Abstract
Background/Aim: The present study aimed to investigate the role of an aggrecan (ACAN) gene variant and proteoglycan levels in the risk of lumbar degenerative disc disease (LDDD). Materials and Methods: A total of 108 patients with LDDD and 103 healthy controls were enrolled. Molecular assessment of the ACAN gene (c.6423T>C) variant was determined by real time-polymerase chain reaction. Proteoglycan levels in serum were measured with enzyme-linked immunosorbent assay. Results: The frequency of all alleles and genotypes in all study groups were distributed according to the Hardy–Weinberg equilibrium. In addition, no association between the ACAN gene (c.6423T>C) variant and presence of risk factors for LDDD was detected. However, proteoglycan levels were significantly lower in patients with LDDD compared to the control group (p<0.00001). Conclusion: Our findings suggest that proteoglycan has emerged as a potential novel biomarker which might be used for prediction of LDDD risk.
Keywords: Lumbar disc degenaration, biomarker, single-nucleotide polymorphism
Lumbar degenerative disc disease (LDDD) is the most common cause of lower back pain and radicular pain. LDDD is generally related to lumbar disc herniation, which is a multifactorial spinal disorder causing workforce loss, disability and increasing healthcare costs (1-4). Lower back pain is mainly related to lumbar disc disease and affects approximately 50% of the population during their lifetime. The incidence of radicular pain is between 13% and 40% (5-7). Some studies demonstrated that surgical treatment was indicated in 20% of patients with LDDD due to persistent and severe radicular pain (8,9). The degenerative process is multifactorial and irreversible and related to mechanical dysfunction (10,11). Gender, age, body mass index of patients, excessive physical activity, smoking habit, and severe stress exposure of the spine are the most common factors involved in the development of LDDD (12). Genetic factors also play a critical role in this process. In light of recent improvements in research, genetic mechanisms have become the major factors considered in the development of LDDD (13-23).
Proteoglycans are glycosylated proteins which are attached to highly anionic glycosaminoglycan with covalent bonds. Proteoglycans are found in different forms within different types of extracellular matrix and connective tissues. Proteoglycans in the nucleus pulposus of intervertebral discs, contain glycosaminoglycan chains of chondroitin sulfate and keratan sulfate attached to a polypeptide with covalent bonds. The keratan sulfate chains form clusters in so-called keratan sulfate-rich regions or in the protein core regions of proteoglycans in new developed hyaline cartilage and immature nucleus pulposus. Proteoglycan distribution in the intervertebral disc is important regarding the streaming potential, swelling and compressive properties of the tissue (24). Because of the decrease of proteoglycan with aging, the disc starts to dehydrate and recurrent damage occurs. As a result of this, the disc enters a degenerative process and herniation of nucleus pulposus arises (25).
Aggrecan encoded by the ACAN gene on chromosome 15q26.1 is the main proteoglycan component in hyaline cartilage and the nucleus pulposus of the intervertebral disc, with multiple functional domains (26). It is directly responsible for the maintenance of disc hydration with the help of osmotic pressure ensured by its chondroitin and keratan sulfate chains (27). Pathological processes in genetic variants of the aggrecan gene may alter the structural and functional characteristics of this extracellular matrix molecule. There are a limited number of studies focused on the correlation of (c.6423T>C) polymorphism of the ACAN gene with disc degeneration and herniation (28,29).
Therefore, we designed a case–control study in a Turkish group and evaluated the correlation between the ACAN (c.6423T>C) polymorphism, proteoglycan levels and development of LDDD.
Materials and Methods
Study participants and clinical investigation. A total of 211 individuals were included in this prospective case–control study. All of the participants were selected from patients followed-up by the Neurosurgery Department of Yeditepe University in Istanbul, Turkey. The study was evaluated and approved by Yeditepe University Ethical Committee (no. 729; 24 May 2017).
A total of 108 out of the 211 participants were patients with lumbar disc herniation and the remaining 103 participants were healthy individuals (control group). None of the healthy individuals had ever experienced symptoms of back pain or radiculopathy. Regarding the clinical evaluation of patients, neurological examination, lumbar magnetic resonance imaging (MRI), visual analog scale (VAS) scoring to define the pain level, and Oswestry Disability Index (ODI) scoring were performed (30). The inclusion criteria of the case group were lower back pain and leg pain related to lumbar disc degeneration, and lumbar intervertebral disc protrusion diagnosed with lumbar MRI. Patients who had infection, trauma, congenital anomalies, osteoporosis, oncological pathologies, spinal stenosis, spondylolisthesis, vertebral fractures, or spinal deformities in their medical history were excluded from the study. During the examination, the level of pain was scored with VAS and quality of life was scored with ODI for each patient. The clinical findings were recorded and followed-up prospectively. The demographic characteristics of the patients and controls were extracted from their medical records. Written informed consent was obtained from all patients and volunteers in the control group. Blood samples were collected 24-48 h before surgical intervention to determine the proteoglycan values. After centrifugation of blood, plasma samples were frozen at −80˚C.
Isolation of DNA. After acquiring informed consent of the participants, peripheral blood samples were drawn into EDTA tubes. DNA extraction was performed with iPrep Purification Instrument (Invitrogen, Life Technologies, Carlsbad, CA, USA) using 350 μl of peripheral blood and Invitrogen iPrep PureLink gDNA blood isolation kit (Invitrogen, Life Technologies). Isolated DNA samples were measured with NanoDrop 2000 (Thermoscientific, Waltham, MA, USA). Optical density ranges of 1.7-1.9 were accepted for genotyping and the final DNA concentration of samples after the dilution was approximately 100 ng/μl.
Identification of ACAN gene (c.6423T>C) polymorphism. Genotyping for ACAN gene rs1042631 polymorphism was performed with Applied Biosystems 7500 Fast Real-Time PCR instrument (Applied Biosystems, Foster City, CA, USA) and TaqMan Genotyping Assay, TaqMan Genotyping Master Mix (TaqMan Reagents, Applied Biosystems) with 100 ng of DNA sample. The reaction mixture and conditions were according to the recommendations of the manufacturer: The reaction conditions were 10 min at 95˚C hold stage and 40 cycles for 15 sec at 92˚C denaturation and 60 sec at 60˚C annealing/extension. Allelic discrimination of samples by collecting and interpreting fluorescence signals of hybridization probes was performed using the software of the 7500 Fast Real-Time PCR device.
Analysis of serum proteoglycan levels. Serum proteoglycan levels were measured with Human Proteoglycan Enzyme-linked Immunosorbent Assay (ELISA) Kit (Bioassay Technology Laboratory, Yangpu Dist., Shangai, China). Serum collection and measurements were performed according to the instructions of the manufacturer. Peripheral blood samples were collected into gel-containing tubes and left to clot for 15 minutes at room temperature. The clotted samples were centrifuged 1,106 × g. All standards and reagents were prepared and brought to room temperature before the procedure. Fifty microliters of each standard were added to a standard well, 40 μl of sample was added to sample wells, and then 10 μl antibody to proteoglycan was added to sample wells. Subsequently, 50 μl streptavidin-horseradish peroxidase was added to the sample and standard wells but not to blank wells and then the plate was covered with a sealer and left to incubate for 60 minutes at 37˚C. After the completion of the incubation, the sealer was removed and the plate was washed five times with a wash buffer then 50 μl of substrate solution A and 50 μl of substrate solution B were added to each well including blank wells and plates were covered with a sealer and left to incubate for 10 minutes at 37˚C. After incubation, 50 μl of stop solution was added to all wells and optical density measurements were performed with a microplate reader (Poweam Medical Co. Nanjing City, Jiangsu Province, China) at a wavelength of 450 nm for 20 minutes following the addition of the stop solution.
Statistical analysis. The statistical analyses were performed with IBM SPSS Statistics version 23 (IBM Corp., Armonk, NY, USA). The differences between the groups regarding genotypes were analyzed with chi-square and Fisher’s exact tests. Comparisons of the demographic information were evaluated with Student’s t-test and one-way analysis of variance (ANOVA). The frequencies of the ACAN alleles were assessed with gene counting methods. For all analyses, differences with p<0.05 were considered statistically significant.
Results
A total of 108 patients and 103 controls were included in the analysis. The mean age of the patients with LDDD and healthy controls were 41.11±9.65 and 39.20±8.98 years respectively. There was no significant difference between the groups considering the mean age (p=0.139). The gender proportion was significantly different between the groups (p=0.457), (45.4% males and 54.6% females in the LDDD group; 50.5% males, 49.5% females in the control group). Mean ODI score was 54.185±20.577, meanwhile VAS scores was 63.333±21.874 for the patient group.
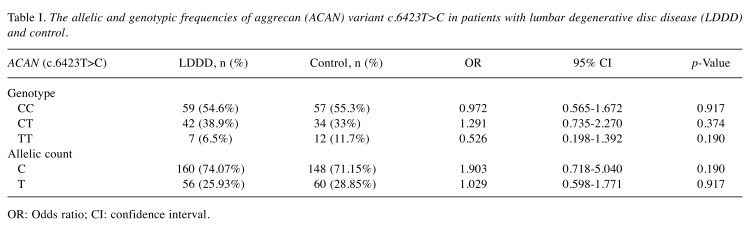
The allelic and genotypic frequencies of the ACAN gene (c.6423T>C) in patients with LDDD and controls are given in Table I. The genotypic frequencies of the ACAN gene (c.6423T>C) in patients with LDDD and controls were consistent with the Hardy–Weinberg equilibrium (χ2=2.075; p=0.354). Furthermore, we did not find any significant difference between the LDDD and control groups regarding the distribution of ACAN (c.6423T>C) alleles (p>0.05).
Table I. The allelic and genotypic frequencies of aggrecan (ACAN) variant c.6423T>C in patients with lumbar degenerative disc disease (LDDD) and control.
OR: Odds ratio; CI: confidence interval.
The serum proteoglycan level was 79.166±92.327 pg/ml in patient group, while it was 162.1971±125.25 pg/ml in the control group. These results indicated that serum proteoglycan levels were significantly lower in patients with LDDD compared to the control group (p<0.00001). However, there was no correlation between VAS, ODI and proteoglycan values (p>0.05). We also evaluated the relationship between the pain scores (VAS and ODI) and genotype but did not detect any significant difference.
Discussion
LDDD is a common disability causing musculoskeletal symptoms and it is the most important cause of mobility limitation and workforce loss throughout the world (31). Intervertebral disc degeneration is considered to be the primary cause of lower back pain, which is an irreversible process via molecular changes in nucleus pulposus (32).
Proteoglycan distribution in the intervertebral disc is important regarding the streaming potential, swelling and compressive properties of the tissue (24,33). Proteoglycans, mainly aggrecan, are the second greatest component of the disc in terms of dry weight after collagen, which constitutes 5-8% of the outer annulus and 11-20% of the inner annulus (34). Impaired regulation of collagen fibrillogenesis by proteoglycans may result in disruption of the lamellar structure and can be related to scoliosis (35). Because of the loss of proteoglycans with increasing age, the disc starts to dehydrate and degenerative processes cause recurrent damage. These processes contribute to disc degeneration and herniation of the nucleus pulposus (25,36). Continuous compressive forces applied to the disc trigger changes in proteoglycan and collagen molecules (37). Hydrostatic pressure induces the production of nitrous oxide in the disc cells, which is one of the mediators that changes proteoglycan synthesis in response to hydrostatic pressure (38). Notochordal cells play a vital role in the maintenance of disc integrity through the stimulation of proteoglycan synthesis (39).
Robinson et al. conducted a study on diabetic and non-diabetic patients with disc herniation. In the disc tissue of diabetic patients, proteoglycans had a lower buoyant density and substantially under-sulfated glycosaminoglycans, which was related to specific neurological damage in these patients, and may lead to an increased susceptibility to disc herniation (40). Cs-Szabo et al. measured mRNA and protein content of aggrecan, bi-glycan, decorin, versican, and fibromodulin in annulus fibrosus and nucleus pulposus. They reported that the mRNA and protein content of all proteoglycans were significantly increased at initial stages of disc degeneration and decreased in severely degenerated tissues. On the contrary, in nucleus pulposus, matrix components were reduced progressively parallel with an increasing degree of degeneration (41).
Keshari et al. evaluated 17 cadaveric disc tissue specimens and biochemical analyses were performed for proteoglycan and collagen in annulus fibrosus and nucleus pulposus. They detected a decline in the concentration of proteoglycans in degenerated nucleus pulposus (42). Iatridis et al. evaluated intervertebral discs and reported that glycosaminoglycan content was between ~40 and 600 μg/mg (dry tissue), with the highest concentration in the nucleus pulposus and lowest in the annulus fibrosus. Usually, posterior regions had higher glycosaminoglycan content than anterior parts of the annulus fibrosus, although values were not as high as in the nucleus pulposus (24). Singh et al. evaluated 46 discs and reported that total proteoglycan and collagen contents in both the annulus fibrosus and nucleus pulposus consistently decreased with increasing age (43). Our study appears to be the first in literature to evaluate proteoglycans in the serum of patients with LDDD. Proteoglycan levels, which were analyzed with ELISA, were lower in patients with disc degeneration than in the controls.
Structural and functional molecules in the disc degradation pathway are suitable candidates for genetic correlation studies. There are only a limited number of studies focused on the correlation between ACAN gene (c.6423T>C) and the risk of LDDD. In the first study, which was conducted by Kawaguchi et al. with 64 individuals, the relationship between this polymorphism and LDDD was examined and a significant difference was determined between the distribution of allele sizes and the severity of MRI-defined LDDD. They investigated a group of Japanese women between 20 and 29 years of age and showed that A18 (18 repeats) and A21 (21 repeats) were over-represented in these females with multilevel and severe degeneration (17). In a Korean population, Kim et al. found that in individuals with degeneration who were younger than 40 years, the A21 allele was over-represented (44). The most important limitation of both studies was that young individuals, who were exposed relatively less to workloads and other stresses, were enrolled so that the results did noticeably reflect the genetic relationships.
Videman et al. showed the T mutant allele of the of the ACAN gene (c.6423T>C) variant to be significantly correlated with the signal intensity of T2-weighted disc signal from MRI, which reflects the water concentration in the disc, and increased the rate of disc herniation in a Finnish population (28). Roughley et al. studied 102 individuals with an average age of 55 years and were not able to find a similar correlation (16). In their study, all of the patients had lower back pain and were diagnosed by lumbar MRI for surgical decompression treatment. The authors suggested that there was an association between ACAN polymorphism and function of the protein, which consists of CS1 and CS2 domains. The gene encoding the CS1 domain displays size polymorphism, with a variable number of tandem repeats. They showed that change in the CS1 domain arising from one short allele of the gene (less than 25 repeats) was not sufficient to impair disc function.
As far as we know, there is no published study focused on the relationship between this variant in the ACAN gene and LDDD risk in the Turkish population. In our study, the ACAN gene (c.6423T>C) variant was not found to have any correlation with the development and severity of LDDD.
In conclusion, this study is the first to evaluate the relevance of the ACAN gene (c.6423T>C) variant and proteoglycan levels in respect to LDDD risk. The assessment of proteoglycan values may provide information which may lead to the usage of proteoglycan as a biomarker for LDDD. Further functional studies are needed to evaluate correlations between genotype and clinical characteristics of patients in large cohorts of various ethnicities.
Conflicts of Interest
On behalf of all Authors, the corresponding Author states that there is no conflict of interest in regard to this study.
References
- 1.Andersson GB. Epidemiological features of chronic low-back pain. Lancet. 1999;354(9178):581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 2.Cervin Serrano S, Gonzalez Villareal D, Aguilar-Medina M, Romero-Navarro JG, Romero Quintana JG, Arambula Meraz E, Osuna Ramirez I, Picos-Cardenas V, Granados J, Estrada-Garcia I, Sanchez-Schmitz G, Ramos-Payan R. Genetic polymorphisms of interleukin-1 alpha and the vitamin D receptor in Mexican Mestizo patients with intervertebral disc degeneration. Int J Genomics. 2014;2014:302568. doi: 10.1155/2014/302568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuceli S. Facet cysts. J Turk Spinal Surg. 2018;29(4):219–221. [Google Scholar]
- 4.Antar V, Baran O, Yüceli S, Erdogan H, Altintas O, Baran GE, Tasdemiroglu E. Assessment of the neuroprotective effects of the acetylcholinesterase inhibitor huperzine A in an experimental spinal cord trauma model. J Neurosurg Sci. 2018;62(2):128–139. doi: 10.23736/S0390-5616.16.03428-7. [DOI] [PubMed] [Google Scholar]
- 5.Cherkin DC, Deyo RA, Loeser JD, Bush T, Waddell G. An international comparison of back surgery rates. Spine. 1994;19(11):1201–1206. doi: 10.1097/00007632-199405310-00001. [DOI] [PubMed] [Google Scholar]
- 6.Frymoyer JW. Lumbar disk disease: Epidemiology. Instr Course Lect. 1992;41:217–223. [PubMed] [Google Scholar]
- 7.McBeth J, Silman AJ, Gupta A, Chiu YH, Ray D, Morriss R, Dickens C, King Y, Macfarlane GJ. Moderation of psychosocial risk factors through dysfunction of the hypothalamic-pituitary-adrenal stress axis in the onset of chronic widespread musculoskeletal pain: Findings of a population-based prospective cohort study. Arthritis Rheum. 2007;56(1):360–371. doi: 10.1002/art.22336. [DOI] [PubMed] [Google Scholar]
- 8.Eser B, Cora T, Eser O, Kalkan E, Haktanir A, Erdogan MO, Solak M. Association of the polymorphisms of vitamin D receptor and aggrecan genes with degenerative disc disease. Genet Test Mol Biomarkers. 2010;14(3):313–317. doi: 10.1089/gtmb.2009.0202. [DOI] [PubMed] [Google Scholar]
- 9.Gu J, Guan F, Guan G, Xu G, Wang X, Zhao W, Ji Y, Yan J. Aggrecan variable number of tandem repeat polymorphism and lumbar disc degeneration: A meta-analysis. Spine. 2013;38(25):E1600–1607. doi: 10.1097/BRS.0000000000000012. [DOI] [PubMed] [Google Scholar]
- 10.Komori H, Shinomiya K, Nakai O, Yamaura I, Takeda S, Furuya K. The natural history of herniated nucleus pulposus with radiculopathy. Spine. 1996;21(2):225–229. doi: 10.1097/00007632-199601150-00013. [DOI] [PubMed] [Google Scholar]
- 11.Yuceli S. Minimally invasive surgery for one level spinal stenosis: Unilateral approach bilateral microdecompression. J Turk Spin Surg. 2018;29(3):189–192. [Google Scholar]
- 12.Battie MC, Videman T, Gibbons LE, Manninen H, Gill K, Pope M, Kaprio J. Occupational driving and lumbar disc degeneration: A case−control study. Lancet. 2002;360(9343):1369–1374. doi: 10.1016/S0140-6736(02)11399-7. [DOI] [PubMed] [Google Scholar]
- 13.Jim JJ, Noponen-Hietala N, Cheung KM, Ott J, Karppinen J, Sahraravand A, Luk KD, Yip SP, Sham PC, Song YQ, Leong JC, Cheah KS, Ala-Kokko L, Chan D. The TRP2 allele of COL9A2 is an age-dependent risk factor for the development and severity of intervertebral disc degeneration. Spine. 2005;30(24):2735–2742. doi: 10.1097/01.brs.0000190828.85331.ef. [DOI] [PubMed] [Google Scholar]
- 14.Paassilta P, Lohiniva J, Goring HH, Perala M, Raina SS, Karppinen J, Hakala M, Palm T, Kroger H, Kaitila I, Vanharanta H, Ott J, Ala-Kokko L. Identification of a novel common genetic risk factor for lumbar disk disease. JAMA. 2001;285(14):1843–1849. doi: 10.1001/jama.285.14.1843. [DOI] [PubMed] [Google Scholar]
- 15.Annunen S, Paassilta P, Lohiniva J, Perala M, Pihlajamaa T, Karppinen J, Tervonen O, Kroger H, Lahde S, Vanharanta H, Ryhanen L, Goring HH, Ott J, Prockop DJ, Ala-Kokko L. An allele of COL9A2 associated with intervertebral disc disease. Science. 1999;285(5426):409–412. doi: 10.1126/science.285.5426.409. [DOI] [PubMed] [Google Scholar]
- 16.Roughley P, Martens D, Rantakokko J, Alini M, Mwale F, Antoniou J. The involvement of aggrecan polymorphism in degeneration of human intervertebral disc and articular cartilage. Eur Cell Mater. 2006;11:1–7. [PubMed] [Google Scholar]
- 17.Kawaguchi Y, Osada R, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T. Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine. 1999;24(23):2456–2460. doi: 10.1097/00007632-199912010-00006. [DOI] [PubMed] [Google Scholar]
- 18.Dong DM, Yao M, Liu B, Sun CY, Jiang YQ, Wang YS. Association between the-1306C/T polymorphism of matrix metalloproteinase-2 gene and lumbar disc disease in chinese young adults. Eur Spine J. 2007;16(11):1958–1961. doi: 10.1007/s00586-007-0454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi M, Haro H, Wakabayashi Y, Kawa-uchi T, Komori H, Shinomiya K. The association of degeneration of the intervertebral disc with 5A/6A polymorphism in the promoter of the human matrix metalloproteinase-3 gene. J Bone Joint Surg Br. 2001;83(4):491–495. doi: 10.1302/0301-620x.83b4.11617. [DOI] [PubMed] [Google Scholar]
- 20.Goupille P, Jayson MI, Valat JP, Freemont AJ. Matrix metalloproteinases: The clue to intervertebral disc degeneration. Spine. 1998;23(14):1612–1626. doi: 10.1097/00007632-199807150-00021. [DOI] [PubMed] [Google Scholar]
- 21.Le Maitre CL, Freemont AJ, Hoyland JA. The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther. 2005;7(4):R732–745. doi: 10.1186/ar1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solovieva S, Kouhia S, Leino-Arjas P, Ala-Kokko L, Luoma K, Raininko R, Saarela J, Riihimaki H. Interleukin 1 polymorphisms and intervertebral disc degeneration. Epidemiology. 2004;15(5):626–633. doi: 10.1097/01.ede.0000135179.04563.35. [DOI] [PubMed] [Google Scholar]
- 23.Solovieva S, Leino-Arjas P, Saarela J, Luoma K, Raininko R, Riihimaki H. Possible association of interleukin 1 gene locus polymorphisms with low back pain. Pain. 2004;109(1-2):8–19. doi: 10.1016/j.pain.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Iatridis JC, MacLean JJ, O’Brien M, Stokes IA. Measurements of proteoglycan and water content distribution in human lumbar intervertebral discs. Spine. 2007;32(14):1493–1497. doi: 10.1097/BRS.0b013e318067dd3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou D, Samartzis D, Bellabarba C, Patel A, Luk KD, Kisser JM, Skelly AC. Degenerative magnetic resonance imaging changes in patients with chronic low back pain: A systematic review. Spine. 2011;36(21 Suppl):S43–53. doi: 10.1097/BRS.0b013e31822ef700. [DOI] [PubMed] [Google Scholar]
- 26.Corps AN, Robinson AH, Movin T, Costa ML, Hazleman BL, Riley GP. Increased expression of aggrecan and bi-glycan mRNA in achilles tendinopathy. Rheumatology. 2006;45(3):291–294. doi: 10.1093/rheumatology/kei152. [DOI] [PubMed] [Google Scholar]
- 27.Urban JPM A, Bayliss MT, Dillon J. Swelling pressures of proteoglycans at the concentrations found in cartilaginous tissues. Biorheology. 1979;16:447–464. doi: 10.3233/bir-1979-16609. [DOI] [PubMed] [Google Scholar]
- 28.Videman T, Saarela J, Kaprio J, Nakki A, Levalahti E, Gill K, Peltonen L, Battie MC. Associations of 25 structural, degradative, and inflammatory candidate genes with lumbar disc desiccation, bulging, and height narrowing. Arthritis Rheum. 2009;60(2):470–481. doi: 10.1002/art.24268. [DOI] [PubMed] [Google Scholar]
- 29.Rajasekaran S, Kanna RM, Senthil N, Raveendran M, Cheung KM, Chan D, Subramaniam S, Shetty AP. Phenotype variations affect genetic association studies of degenerative disc disease: Conclusions of analysis of genetic association of 58 single nucleotide polymorphisms with highly specific phenotypes for disc degeneration in 332 subjects. Spine J. 2013;13(10):1309–1320. doi: 10.1016/j.spinee.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Williams MG, Wafai AM, Podmore MD. Functional outcomes of laminectomy and laminotomy for the surgical management lumbar spine stenosis. J Spine Surg. 2017;3(4):580–586. doi: 10.21037/jss.2017.10.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woolf AD. The bone and joint decade 2000-2010. Ann Rheum Dis. 2000;59(2):81–82. doi: 10.1136/ard.59.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13(3):318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Mulligan KR, Ferland CE, Gawri R, Borthakur A, Haglund L, Ouellet JA. Axial t1rho mri as a diagnostic imaging modality to quantify proteoglycan concentration in degenerative disc disease. Eur Spine J. 2015;24(11):2395–2401. doi: 10.1007/s00586-014-3582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith LJ, Fazzalari NL. The elastic fibre network of the human lumbar annulus fibrosus: Architecture, mechanical function and potential role in the progression of intervertebral disc degeneration. Eur Spine J. 2009;18(4):439–448. doi: 10.1007/s00586-009-0918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhtar S, Davies JR, Caterson B. Ultrastructural immuno-localization of alpha-elastin and keratan sulfate proteoglycan in normal and scoliotic lumbar disc. Spine. 2005;30(15):1762–1769. doi: 10.1097/01.brs.0000171912.44625.29. [DOI] [PubMed] [Google Scholar]
- 36.Ellingson AM, Nagel TM, Polly DW, Ellermann J, Nuckley DJ. Quantitative t2* (t2 star) relaxation times predict site specific proteoglycan content and residual mechanics of the intervertebral disc throughout degeneration. J Orthop Res. 2014;32(8):1083–1089. doi: 10.1002/jor.22633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutton WC, Toribatake Y, Elmer WA, Ganey TM, Tomita K, Whitesides TE. The effect of compressive force applied to the intervertebral disc in vivo. A study of proteoglycans and collagen. Spine. 1998;23(23):2524–2537. doi: 10.1097/00007632-199812010-00007. [DOI] [PubMed] [Google Scholar]
- 38.Liu GZ, Ishihara H, Osada R, Kimura T, Tsuji H. Nitric oxide mediates the change of proteoglycan synthesis in the human lumbar intervertebral disc in response to hydrostatic pressure. Spine. 2001;26(2):134–141. doi: 10.1097/00007632-200101150-00005. [DOI] [PubMed] [Google Scholar]
- 39.Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: Regulation of proteoglycan synthesis. Exp Cell Res. 1999;246(1):129–137. doi: 10.1006/excr.1998.4287. [DOI] [PubMed] [Google Scholar]
- 40.Robinson D, Mirovsky Y, Halperin N, Evron Z, Nevo Z. Changes in proteoglycans of intervertebral disc in diabetic patients. A possible cause of increased back pain. Spine. 1998;23(8):849–856. doi: 10.1097/00007632-199804150-00001. [DOI] [PubMed] [Google Scholar]
- 41.Cs-Szabo G, Ragasa-San Juan D, Turumella V, Masuda K, Thonar EJ, An HS. Changes in mRNA and protein levels of proteoglycans of the annulus fibrosus and nucleus pulposus during intervertebral disc degeneration. Spine. 2002;27(20):2212–2219. doi: 10.1097/00007632-200210150-00006. [DOI] [PubMed] [Google Scholar]
- 42.Keshari KR, Lotz JC, Kurhanewicz J, Majumdar S. Correlation of hr-mas spectroscopy derived metabolite concentrations with collagen and proteoglycan levels and Thompson grade in the degenerative disc. Spine. 2005;30(23):2683–2688. doi: 10.1097/01.brs.0000188256.88859.9c. [DOI] [PubMed] [Google Scholar]
- 43.Singh K, Masuda K, Thonar EJ, An HS, Cs-Szabo G. Age-related changes in the extracellular matrix of nucleus pulposus and annulus fibrosus of human intervertebral disc. Spine. 2009;34(1):10–16. doi: 10.1097/BRS.0b013e31818e5ddd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim NK, Shin DA, Han IB, Yoo EH, Kim SH, Chung SS. The association of aggrecan gene polymorphism with the risk of intervertebral disc degeneration. Acta Neurochir. 2011;153(1):129–133. doi: 10.1007/s00701-010-0831-2. [DOI] [PubMed] [Google Scholar]