Abstract
In view of the complex procedure of nucleic acid extraction, there exists a huge challenge for the widespread use of point-of-care diagnostics for nucleic acid testing. To achieve point-of-care applications in a more rapid and cost-efficient manner, we designed a snake pipe-shaped microfluidic chip so as to accomplish reagents-prestored, time-saving, operation-simple nucleic acid extraction. All reagents needed for this process, including lysis buffer, wash buffer, elution buffer, and so on, were preloaded in the snake pipe and securely isolated by membrane valves, without the need for using any specialized equipment. By an integrated chip and a powerful ultrasonic, this device could complete virus nucleic acid extraction from sophisticated serum samples in less than 1 min. We used hepatitis B virus (HBV) and human immunodeficiency virus (HIV) mixed with different sources of serum as samples to be extracted. The coefficient of variation of HBV and HIV extraction on-chip was 1.32% and 2.74%, respectively, and there were no significant differences between on-chip and commercial instrument extraction (P > 0.05, α = 0.05) in different dilution ratios, which showed that the extraction device we established had excellent stability and sensitivity.
I. INTRODUCTION
Following the criteria of point-of-care (POC) testing introduced by WHO, known as ASSURED (affordable, sensitive, specific, user-friendly, robust & rapid, equipment-free, and deliverable), efforts have been made to realize simple nucleic acid extraction (NAE).1,2 Nevertheless, due to the complex process of nucleic acid extraction, portable and on-site nucleic acid extraction methods are still nonexistent. In recent years, to realize an efficient nucleic acid extraction for applications of resource limited settings, focused attention has been given for the paper-based nucleic acid extractions.3–6 With advantages of simplicity and biocompatibility, paper with cellulose fibers impregnated with chemicals could capture, stabilize, and protect nucleic acid. Nevertheless, paper-based devices may unspecifically absorb nucleic acid variously, leading to a decreased detection limit; thus, a breakthrough still has to be achieved so that NAE protocols could leave the laboratory settings to the “real world” of point-of-care diagnostics (POC-Dx).7–9 Microfluidic technology possesses the characteristics of miniaturization and integration, which highly meet the requirements of POC-Dx.10–12 Recently, microfluidic chip designs based on polymers (PDMS or PMMA), glass, and silicon have shown the potential to establish a platform for sensitive and accurate NAE. Additionally, automatic processing process and fully sealed chips not only provide superior qualities of reduced human-intervention, time-saving, and labor-economics, but also achieve improvements in bio-safety, avoiding cross contamination, and minimizing environmental pollution.13,14 Nowadays, fully automatic and integrated molecular diagnostic systems that can accomplish “sample in, answer out” have become an important research area in the development of POC molecular diagnostics.4,15 While the total analysis system is difficult to achieve due to technological barriers,16 the promotion of integration can be obtained through the breakthrough in research on independent functional modules17 and then assembling the components to form a complete integrated system containing nucleic acid extraction, amplification, and detection. Under the guidance of this thought, we first designed a simple device for NAE.
In this work, we innovatively demonstrated a microfluidic chip for nucleic acid extraction from sophisticated serums based on a solid-phase extraction technique using silica membrane adsorbing nucleic acids specifically18,19 and ultrasonic assisted lysis and mixture. Compared with the existing microfluidics approaches, the device we designed may be more applicable in developing countries (Table I). To evaluate the practical utility of the nucleic acid extraction platform we developed, hepatitis B virus (HBV) and human immunodeficiency virus (HIV) were chosen to represent DNA and RNA, respectively, and a commercial instrument reference was set as the control compared with on-chip extraction. Extracted nucleic acids were transferred to be detected by quantitative real-time polymerase chain reaction (qRT-PCR), and the amount of yield was determined by the cycle threshold (Ct) value. Through a series of optimizations such as chip structure and purification times, the on-chip extraction we investigated could guarantee a fair amount of stability and sensitivity.
TABLE I.
Comparison between different nucleic acid extraction methods.
| Conventional extraction | FTA (Flinders Technology Associates) card | Membrane | Cepheid® (microfluidic) | The article (microfluidic) | |
|---|---|---|---|---|---|
| Lysis | Chemicals | Chemicals | Chemicals | Chemicals and Ultrasound | Chemicals and Ultrasound |
| Purification | Phenol-chloroform | Filtration isolation | Filtration isolation | Magnetic beads | Magnetic beads |
| Time | 2 h | 1 h | <10 min | <1 min | <1 min |
| Cost | $3.5/test | <$1.5/test | <$1.0/test | >$10/test | $2.5/test |
| Application | Laboratory | POCT (point of care testing) | POCT | POCT | POCT |
| Stage | Commercial | Commercial | Commercial | Commercial | Under development |
II. MATERIALS AND METHODS
A. Chip preparation
1. Chip design
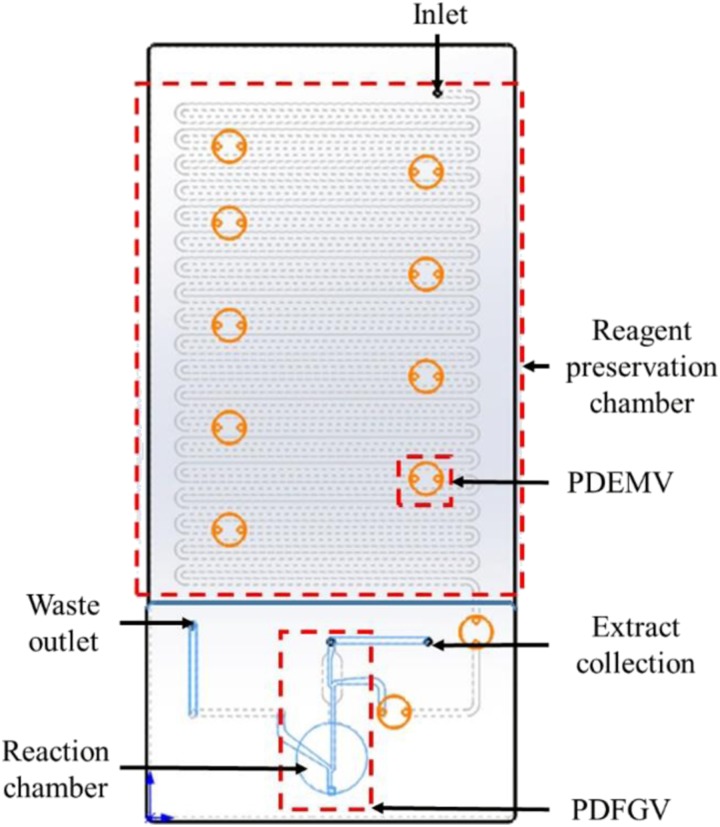
The chip was integrated with three major functioning components, which were identified as a reagent preservation chamber, a reaction chamber, and a fluid control valve (Fig. 1). The breadth of the channel was 1 mm, the radius of the valve was 2.5 mm, the radius of the reaction chamber was 5.5 mm, and the whole chip was 120 * 55 mm2. Two kinds of fluid control valves were used in this chip design, each having different working principles and functioning purposes. The first one was named as the pressure driven elastic membrane valve (PDEMV); it served as an on-off switch for the reagent flow. The PDEMV isolated different reagents from each other in the chip and controlled the flow of the fluid. The valve was sealed by the elastic membrane, with no outer pressure being applied. After applying air pressure from the inlet, the elastic membrane will be deformed and channels will be connected. This working principle is shown in Fig. 2(b). The second valve is the pressure driven fluid guiding valve (PDFGV), which is used to guide the fluid direction while the chip is working. The reaction chamber, the reagent preservation chamber, and the waster outlet were connected by the PDFGV, and the reagent was guided into different chambers based on the pressure differences between channels. The working principle of the PDFGV is shown in Fig. 2(c).
FIG. 1.
Two dimensional schematic diagram of the chip.
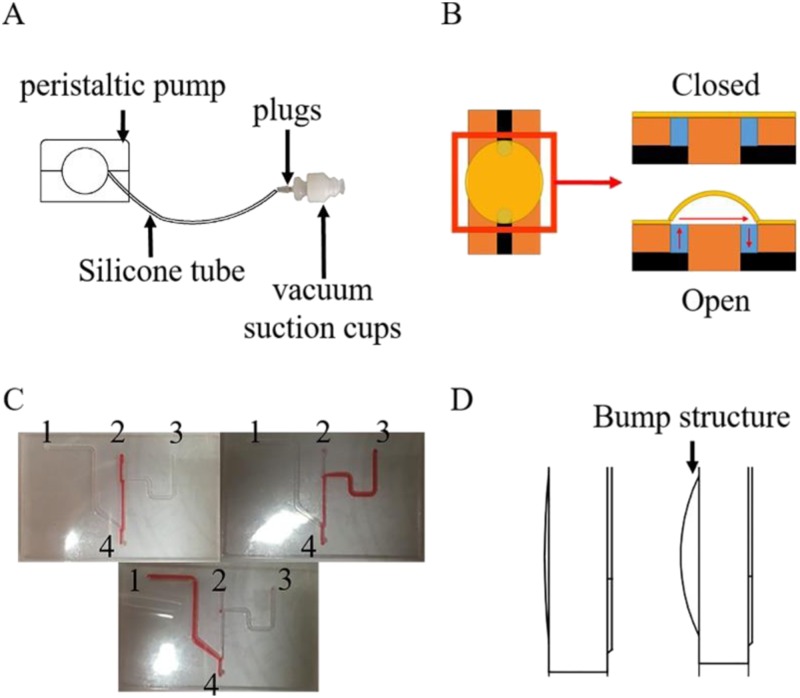
FIG. 2.
(a) Air and suction device. (b) Working principle of the PDEMV valve. (c) Working principle of the PDFGV valve. (d) No bump structure (left) vs bump structure (right).
A snake-shaped channel which was divided into several isolated chambers by multiple breakpoints was used for reagent preservation. The break point was sealed by PDEMV while preserving the reagents isolated in their own chambers. While the chip is working, the PDEMV will be opened by the driven air pressure applied from the inlet. The chambers are connected with each other and the reagents will be allowed to flow through the channel to reach the reaction chamber.
2. Reagents preloading
All reagents were preloaded in the pipe, including the lysis buffer, wash buffer, and elution buffer, which could be released in a sequence by a peristaltic pump [Fig. 2(a)]. In particular, reagents were filled in several pipelines and then sealed by the respective membrane valves. Figure 2(b) illuminates the working principle of the PDEMV. While closed, the two chambers are isolated; while the vacuum cup begins to blow air produced by the peristaltic pump to provide a positive pressure, the valve opens, and then the reagents could be transferred from one chamber to another. Figure 2(c) illuminates the working principle of the PDFGV. While two of #1, 2, and 3 holes are sealed, the reagents can be outflowed from other holes. The cost of the reagents used in the chip for NAE is about $1 each.
3. Chip fabrication
The chip was fabricated by applying a standard PMMA-double tape fabrication technique.20–22 For a brief description, a 250 * 250 * 1 mm3 PMMA plate was cut by a laser cutter (Legend Technology, Guangzhou, China) according to the design generated by AutoCAD 2016 software (Autodesk, USA). Each layer was stacked in order and fixed by the double tape preliminarily; the bonding was reinforced by low temperature heat pressing in a vacuumed environment provided by a vacuum thermocompression bonding machine (WH-2000A, Wenhao, Suzhou, China). The pressing process was completed in half an hour at 70 °C and 0.5 Mpa. The production and fabrication price for each chip is about $1.5.
B. Preparation of samples
Serum exclusive of HBV and HIV was donated by the Central Blood Station (Xiamen, China) and stored at −20 °C. HBV and HIV were obtained from the laboratory cultured in a virus culture medium, with a water bath heated at 60 °C for half an hour to be inactivated and then stored at −20 °C (the viral load of HBV is 106 copies/ml, the viral load of HIV is 105 copies/ml). Samples were simulated by virus diluted with serum.
All experiments were performed in compliance with the hospital guidelines. [Independent ethics committee approval was obtained from the Ethics Committee of the National Institute of Diagnostics and Vaccine Development in Infectious Diseases (NIDVD)].
C. Operation of reference and on-chip extraction
Viral DNA/RNA were extracted from mimics by using a commercial DNA/RNA extraction kit based on the magnetic beads method (GenMag Bio, Beijing, China). Prior to each experiment, serum and virus were balanced to room temperature, and inactivated virus was diluted by 10 times the ratio of the serum to obtain viruses of different concentrations as specimens to be extracted.
As a reference control, the automated extraction instrument (Biomek NX) purchased from Beckman Coulter (USA) was used according to the manufacturer's instruction. For a brief description, first, 50 μl specimens, 75 μl lysis buffer, 5 μl magnetic beads, 2.5 μl proteinase K, and 1 μl acryl carrier were added to a deep-well plate (nuclease free) and then mixed at 3000 rpm for 9 min in an oscillator to render the nucleic acid to make full contact with the magnetic beads; the next step was purification, to be specific, pulling the beads to the bottom of the plate using a magnet for 1 min and discarding the supernatant, then adding wash buffer I and wash buffer II 200 μl to suspend the magnetic beads again and then to discard liquids as well, and separately, wash buffer III should avoid the resuspension of magnetic beads in the event of any loss of nucleic acid; lastly, 100 μl elution buffer was added to elute the purified DNA/RNA from the beads’ surface for 4 min in an oscillator, and then the beads were fixed by the magnet; the eluate with the purified DNA/RNA was transferred to a fresh tube and stored at −20 °C.
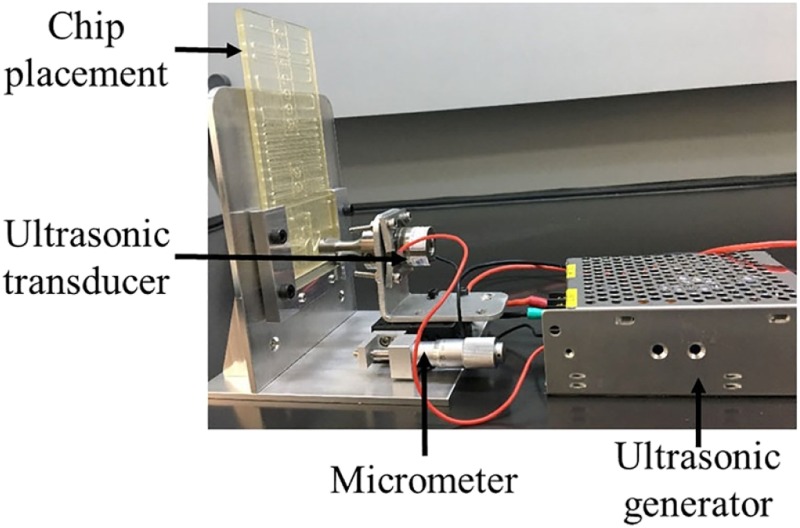
The extraction protocol is shown in Fig. 3, which is similar to the reference control mentioned above. The reagents’ dosage and releasing order were the same, but the ultrasonic blending method was different, where powerful energy in the associated ultrasonic wave produced by an ultrasonic generator (HNH-4020-SS-220VAC, Hainertec, Suzhou, China) and an ultrasonic transducer (HND-4SE-1940,40 KHz, Hainertec, Suzhou, China) could help with the lysis and mixture at the same time. The ultrasonic horn is located at the chip surface while in use (Fig. 3). Beyond this, compared with the manual control, the lysis time reduced from 9 min to 10 s, and the elution time shortened to 5 s, thus accelerating the extraction process to a large extent. It should be noted that intermittent ultrasound is better than the continuous one.
FIG. 3.
Ultrasonic device for nucleic acid extraction.
D. Quantification by real-time PCR (QRT-PCR) amplification
DNA and RNA yielded from manual reference and on-chip extraction were quantified by real-time PCR and real-time reverse transcription PCR, respectively. The HIV reaction system contains 2.5 μl 10× FB I buffer (Takara, Beijing, China), 0.4 mM dNTPs (Takara, Beijing, China), 0.2 μM forward primer, reverse primer, probe, respectively, 0.08 U Polymerase, 0.64 U reverse transcriptase, 5 μl template (total 25 μl); The HBV reaction system contains 2.5 μL 10× FB I buffer (Takara, Beijing, China), 0.4 mM dNTPs (Takara, Beijing, China), 0.2 μM forward primer, reverse primer, probe, respectively, 0.08 U Polymerase, 5 μl template (total 25 μl); The primers and probes were synthesized by Sangon Biotech (Shanghai, China). For HIV, the forward primer sequence (5′ to 3′) is ACAATGAGACACCAGGGATTAG, the reverse primer sequence is AGGCTCTAAGATTGTTGTCATG, and the probe sequence is TGGTGATCCTTTCCATCCCTGTGGAAG. For HBV, the forward primer sequence (5′ to 3′) is GCTGCTAGGTTGTACTGCCA, the reverse primer sequence is AGAAGGGGACGAGAGAGTCC, and the probe sequence is AATGCAGGGCAGCCGCGACTT. SpeedStar® HS DNA Polymerase was purchased from Takara Biomedical Technology (Beijing, China), TransScript Reverse Transcriptase was obtained from Transgen Biotech (Beijing, China), and DEPC water (DNAse/RNAse free) was made in the laboratory. Bio-Rad CFX96 thermocycler (Bio-Rad, USA) was used to conduct the PCR reaction in this research. A thermal cycling protocol for HBV DNA amplification started with predegeneration (10 min/95 °C), 40 cycles of denaturation (15 s/95 °C), and annealing/extension (50 s/55 °C). The thermal cycling protocol for HIV RNA amplification started with reverse transcription (5 min/50 °C) followed by hot-start (10 min/95 °C) and 40 cycles of denaturation (15 s/95 °C) and annealing/extension (50 s/55 °C).
III. RESULTS AND DISCUSSION
A. Reagents releasing and device operation
To realize automated reagents to release during the on-chip extraction process, we used a simple valve actuator customized in our laboratory. The main components of the valve actuator included two vacuum suction cups (8 mm in diameter), a silicone tube (3 mm in diameter), and a peristaltic pump [Fig. 2(a)]. The vacuum suction cup connected to the peristaltic pump can provide positive or negative pressure, which controls the flow of the fluids and switches the valves. The movement of the ultrasonic horn and the magnet are controlled by the positioning slides; the ultrasonic horn or magnet needs to be in contact with the chip, moving forward until it fits closely, moving backward otherwise.
B. Verification of chip absorption
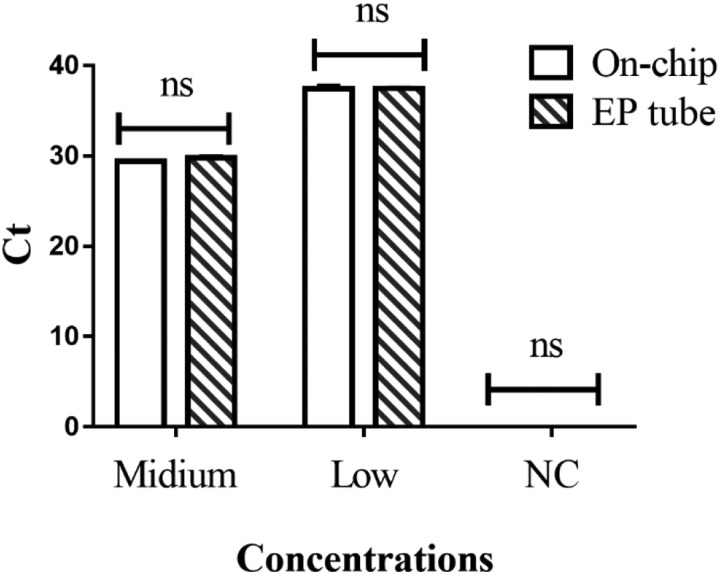
The surface of chip (PMMA) may adsorb nucleic acid, which could lead to a decrease in yield. The nucleic acid extracts were packed into a chip reaction chamber and an EP tube separately and taken out for a PCR reaction after 30 min incubation under room temperature; the result showed no significant difference between the chip chamber and the EP (Eppendorf) tube (Fig. 4), indicating that the chip surface adsorption effect could be ignored.
FIG. 4.
Nucleic acid adsorption assay on-chip.
C. Reaction parameters’ determination
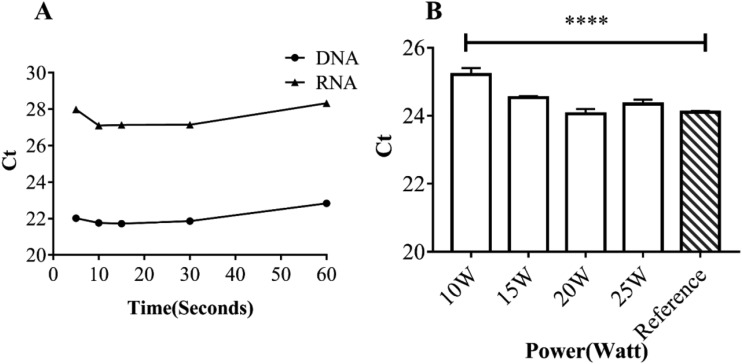
The power and period of the ultrasonic assisted reaction are correlated with the yield of nucleic acid extraction. To optimize the yield, we compared ultrasonic extraction effects under 5 s, 10 s, 15 s, 30 s, and 60 s of ultrasonic agitation separately [Fig. 5(a)]. The results indicated that the yield of extraction could reach a peak level in just 10 s for HBV or HIV virus spiked serum samples. (Ct value refers to the number of cycles experienced when the fluorescence signal in each reaction tube reaches the set threshold; the lower the size of Ct value, the higher the yield of extraction.) As the extended ultrasonic period could not exert a significant influence on the yield, 10 s was chosen as the lysis time in the following assays. Similarly, in the process of exploring the appropriate ultrasonic power [Fig. 5(b)], 20 W was found to have a better yield than 15 W by using HIV; however, 20 W power was so high that there was a possibility of great damage to the chip, damage characterized as crack and fluid leakage. Also, compared with 10 W (25.21 ± 0.09), 15 W has a higher yield of 24.53 ± 0.02 (P < 0.05). Therefore, we chose 15 W as the power for subsequent experiments to make sure that the experiment could be carried out normally.
FIG. 5.
(a) Exploration of optimal ultrasonic time at 15 W (5 s, 10 s, 15 s, 30 s, and 60 s). (b) Exploration of optimal ultrasonic power (10 W, 15 W, 20 W, and 25 W).
D. Improvement of on-chip extraction yield
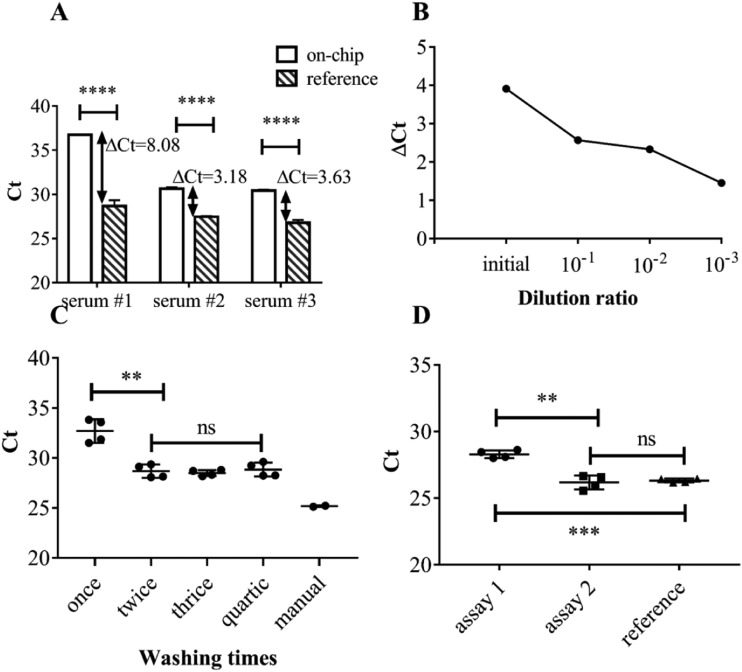
Polymerase chain reaction (PCR) assays could be inhibited severely by proteins, growth factors, inorganic metal ions, and other substances contained in the serum.23,24 As shown in Fig. 6(a), a significant difference is observed in the yield of on-chip extraction from viruses diluted in different serums. As shown in Fig. 6(b), with the increase in dilution ratio, the inhibition effect is gradually relieved, which confirmed the presence of inhibitors. Inhibitors could be eliminated basically [Fig. 6(c)] by washing twice. After eliminating the inhibitors, a difference was still observed between on-chip extraction and commercial reference. To enhance the on-chip extraction efficiency, doubling the dosage of lysis buffer and magnetic beads on-chip could generate a significant improvement [Fig. 6(d)]. After a series of optimizations, the on-chip extraction yield showed a significant improvement.
FIG. 6.
(a) Yield of on-chip vs commercial reference extraction while using different sources of serum as dilutions. (b) Gradient dilution assay to verify the existence of the PCR inhibitors. ΔCt = (Ct of on chip) − (Ct of manual reference). (c) Scavenging of inhibitors by increasing washing times. (d) Yield of different assays: assay 1 denotes normal dosage of lysis buffer and magnetic beads, assay 2 denotes double dosage of lysis buffer and magnetic beads, and assay 3 denotes commercial reference.
E. Stability and repeatability verification
Coefficient of variation (CV) has been used as the criterion to evaluate the stability and repeatability of the on-chip extraction method. To optimize the energy transduction between the ultrasonic horn and the reaction chamber, a bump structure [Fig. 2(d)] was designed to allow the ultrasonic horn to be in full contact with the chip. At the same time, the bump could make the contact site between the ultrasonic horn and the chip more fixed, which would improve the reproducibility effectively. As shown in Fig. 7, the coefficient of variation of HBV and HIV extraction on-chip is 1.32% and 2.74%, respectively.
FIG. 7.
Stability and repeatability evaluation of on-chip extraction. The coefficient of variation of HBV and HIV extraction on-chip is 1.32% and 2.74%, respectively.
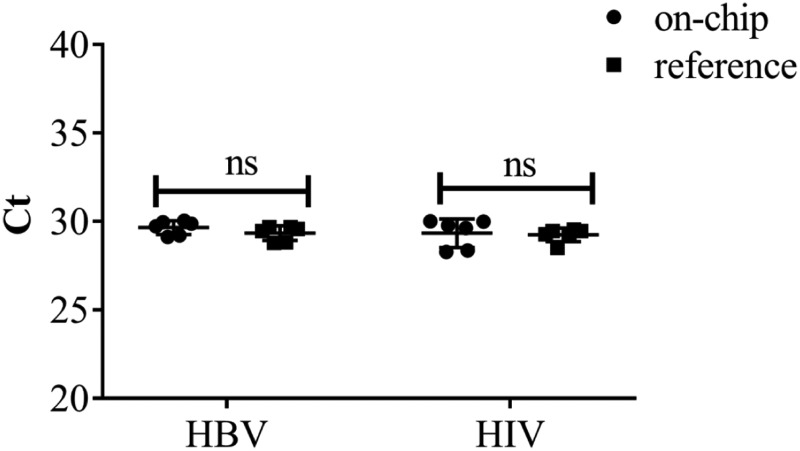
F. Sensitivity evaluation
To prove that on-chip extraction could extract nucleic acid at a low viral load, we compared the yields of on-chip vs commercial reference extraction from different HBV and HIV dilutions, respectively. We used Welch's t test to assess whether there were any differences between on-chip extraction and manual reference. As shown in Table II, there are no significant differences between them (P > 0.05, α = 0.05), which showed the high sensitivity of the on-chip extraction method. The approximate LOD (limit of detection) of our system for HBV was 103 copies/ml, and for HIV, it was 5 × 103 copies/ml.
TABLE II.
Comparison of yields between on-chip and commercial reference of HBV and HIV virus dilutions depicted as mean ± standard error of mean, respectively.
| Dilution ratio | DNA (HBV) | RNA (HIV) | ||||
|---|---|---|---|---|---|---|
| Commercial ref. (n = 3) | On-chip (n = 3) | Welch's t test P value | Commercial ref. (n = 3) | On-chip (n = 3) | Welch's t test P value | |
| 10−1 | 29.39 ± 0.15 | 29.18 ± 0.19 | 0.4135 | 31.76 ± 0.24 | 31.67 ± 0.21 | 0.8002 |
| 10−2 | 32.72 ± 0.12 | 32.79 ± 0.04 | 0.5881 | 34.96 ± 0.17 | 34.69 ± 0.18 | 0.3038 |
| 10−3 | 35.71 ± 0.13 | 35.43 ± 0.19 | 0.2719 | 37.68 ± 0.18 | 38.14 ± 0.35 | 0.2806 |
IV. CONCLUSIONS
We developed a simple platform based on a microfluidic technique, which could complete NAE in less than 1 min. In this paper, NAE was innovatively performed in an integrated microfluidic chip with easy fabrication, easy operation, and high integration. This platform removed the contradiction between simplicity and sensitivity and addressed the shortcomings of time consumption and labor intensiveness.
The POC technique was a platform which was identified as highly integrated, easy to perform, and low cost. Although our platform required a pump, a linear stage, and an ultrasonic system to perform the test, all these mechanical components are reusable and can be integrated into the machine, and the only disposable component in this system is the testing chip. On the other hand, the platform requires manual operations at present, but it was designed for automated operations. To meet with the requirements of POC, future work in this area will primarily focus on the following three aspects: the first is to achieve the industrial production of the chip. Snake-shaped pipeline and valves were designed to achieve the storage and releasing of reagents, but the current structure is too complex for molding. Designs should be simplified to reduce the complexity of production. The second is to extend the range of NAE to other pathogens such as bacteria or fungi. By means of the cavitation effect and mechanical action during ultrasonic propagation, the disruption or disintegration of the cellular membrane could be carried out rapidly accompanied by the full mixing of magnetic beads and liquids,25–27 and the extraction process for the virus could be as shorter as 1 min compared with commercial references as long as 20 min; NAE for other pathogens needs to be tested. The last is to integrate modules for amplification and detection, thus achieving a nucleic acid molecular diagnostic system. Specifically, we may develop a rapid one-step nucleic acid dipstick assay28,29 or design an amplification chamber linked to the current chip. Additionally, real patient samples should be evaluated in future studies, possibly as part of a clinical trial.
Microfluidics has become a new promising strategy for the implementation of point-of-care NAE. Previous attempts to develop microfluidic chip-based platforms for NAE did not take both simplicity and sensitivity into account, but the portable device we developed could guarantee both. There exists a high risk of contamination for a high concentration of nucleic acid extract, which restrains nucleic acid testing (NAT) to specialized laboratories. But, it could be solved by an all-in-one chip.30–33 The chip we designed for NAE in this paper has laid a solid foundation for the “sample in, answer out” pattern. More efforts are needed to form a complete integrated system containing nucleic acid extraction, amplification, and detection, thus realizing point-of-care diagnostics for nucleic acid testing (NAT).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NNSFC) (Grant No. 81871505), the Infectious Diseases Major Project of China (Grant No. 2017ZX10302101-001), the Key Research and Development Plan of China (Grant No. 2018YFC1200100), and the Natural Science Foundation of Fujian Province (Grant No. 2017J05136).
References
- 1.Peeling R. W., Holmes K. K., Mabey D., and Ronald A., Sex. Transm. Infect. 82, v1–v6 (2006). 10.1136/sti.2006.024265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majors C. E., Smith C. A., Natoli M. E., Kundrod K. A., and Richards-Kortum R., Lab Chip 17, 3351–3387 (2017). 10.1039/C7LC00374A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi J. R., Hu J., Tang R., Gong Y., Feng S., Ren H., Wen T., Li X., Wan Abas W. A., Pingguan-Murphy B., and Xu F., Lab Chip 16, 611–621 (2016). 10.1039/C5LC01388G [DOI] [PubMed] [Google Scholar]
- 4.Choi J. R., Tang R., Wang S., Wan Abas W. A., Pingguan-Murphy B., and Xu F., Biosens. Bioelectron. 74, 427–439 (2015). 10.1016/j.bios.2015.06.065 [DOI] [PubMed] [Google Scholar]
- 5.Fu Y., Zhou X., and Xing D., Sens. Actuators B 261, 288–296 (2018). 10.1016/j.snb.2018.01.165 [DOI] [Google Scholar]
- 6.Tang R., Yang H., Gong Y., You M., Liu Z., Choi J. R., Wen T., Qu Z., Mei Q., and Xu F., Lab Chip 17, 1270–1279 (2017). 10.1039/C6LC01586G [DOI] [PubMed] [Google Scholar]
- 7.Ali N., Rampazzo R. C. P., Costa A. D. T., and Krieger M. A., Biomed Res. Int. 2017, 9306564 (2017). 10.1155/2017/9306564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Causer L. M., Guy R. J., Tabrizi S. N., Whiley D. M., Speers D. J., Ward J., Tangey A., Badman S. G., Hengel B., Natoli L. J., Anderson D. A., Wand H., Wilson D., Regan D. G., Shephard M., Donovan B., Fairley C. K. and Kaldor J. M., Sex. Transm. Infect. 94, 340–345 (2018). 10.1136/sextrans-2017-053443 [DOI] [PubMed] [Google Scholar]
- 9.Chin C. D., Linder V., and Sia S. K., Lab Chip 12, 2118–2134 (2012). 10.1039/c2lc21204h [DOI] [PubMed] [Google Scholar]
- 10.Ahrberg C. D., Manz A., and Chung B. G., Lab Chip 16, 3866–3884 (2016). 10.1039/C6LC00984K [DOI] [PubMed] [Google Scholar]
- 11.Kim T. H., Park J., Kim C. J., and Cho Y. K., Anal. Chem. 86, 3841–3848 (2014). 10.1021/ac403971h [DOI] [PubMed] [Google Scholar]
- 12.Thompson B. L., Birch C., Li J., DuVall J. A., Le Roux D., Nelson D. A., Tsuei A. C., Mills D. L., Krauss S. T., Root B. E., and Landers J. P., Analyst 141, 4667–4675 (2016). 10.1039/C6AN00209A [DOI] [PubMed] [Google Scholar]
- 13.Foudeh A. M., Fatanat Didar T., Veres T., and Tabrizian M., Lab Chip 12, 3249–3266 (2012). 10.1039/c2lc40630f [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Zhao Y., Qin Y., Du X., Wang Q., and Lyu J., RSC Adv. 6, 13399–13406 (2016). 10.1039/C5RA26225A [DOI] [Google Scholar]
- 15.Zhang L., Zhang Y., Wang C., Feng Q., Fan F., Zhang G., Kang X., Qin X., Sun J., Li Y., and Jiang X., Anal. Chem. 86, 10461–10466 (2014). 10.1021/ac503072a [DOI] [PubMed] [Google Scholar]
- 16.Xu Y., Jang K., Yamashita T., Tanaka Y., Mawatari K., and Kitamori T., Anal. Bioanal. Chem. 402, 99–107 (2012). 10.1007/s00216-011-5296-5 [DOI] [PubMed] [Google Scholar]
- 17.Marshall L. A., Wu L. L., Babikian S., Bachman M., and Santiago J. G., Anal. Chem. 84, 9640–9645 (2012). 10.1021/ac302622v [DOI] [PubMed] [Google Scholar]
- 18.Wen J., Legendre L. A., Bienvenue J. M., and Landers J. P., Anal. Chem. 80, 6472–6479 (2008). 10.1021/ac8014998 [DOI] [PubMed] [Google Scholar]
- 19.Esser K.-H., Marx W. H., and Lisowsky T., Nat. Methods 3, i–ii (2006). 10.1038/nmeth845 [DOI] [Google Scholar]
- 20.Khashayar P., Amoabediny G., Larijani B., Hosseini M., Van Put S., Verplancke R., and Vanfleteren J., J. Braz. Soc. Mech. Sci. Eng. 39(5), 1469–1477 (2016). 10.1007/s40430-016-0684-6 [DOI] [Google Scholar]
- 21.Yang J., Liu Y., Rauch C. B., Stevens R. L., Liu R. H., Lenigk R., and Grodzinski P., Lab Chip 2(4), 179–187 (2002). 10.1039/b208405h [DOI] [PubMed] [Google Scholar]
- 22.Wu Z. and Nguyen N.-T., Sens. Actuators B 107(2), 965–974 (2005). 10.1016/j.snb.2004.11.014 [DOI] [Google Scholar]
- 23.Kosch T. A. and Summers K., Mol. Ecol. Resour. 13(2), 230–236 (2013). 10.1111/1755-0998.12041 [DOI] [PubMed] [Google Scholar]
- 24.Queipo-Ortuno M. I., De Dios Colmenero J., Macias M., Bravo M. J., and Morata P., Clin. Vaccine Immunol. 15(2), 293–296 (2008). 10.1128/CVI.00270-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azimi S. M., Nixon G., Ahern J., and Balachandran W., Microfluid. Nanofluid. 11(2), 157–165 (2011). 10.1007/s10404-011-0782-9 [DOI] [Google Scholar]
- 26.Liu C. J., Lien K. Y., Weng C. Y., Shin J. W., Chang T. Y., and Lee G. B., Biomed. Microdevices 11(2), 339–350 (2009). 10.1007/s10544-008-9240-1 [DOI] [PubMed] [Google Scholar]
- 27.Xu G., Gunson R. N., Cooper J. M., and Reboud J., Chem. Commun. 51(13), 2589–2592 (2015). 10.1039/C4CC08389J [DOI] [PubMed] [Google Scholar]
- 28.Zhang S., Xue M., Zhang J., Chen Q., Chen J., Wang Z., Zhou W., Chen P., Xia N., and Ge S., Clin. Biochem. 46(18), 1852–1856 (2013). 10.1016/j.clinbiochem.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 29.Qiu X., Zhang S., Xiang F., Wu D., Guo M., Ge S., Li K., Ye X., Xia N., and Qian S., Sens. Actuators B 243, 738–744 (2017). 10.1016/j.snb.2016.12.058 [DOI] [Google Scholar]
- 30.Jung W., Han J., Choi J.-W., and Ahn C. H., Microelectron. Eng. 132, 46–57 (2015). 10.1016/j.mee.2014.09.024 [DOI] [Google Scholar]
- 31.Shaw J. L. V., Pract. Lab. Med. 4, 22–29 (2016). 10.1016/j.plabm.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y., Huang Y. Y., Liu X., Zhang X., Ferrari M., and Qin L., Trends Biotechnol. 32(3), 132–139 (2014). 10.1016/j.tibtech.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarei M., Biosens. Bioelectron. 98, 494–506 (2017). 10.1016/j.bios.2017.07.024 [DOI] [PubMed] [Google Scholar]