dox loading of less than 1 pg cell−1 at the same conditions (Figure S4a).
Figure 2.
DSN-52 nanoparticles uptake by macrophages (RAW264.7 cells). a) Intracellular Dox contents, measured at 1, 2, and 4 hours’ incubation with DSN-52 nanoparticles. ***, p<0.001. NS, not significant. b) Intracellular Dox contents, measured when the initial DSN-52 Dox concentration was 0, 10, 20, and 40 μg mL−1. The incubation time was fixed at 2 h. c) Cell viability at 12 h via MTT assay. The cells were first incubated with DSN-52 at 0, 10, 20, and 40 μg mL−1 (Dox concentration) for 2 h. After PBS washing, fresh growth medium was added, and cell viability was measured at 12 h by MTT assay. d) Cell viability at 24 h via MTT assay. Dox (black curve), RAW264.7 cells were incubated with free Dox for 24 h. DSN-52 (blue curve), RAW264.7 cells were laden with DSN-52 and then incubated in normal growth medium for 24 h. e) Live and Dead cell assay results of DSN-MF and MF cells at 2 h. Green, living cells; red, dead cells. Scale bars, 100 μm. f) Transmigration assay. DSN-MF or MF cells were loaded onto the top of a transwell chamber, whilst U87MG cells were seeded at the bottom. Macrophages were stained into blue color via Giemsa staining. Scale bars, 100 μm. g) Fluorescence microscopic images of invaded/migrated DSN-MF cells, the experimental conditions were the same as those in f. Scale bars, 100 μm. Percentages of DSN-MF and MF cells that had h) migrated and i) invaded. NS, not significant.
After 2 h incubation, we replenished the incubation medium and analyzed the viability of the DSN-laden cells. When initial DSN-52 concentration was 20 μg Dox mL−1 or below, cell viability maintained at ~70% or above at 12 h (Figure 2c). This is striking considering that the IC50 of free Dox is 1.5 μg mL−1 (Figure 2d). Based on these observations, we chose 20 μg Dox mL−1 and 2 h incubation for drug loading. Under this condition, macrophages bore a stunning Dox content at 16.6 ± 4.8 pg Dox cell−1. Calcein AM/EthD-III assay showed that 99.2% of DSN-52 loaded macrophages (DSN-MF) were heathy at the completion of nanoparticle incubation (Figure 2e). Incubation at 4˚C in the presence of 0.1 wt.% NaN3 led to ~80% decrease of cellular uptake, suggesting that the nanoparticle uptake was mainly mediated by endocytosis (Figure S3).
One concern is that the intracellular Dox, while not lethal, may affect cell functions. In particular, the nanoparticle loading may compromise cells’ chemotactic migration toward cancer cells. We examined this in a transwell experiment, where U87MG glioblastoma cells were seeded onto the bottom chamber of the device, and DSN-MF loaded onto the top. We found that DSN-MF could efficiently transmigrate the well (Figure 2f, 2g), with both invasion and migration percentages comparable to untreated RAW264.7 cells (referred as MF onward, Figure 2h, 2i). Meanwhile, when U87MG cells were absent, there was no cell transmigration (Figure S5a).
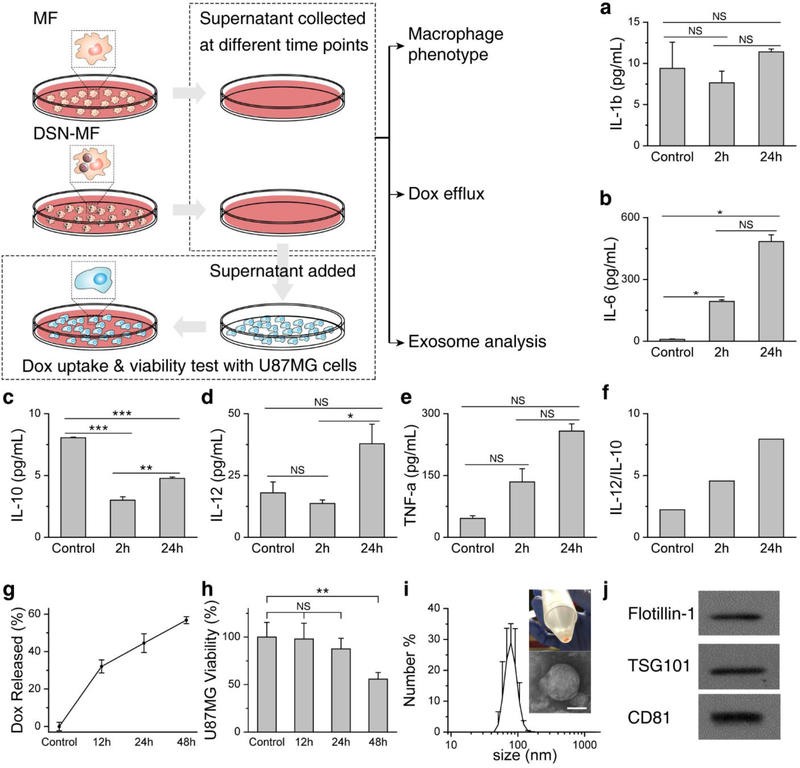
We also examined the impact of DSN-52 loading on macrophage phenotype changes. Specifically, we analyzed the amounts of cytokines, including IL-1β, IL-6, IL-12, TNF-α, and IL-10, that were secreted from DSN-MF. Except for IL-1β, which showed comparable secretion relative to the control, other pro-inflammatory markers, including IL-6, IL-12 and TNF-α, all showed significantly elevated secretion (Figure 3a–e). In particular, the IL-6 level was drastically increased from 9.1 pg mL−1 in untreated macrophages to 484.2 pg mL−1 in DSN-MF at 24 h (Figure 3b). On the contrary, the level of IL-10, an anti-inflammation marker, was reduced from 8.1 pg mL−1 in the control to 4.8 pg mL−1 DSN-MF at 24 h (p < 0.001, Figure 3c). Accompanied with it, the IL-12/IL-10 ratio was increased from 2.2 in MF to 8.0 in DSN-MF (Figure 3f). These results indicate that RAW264.7 cells after DSN-52 loading were polarized toward the pro-inflammation M1 phenotype.[29,30]
Figure 3.
Impact of DSN loading on macrophage phenotypes. Secretion of a) IL-1β, b) IL-6, c) IL-10, d) IL-12, and e) TNF-α from DSN-MF at 2 and 24 h. MF (untreated RAW264.7 cells) served as controls. f) IL-12/IL-10 ratio at 2 and 24 h. g) Percentage of Dox released from DSN-MF at different times (Dox retained in cell debris is excluded by centrifugation). h) Cell viability assay results with U87MG cells. Supernatants taken from DSN-MF culture dishes at different time points were added to a separate plate grown with U87MG cells. Cell viability was measured after 48 h incubation. *, P<0.05; **, P<0.01; ***, P<0.001; NS, not significant. i) Hydrodynamic size of exosomes via DLS analysis (z-average size = 97.35 nm, PDI = 0.127). Exosomes were collected from DSN-MF supernatant at 48 h via centrifugations. An inset photograph of the resulting exosomes and a negative-stained TEM image were also shown. Scale bar, 50 nm. j) Western blot analysis of exosome lysates. Flotilin-1, TSG101, and CD81, three markers of exosomes, were detected.
We then analyzed Dox efflux. We observed time-dependent increase of Dox content in the supernatant of DSN-MF, which released over 50% of the loaded Dox within 48 h (Figure 3g, Table S4). We took the supernatants from different time points and added them to the incubation media of U87MG cells cultured in separate plates (Figure 3a). For the 48-h conditioned medium, incubation with U87MG led to extensive cell uptake of Dox (Figure S5b), which eventually led to cell death (Figure 3h). Notably, conditioned medium taken at 12 h caused little U87MG cell viability drop (Figure 3h), which is attributed to the stalled drug release of the nanocapsules.
Interestingly, Dox was not released entirely in the form of free molecules. When analyzing DSN-MF conditioned medium, we found many nanoparticles with a size of 50–150 nm in the supernatant (negative staining TEM, Figure 3i). Through a series of centrifugation (Supplementary Information), we were able to enrich these nanoparticles (Figure 3i). Further Western blotting analysis found high contents of Flotilin-1, TSG101, and CD81 in these nanoparticles (Figure 3j), suggesting that these were exosomes secreted by macrophages.[31] Spectroscopy analysis revealed that a 16.5% of the released Dox was entrapped within the secreted exosomes (Figure 3i–j, Figure S5c, Table S4). Considering possible exosome loss during differential centrifugation, the actual percentage of Dox released in exosomes could be even higher. Unlike artificial liposomes or micelles, exosomes present on their surface adhesion proteins, integrins, and tetraspanins, which may facilitate cancer cell uptake.[32,33] It is envisioned that during therapy, DSN-MF produces Dox-laden exosomes in situ inside tumors, further improving the selectivity and efficiency of the delivery approach.
In vivo bio-distribution studies
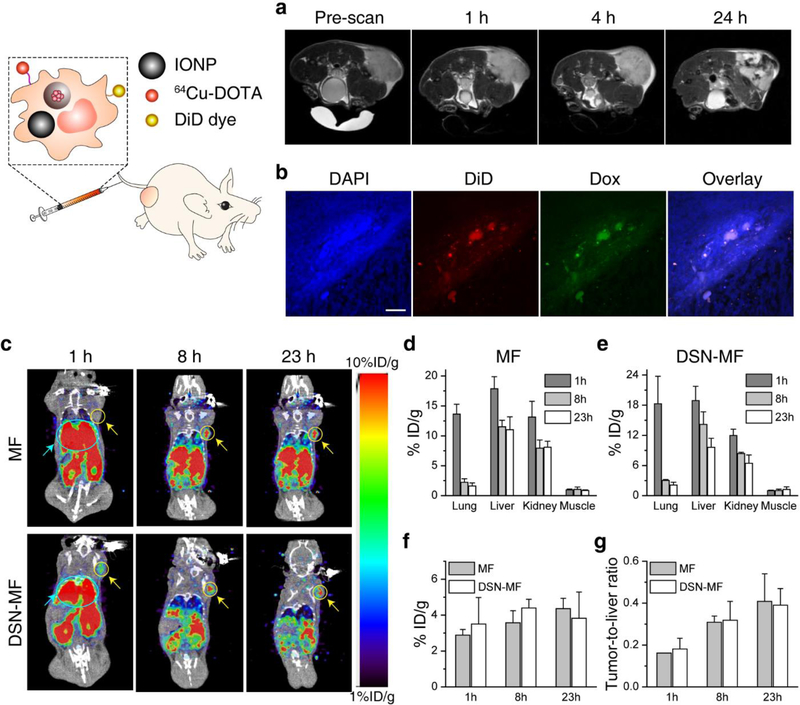
We studied the tumor tropic properties of DSN-MF in U87MG tumor bearing nude mice. To keep track of the cells, we co-loaded 50 nm iron oxide nanoparticles (IONPs)[34,35] into DSN-MF and i.v. injected 2 × 106 of the cells into each animal (n = 3). T2-weighted magnetic resonance imaging (MRI) found minimal signal changes in tumors at 4 h, but extensive hypointensities at 24 h (Figure 4a). To verify cell migration, we also labeled the cell membrane with DiD, and examined the tumor samples by histopathology at 24 h post i.v. injection. We observed positive Prussian blue staining (Figure S6) within tumors, along with signals from Dox and DiD dye (Figure 4b), confirming that macrophages as a vehicle can deliver Dox to tumors.
Figure 4.
In vivo tumor targeting of DSN-MF, evaluated in nude mice bearing subcutaneously inoculated U87MG tumors. a) Axial T2 MR images, acquired at 0, 1, 4, and 24 h post i.v. injection of DSN-MF cells. The cells were pre-loaded with iron oxide nanoparticles. b) Confocal microscopic images of tumor cryo-sections using the z-stack scan mode (step = 2 μm). DSN-MF cells were pre-labeled with DiD. Red, DiD; green, Dox; blue, cell nuclei. Scale bars, 50 μm. c) Decay-corrected whole-body coronal PET images, acquired at 1, 8, and 23 h post injection. DSN-MF or MF cells were labeled with 64Cu-PTSM. Tumor area was highlighted with yellow cycles; lung area was highlighted using cyan cycle. d, e) Distribution of d) MF cells and e) DSN-MF cells in the lung, liver, kidney, and muscle at different time points. f) Tumor uptake of MF and DSN-MF cells at different times. g) Tumor-to-liver ratios of MF and DSN-MF cells, based on images results in c.
For quantitative analysis, we also labeled DSN-MF and untreated RAW264.7 cells (MF) with 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone, 64Cu-PTSM) and monitored cell migration by positron emission tomography (PET).[36] We found that the majority of macrophages were initially accumulated in the lung (Figure 4c). This was attributed to the pulmonary first-pass effect, which is commonly seen with i.v. injected cells.[37,38] Specifically, the lung uptake at 1 h was 18.30 and 13.64 %ID g−1, respectively, for DSN-MF and MF (Figure 4d, 4e). Between 1 and 8 h, there was a significant decrease of radioactivity in the lung. Meantime, tumor accumulation was significantly increased (Figure 4f), suggesting chemotactic migration of macrophages to tumors. For DSN-MF, the decay-adjusted tumor-to-liver ratio (TLR) was increased from 0.18 at 1 h, to 0.32 and 0.39, respectively, at 8 and 23 h (Figure 4g). These values were not significantly different from the MF group (p>0.05). In general, DSN-MF showed comparable pharmacokinetics to untreated macrophages (Figure 4c–g), suggesting negligible impact of DSN loading on tumor migration.
Therapy studies
The therapy study was also conducted in U87MG subcutaneous tumor models. We started therapy when the tumors reached to a size of ~100 mm3. The animals were randomly sorted to receive the following treatments (n=5) via intravenous injection on Day 0: i) PBS; ii) free Dox (3 mg Dox kg−1); iii) DSN-52 only (3 mg Dox kg−1); iv) untreated RAW264.7 cells (MF, ~4 × 106 cells per mouse); v) DSN-MF (3 mg Dox kg−1, ~4 × 106 cells per mouse). Only one dose was given to the animals.
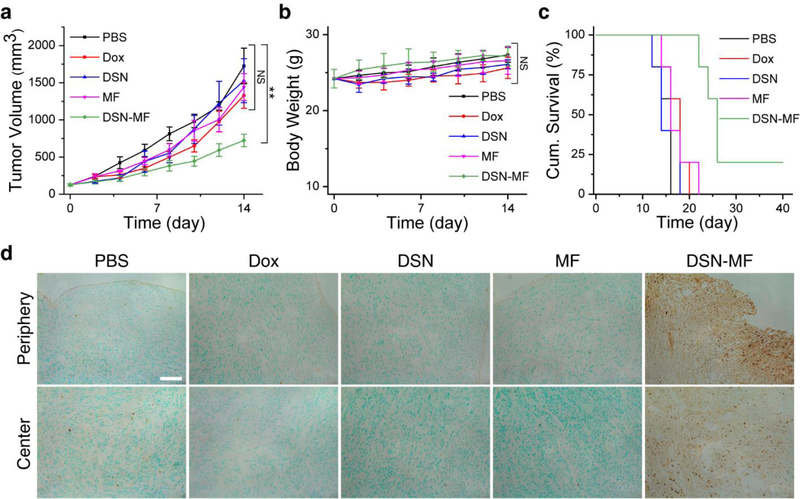
For the Dox, DSN-52, and MF groups, we observed marginal tumor suppression. On Day 14, the average tumor volumes were 1328.6, 1528.47, 1442.27 mm3, respectively, for the three control groups. Relative to the PBS group, the tumor growth inhibition (TGI) rates were 24.84%, 12.35%, and 17.73%, but the changes were insignificant (p = 0.17, 0.80, and 0.32, respectively, Figure 5a, Figure S7a). There was also no benefit in survival. The median survival was 16, 14, and 14 days, respectively, for the Dox, DSN-52, and MF groups, compared to that of 14 days for the PBS control (Figure 5c). As a comparison, the DSN-MF group showed an impressive TGI rate of 62.66% on Day 14, and the treatment significantly extended the animal median survival to 26 days. No body weight loss was observed during the whole treatment process (Figure 5b).
Figure 5.
Therapy studies with U87MG tumor bearing mice. Animals were randomized to receive one dose i.v. injection of either PBS, free Dox (3 mg Dox kg−1), DSN-52 (3 mg Dox kg1), RAW264.7 cells (MF, ~4 × 106 cells per mouse), or DSN-MF (3 mg Dox kg−1, ~4 × 106 cells per mouse). a) Tumor growth curves. b) Body weight changes. c) Kaplan-Meier plot of animal survival. d) In situ Apoptosis staining (Abcam) analysis of cryo-sectioned tumor tissues at 24 h post treatments. Cytoplasm region was counterstained into green color by methyl green; nuclei of apoptotic cells were counterstained into dark brown dots by diaminobenzidine. Scale bar, 50 μm.
In separate studies, we euthanized mice 24 h after treatment, and performed in situ apoptosis staining (Abcam) on the tumor tissues (Figure 5d, Figure S7b). DSN-MF treatment led to extensive cell apoptosis. The positive staining was found at both the peripheral and central tumor areas and occupied 17.83% area of the whole tissue region, which was significantly higher than the controls (Figure S7c). This is again attributed to the capacity of macrophages to pass biological barriers and migrate to inflamed sites. As a comparison, we observed only sporadic positive staining in the control groups (Figure 5d).
Toxicity studies
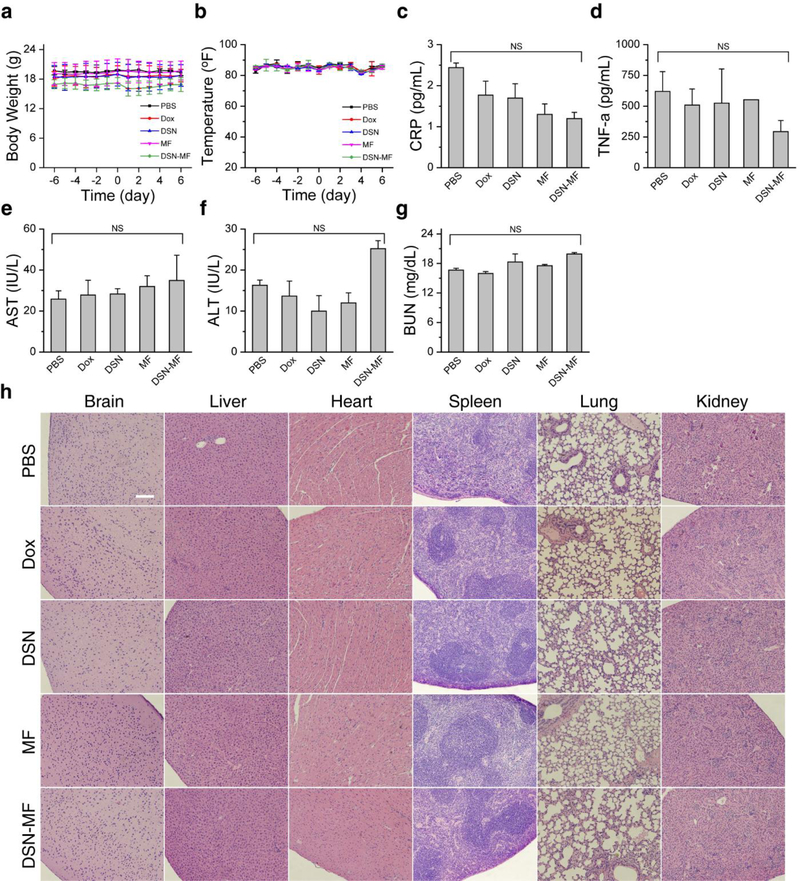
We also investigated whether DSN-MF induces systematic toxicity in normal balb/c mice (n = 3). In all Dox related groups (Dox, DSN-52, and DSN-MF), animals showed a small degree of body weight loss on Day 2, but the loss was recovered after 3–5 days (Figure 6a). Meanwhile, there was no detectable change in rectal temperature for all the treatment groups throughout the study (Figure 6b). After 7 days, we euthanized the animals and examined major organ tissues by H&E staining. We found a minor elevation of leukocyte infiltration in the alveolar areas in the DSN-MF group, which is likely attributed to the accumulation of exogenous macrophages.[38] No pathological changes were observed in all other organs (Figure 6h). These include no detection of cardiotoxicity, which is commonly associated with doxorubicin-based treatments. One concern was that too many activated macrophages may cause increased hemophagocytosis, but there was no evidence of this in the spleen (Figure 6h).
Figure 6.
Toxicity studies. a) Animal body weight changes. b) Animal rectal temperature changes. There was a small degree of weight loss in Dox, DSN-52, and DSN-MF group, which was recovered within 5 days. Mice were euthanized on Day 7 for H&E and plasma protein marker analysis: c) Plasma CRP, d) TNF-α, e, f) AST, ALT levels, and g) BUN levels. For DSNMF, all the indices were in the normal range. h) H&E staining of major organs, which were collected on Day 7 post treatments. Except for a small degree of elevated leukocyte infiltration, no pathological changes were observed for the DSN-MF group. Scale bar, 100 μm.
We also examined blood samples taken from the treated animals. For the DSN-MF group, complete blood count (CBC) analysis found that all indices were in the normal ranges. As a comparison, the Dox group showed abnormalities including elevated white blood cell and red blood cell counts, high hemoglobin and mean corpuscular hemoglobin levels, low mean corpuscular volumes, low mature neutrophil counts, and elevated immature neutrophil counts (Table S5). These impacts come from hematotoxicity of Dox,[39] which were prevented by selective delivery via macrophages. In addition, we also analyzed protein markers related to inflammation (CRP and TNF-α in Figure 6c, 6d),[40] liver function (AST and ALT in Figure 6e, 6f), and kidney function (BUN in Figure 6g). Compared to the control, the ALT level was increased in the DSN-MF group (Figure 6f), though it was still within the normal range.[41] All the other markers are also within the normal ranges.[41] Overall, the histopathology and blood tests confirmed that DSN-MF induced little systematic toxicity to animals.
Discussions and conclusion
Inflammation has long been associated with tumor promotion and progression.[42,43] The fact that macrophages or monocytes can respond to chemotactic cues and migrate to inflammation sites has made macrophages a potentially attractive drug delivery vehicle.[22,24] It is envisioned that macrophages can carry therapeutics to tumors, including metastatic sites and tumor central areas, in a highly selective manner. The main challenge of the approach is that it is difficult to load sufficient amounts of drugs onto macrophages. The current study provides a solution. Our drug-nanocapsule minimally releases therapeutics in the first 6–12 h of cell entry, permitting us to hijack macrophages as an efficient vehicle to enrich drugs in tumors without killing them pre-maturely. In this context, the phagocytic property of macrophages becomes an advantage, allowing for a very high drug loading (e.g. 16.6 pg cell−1) not possible with the conventional “backpack” approach. DSN-MF i.v. injected are first trapped in the lung but afterward gradually migrate to tumors, with a tumor migration rate comparable to untreated macrophages.[44] While many have attempted to load drugs onto live cells for adoptive cell transfer (ACT)[2,7,45,46] (including neutral stem cells and T cells), the more successful examples are seen with using the approach to improve carrier cell survival and functions.[12–14,47] Due to limited drug loading, exploiting ACT to systematically deliver therapeutic drugs has been a challenge. Here we show that DSN-MF can be injected at a clinically relevant chemotherapeutic dose (3 mg kg−1), which again is due to the high drug loading our nanocapsule approach permits. Post-mortem analysis found extensive cell death in tumors, including the central mass (Figure 5d, Figure S7b–c), confirming the benefits of macrophages-based tumor tropism.
For nanoparticle-based drug delivery, drug accumulation in a tumor and its distribution within it rely almost entirely on passive diffusion. Many nanoparticles after extravasation stay in the tumor peripheral region, never reaching the avascular tumor center. Despite a compromised lymphatic system, these nanoparticles are over time drained into the lymphatic system and cleared from the site. Weissleder et al. observed that in many tumors, nanoparticles are first taken up by local macrophages which serve as a depot for continuous drug release.[48] The group also showed that elevating numbers of macrophages in tumors, for instance by external irradiation, provide strongholds for nanoparticles in tumors, leading to improved drug retention and enhanced therapeutic outcomes.[49] In our strategy, drugs are loaded into macrophages ex vivo but similar stronghold effects should have contributed to the treatment.
While the current study is focused on Dox, we anticipate that the platform can be easily extended other therapeutics. Given the high loading capacity of macrophages, it is even possible to use the strategy to deliver a multitude of therapeutics to tumors for combination therapy. Moreover, applications are not limited to cancer therapy. Many other diseases, such as tuberculosis, atherosclerosis, and stroke, are also associated with acute or chronic inflammation. Macrophage-based drug delivery may also hold advantages in the treatment of these diseases. It will be also interesting to test the approach with macrophages derived from autologous monocytes, which is more clinically relevant. It is possible to load nanocapsules into other cell types such as T cells, neural stem cells, and dendritic cells for drug delivery. These possibilities will be explored in future studies.
Supplementary Material
Acknowledgements
This work was also supported by two National Institutes of Health grants (R01EB022596, J.X., and R01NS093314, J.X.), one Congressionally Directed Medical Research Programs grant (CA140666, J.X.), and one National Science Foundation grant (NSF1552617, J.X.). W. Zhang and M. Wang contributed equally to this work.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Weizhong Zhang, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Mengzhe Wang, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Wei Tang, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Ru Wen, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Shiyi Zhou, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Chaebin Lee, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Hui Wang, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Wen Jiang, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Ian Delahunty, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Zipeng Zhen, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Hongmin Chen, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Matthew Chapman, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
Zhanhong Wu, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Elizabeth W. Howerth, Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, Georgia 30602, USA
Houjian Cai, Department of Pharmaceutical and Biomedical Sciences, College of Pharmacy, University of Georgia Athens, Athens, Georgia 30602, USA.
Zibo Li, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Jin Xie, Department of Chemistry, University of Georgia, Athens, GA 30602, USA.
References
- [1].Wang Q, Cheng H, Peng H, Zhou H, Li PY, Langer R, Adv. Drug Deliv. Rev 2015, 91, 125. [DOI] [PubMed] [Google Scholar]
- [2].Su Y, Xie Z, Kim GB, Dong C, Yang J, ACS Biomater. Sci. Eng 2015, 1, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Anselmo AC, Mitragotri S, J. Control. Release 2014, 190, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yi L, Xiao H, Xu M, Ye X, Hu J, Li F, Li M, Luo C, Yu S, Bian X, Feng H, J. Neuroimmunol 2011, 232, 75. [DOI] [PubMed] [Google Scholar]
- [5].Singh A, Talekar M, Raikar A, Amiji M, J. Control. Release 2014, 190, 515. [DOI] [PubMed] [Google Scholar]
- [6].Li S, Feng S, Ding L, Liu Y, Zhu Q, Qian Z, Gu Y, Int. J. Nanomedicine 2016, 11, 4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ayer M, Klok HA, J. Control. Release 2017, 259, 92. [DOI] [PubMed] [Google Scholar]
- [8].Cheng H, Kastrup CJ, Ramanathan R, Siegwart DJ, Ma M, Bogatyrev SR, Xu Q, Whitehead KA, Langer R, Anderson DG, ACS Nano 2010, 4, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li L, Guan Y, Liu H, Hao N, Liu T, Meng X, Fu C, Li Y, Qu Q, Zhang Y, Ji S, Chen L, Chen D, Tang F, ACS Nano 2011, 5, 7462. [DOI] [PubMed] [Google Scholar]
- [10].Mooney R, Weng Y, Garcia E, Bhojane S, Smith-Powell L, Kim SU, Annala AJ, Aboody KS, Berlin JM, J. Control. Release 2014, 191, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mooney R, Weng Y, Tirughana-Sambandan R, Valenzuela V, Aramburo S, Garcia E, Li Z, Gutova M, Annala AJ, Berlin JM, Aboody KS, Futur. Oncol 2014, 10, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mitchell MJ, Wayne EC, Rana K, Schaffer CB, King MR, Proc. IEEE Annu. Northeast Bioeng. Conf. NEBEC 2014, 2014–Decem, 1. [Google Scholar]
- [13].Wayne EC, Chandrasekaran S, Mitchell MJ, Chan MF, Lee RE, Schaffer CB, King MR, J. Control. Release 2016, 223, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chandrasekaran S, Chan MF, Li J, King MR, Biomaterials 2016, 77, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Anselmo AC, Gilbert JB, Kumar S, Gupta V, Cohen RE, Rubner MF, Mitragotri S, J. Control. Release 2015, 199, 29. [DOI] [PubMed] [Google Scholar]
- [16].Tang W, Zhen Z, Wang M, Wang H, Chuang Y-J, Zhang W, Wang GD, Todd T, Cowger T, Chen H, Liu L, Li Z, Xie J, Adv. Funct. Mater 2016, 26, 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang B, Abraham WD, Zheng Y, Bustamante López SC, Luo SS, Irvine DJ, Sci. Transl. Med 2015, 7, 291ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stephan MT, Moon JJ, Um SH, Bershteyn A, Irvine DJ, Nat. Med 2010, 16, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parodi A, Quattrocchi N, van de Ven AL, Chiappini C, Evangelopoulos M, Martinez JO, Brown BS, Khaled SZ, Yazdi IK, Enzo MV, Isenhart L, Ferrari M, Tasciotti E, Nat. Nanotechnol 2013, 8, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Molinaro R, Corbo C, Martinez JO, Taraballi F, Evangelopoulos M, Minardi S, Yazdi IK, Zhao P, De Rosa E, Sherman MB, De Vita A, Toledano Furman NE, Wang X, Parodi A, Tasciotti E, Nat. Mater 2016, 15, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Choi J, Kim HY, Ju EJ, Jung J, Park J, Chung HK, Lee JS, Lee JS, Park HJ, Song SY, Jeong SY, Choi EK, Biomaterials 2012, 33, 4195. [DOI] [PubMed] [Google Scholar]
- [22].Pang L, Qin J, Han L, Zhao W, Liang J, Xie Z, Yang P, Wang J, Oncotarget 2016, 7, 37081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang WC, Chiang WH, Cheng YH, Lin WC, Yu CF, Yen CY, Yeh CK, Chern CS, Chiang CS, Chiu HC, Biomaterials 2015, 71, 71. [DOI] [PubMed] [Google Scholar]
- [24].Xie J, Wang J, Niu G, Huang J, Chen K, Li X, Chen X, Chem. Commun. (Camb) 2010, 46, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Scarpa E, Bailey JL, Janeczek AA, Stumpf PS, Johnston AH, Oreffo ROC, Woo YL, Cheong YC, Evans ND, Newman TA, Sci. Rep 2016, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vandenbranden M, De Gand G, Brasseur R, Defrise-Quertain F, Ruysschaert JM, Biosci. Rep 1985, 5, 477. [DOI] [PubMed] [Google Scholar]
- [27].Zhang S, Chu Z, Yin C, Zhang C, Lin G, Li Q, J. Am. Chem. Soc 2013, 135, 5709. [DOI] [PubMed] [Google Scholar]
- [28].DiCiccio JE, Steinberg BE, J. Gen. Physiol 2011, 137, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mosser DM, Edwards JP, Nat. Rev. Immunol 2008, 8, 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mantovani A, Sozzani S, Locati M, Allavena P, Sica A, Trends Immunol 2002, 23, 549. [DOI] [PubMed] [Google Scholar]
- [31].Soekmadji C, Nelson CC, Biomed Res. Int 2015, 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W, J. Hematol. Oncol 2015, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Azmi AS, Bao B, Sarkar FH, and Azmi1 F. H. S. Asfar S., Bao Bin, Cancer Metastasis Rev 2014, 32, 1. [Google Scholar]
- [34].Quan Q, Xie J, Gao H, Yang M, Zhang F, Liu G, Lin X, Wang A, Eden HS, Lee S, Zhang G, Chen X, Mol. Pharm 2011, 8, 1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen H, Zhang W, Zhu G, Xie J, Chen X, Nat. Rev. Mater 2017, 2, 17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Adonai N, Adonai N, Nguyen KN, Walsh J, Iyer M, Toyokuni T, Phelps ME, McCarthy T, McCarthy DW, Gambhir SS, Proc. Natl. Acad. Sci 2002, 99, 3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dou H, Destache CJ, Morehead JR, Mosley RL, Boska MD, Kingsley J, Gorantla S, Poluektova L, Nelson J. a., Chaubal M, Werling J, Kipp J, Rabinow BE, Gendelman HE, Blood 2006, 108, 2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fischer UM, Harting MT, Jimenez F, Monzon-Posadas WO, Xue H, Savitz SI, Laine GA, Cox CS, Stem Cells Dev. 2009, 18, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Eppstein DA, Kurahara CG, Bruno NA, Terrell TG, Cancer Res. 1989, 49, 3955. [PubMed] [Google Scholar]
- [40].MOUSE C-REACTIVE PROTEIN ( CRP ) ELISA Life Diagnostics, Inc. (lifediagnostics.com)
- [41].Chen S, Li G, Zhu K, Sun P, Wang R, Zhao X, Oncol. Lett 2014, 7, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Coussens LM, Werb Z, Nature 2002, 420, 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dvorak HF, Cancer Immunol. Res 2015, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dou H, Destache CJ, Morehead JR, Mosley RL, Boska MD, Kingsley J, Gorantla S, Poluektova L, Nelson J. a., Chaubal M, Werling J, Kipp J, Rabinow BE, Gendelman HE, Blood 2006, 108, 2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Xie Z, Su Y, Kim GB, Selvi E, Ma C, Aragon-Sanabria V, Hsieh J-T, Dong C, Yang J, Small 2017, 13, 1603121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Batrakova EV, Gendelman HE, Kabanov AV, Expert Opin Drug Deliv 2011, 8, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lee S, Choi E, Cha M-J, Hwang K-C, Oxid. Med. Cell. Longev 2015, 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim J, Lee JE, Lee J, Yu JH, Kim BC, An K, Hwang Y, Shin C-H, Park J-G, Kim J, Hyeon T, J. Am. Chem. Soc 2006, 128, 688. [DOI] [PubMed] [Google Scholar]
- [49].Miller MA, Chandra R, Cuccarese MF, Pfirschke C, Engblom C, Stapleton S, Adhikary U, Kohler RH, Mohan JF, Pittet MJ, Weissleder R, Sci. Transl. Med 2017, 9, eaal0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.