Abstract
Background and Purpose
Graph theory analysis of brain connectivity data is a promising tool for studying the function of healthy and diseased brain. The consistency of resting state functional MRI (rsfMRI) connectivity measures across multiple scanner types is an important factor in designing multi-institutional research studies and has important implications for potential use of this technique in a heterogeneous clinical setting. We sought to quantitatively study the inter-scanner variability of rsfMRI graph theory metrics obtained from healthy volunteers scanned on three different scanner platforms.
Methods
In this prospective IRB approved study, 9 healthy volunteers were enrolled for brain MRI on 3 3T scanners (Magnetom Prisma, Skyra, and Trio, Siemens, Erlangen, Germany) in 3 separate scan sessions within approximately 1 week. Standard pre-processing of rsfMRI was performed with SPM12. Subject scans were normalized to MNI space, and connectivity of 116 ROIs based on the automated anatomic labeling (AAL) atlas were calculated using Conn toolbox. Whole-network graph theory metrics were calculated using Brain Connectivity Toolbox, and intra-class correlation (ICC) across three scan sessions was assessed.
Results
A total of 25 rsfMRI exams were completed in 9 subjects with a median-intersession time of 3 days. Among all three sessions, there was good to excellent agreement in characteristic path length and global efficiency (ICC 0.79, 0.79) and good agreement in transitivity, local efficiency and clustering coefficient (ICC 0.72, 0.69, 0.62).
Conclusions
There was high consistency of graph theory metrics of rsfMRI connectivity networks among healthy volunteers scanned on three different generation 3T MRI scanners.
Keywords: resting state functional MRI, graph theory, variability
Introduction:
Resting state functional magnetic resonance imaging (rsfMRI) has emerged in the last decade as an important research tool for evaluation of the brain in health and disease. First described in 1995 by Biswal et al, spontaneous slow (<0.1 Hz) fluctuations of blood oxygen level dependent (BOLD) signal in spatially distinct brain regions is used to infer the connectivity between these regions.1 There are a multitude of approaches to rsfMRI data analysis, including seed-based analysis, independent component analysis (ICA), and graph theory analysis.2–4 The strength of graph theory lies in its established mathematical foundation and general applicability to various networks, including both functional and structural brain networks constructed using data from rsfMRI, magnetoencephalography, and diffusion tensor imaging.5 The local and global properties of networks can be summarized as various metrics to provide insight into the interaction between different brain regions.
Numerous recent studies using graph theoretical analysis have demonstrated group level differences between disease states; however, there are yet several barriers to clinical translation.6–8 Importantly, the test-retest reliability of any measure must be defined before incorporating it in trials or using it as a diagnostic test. Reproducibility of graph theory analysis in functional imaging has been studied in several prior works and summarized in two recent reviews.9,10 While the influence of various acquisition and processing factors have been studied, no study to our knowledge has evaluated the variability of graph theoretical networks across different MRI platforms.11–13 Cross-platform consistency is critical for conducting and comparing multi-institutional studies and for applicability to a heterogeneous clinical environment with many scanner types. The goal of this study is to evaluate the variability of graph theoretical metrics in normal subjects by performing three scanning sessions on each subject on different 3T MRI platforms.
Methods
Participants
This prospective study was approved by the local Institutional Review Board, and individual consent was obtained from each subject. Healthy volunteers aged 18 or older were recruited from the community to be included in the study. Exclusion criteria were known current or prior neurological or psychiatric disorder, current substance use disorder and contraindication to undergo MRI such as metallic implants. The median age of the patients completing the study was 35 (interquartile range 32 – 41), with 5 females and 4 males.
MR Scanning
Each patient underwent brain MRI on three 3T MRI scanners in separate sessions within approximately a week (Magnetom Trio, Magnetom Skyra, Magnetom Prisma, Siemens Healthineers, Erlangen, Germany). The three platforms differ in gradient performance, bore size, option of coils and electronic systems, and are the most commonly used 3-Telsa scanners for research purposes that are manufactured by Siemens. Seven of the 9 subjects completed all three sessions, while 2 subjects did not complete the scan on the Magnetom Skyra due to software malfunction. Median intersession time was 3 days (interquartile range 2–5 days).
RsfMRI was acquired with subjects remaining awake and eyes fixed on a stationary marker. Multiband echo planar imaging (EPI) blood oxygen level dependent (BOLD) acquisition was performed using 64 channel coils for both Prisma and Skyra scanners and a 32-channel coil for the Trio scanner with the following parameters. Scanner 1 (Magnetom Prisma), subjects 1–7: TR 720 msec, TE 37 msec, acquisition time 7.2 min, voxel size 2 mm, matrix 104 × 104, FOV 208 × 208 mm, MB 8; subjects 8–9: TR 750 msec, TE 37 msec, acquisition time 8.25 min, voxel size 2.5 mm, matrix 88 × 88, FOV 220 × 220 mm, MB 6. Scanner 2 (Magnetom Trio): TR 750 msec, TE msec 37, acquisition time 8.25 min, voxel size 2.5 mm, matrix 88 × 88, FOV 220 × 220 mm, MB 6. Scanner 3 (Magnetom Skyra): TR 747 msec, TE 37 msec, acquisition time 8.25 min, voxel size 2.5 mm, matrix 88 × 88, FOV 220 × 220 mm, MB6. Additional MRI sequences were acquired at each session, including structural T1 MPRAGE data using 1 mm isotropic voxels.
Pre-processing
Pre-processing of functional and structural data was performed using commonly used open-source software as follows. First, EPI distortion correction of the functional data was performed on opposing phase encoding direction BOLD acquisitions using FMRIB Software Library Topup (FSL v6.0, FMRIB, Oxford, UK). Next, Statistical Parametric Mapping (SPM12, Wellcome Centre for Human Neuroimaging, London, UK), based pre-processing pipeline in Conn Toolbox 2017 (McGovern Institute for Brain Research, MIT, Cambridge, MA) was used to perform functional realignment, motion estimation, structural segmentation and normalization, functional normalization to Montreal Neurologic Institute (MNI) space, functional outlier detection, band-pass filter of 0.01 to 0.1 Hz, and regression of confounding affects (white matter, CSF, realignment, artifact scrubbing).14 Processed region-wise BOLD times-series data from 116 brain regions as defined by the Automated Anatomic Labeling (AAL) atlas was exported for further analysis.15
Correlation matrix
Intrinsic brain connectivity between the regions in the AAL atlas were calculated from the extracted BOLD time-series data using partial correlations.16 Partial correlation measures the direct connectivity between two nodes by estimating their correlation after regressing out effects from all the other nodes in the network, hence avoiding spurious effects in network modeling. We estimated the partial correlation matrix using the DensParcorr R package which is based on an efficient and reliable statistical methods for estimating functional connectivity via partial correlation in large scale brain networks.17 Subject/scanner-specific connectivity matrix of the AAL atlas was estimated based on each subject’s rs-fMRI scans obtained at each of the 3 MRI scanners.
Graph theory analysis
A weighted undirected graph was constructed from the partial correlation matrix using Brain Connectivity Toolbox (Mikail Rubinov and Olaf Sporn) implemented in Matlab (Mathworks, Natick, MA). The nodes were defined by the AAL regions and edge weights were defined by partial correlation coefficients, with a graph density of 0.4 (preserving the weights of the top 40% of partial correlations). We calculated common global network measures including global efficiency, transitivity and characteristic path length, and local network measures including local efficiency and clustering coefficient. Characteristic path length is the average of the shortest length between nodes in the network. Global efficiency is the average inverse shortest path length in the network, and local efficiency is the same metric calculated in node neighborhoods. Clustering coefficient is the fraction of triangles around a node and reflects how many of an individual node’s neighbors are neighbors of each other, whereas the related measure transitivity is the ratio of the triangles to triplets within the entire network.5
Intraclass correlation
We then assessed the agreement/reliability among the network measures across the 3 MRI scanners using intra-class correlation (ICC). Specifically, we used a two-way mixed single score ICC which is based on two-way ANOVA with the three scanners as fixed effects and subjects as random effects, and the unit of analysis is measurements obtained on every single scanner.18,19
Results
The overall ICC across three scanners for global efficiency, local efficiency, transitivity, clustering coefficient and characteristic path length ranged from good to excellent, and were 0.79, 0.69, 0.71, 0.62 and 0.79 respectively. Table 1 includes overall as well as pairwise ICC results. Seven subject data were used to calculate overall ICC results and pairwise ICC for scanner1 vs 3 and scanner 2 vs 3, due to missing data from subjects 1 and 3. Data from all nine subjects were used to calculate pairwise ICC for scanner 1 vs 2.
Table 1.
Intra-class correlation between measures overall and pair-wise between each two scanners
| Measure | Overall | Scanner 1 vs 2 | Scanner 1 vs 3 | Scanner 2 vs 3 |
|---|---|---|---|---|
| Global efficiency | 0.79 | 0.83 | 0.81 | 0.75 |
| Transitivity | 0.72 | 0.81 | 0.68 | 0.67 |
| Characteristic path length | 0.79 | 0.79 | 0.80 | 0.79 |
| Local efficiency | 0.69 | 0.76 | 0.67 | 0.66 |
| Clustering coefficient | 0.62 | 0.70 | 0.58 | 0.60 |
Scanner 1, Magnetom Prisma; Scanner 2, Magnetom Trio; Scanner 3, Magnetom Skyra.
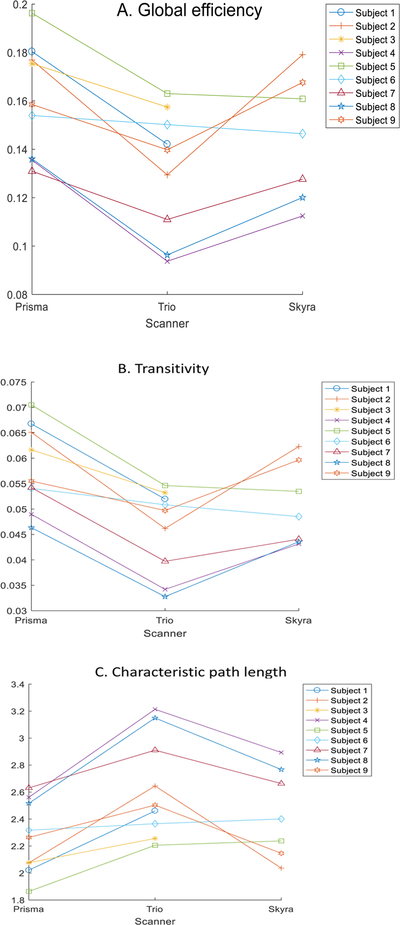
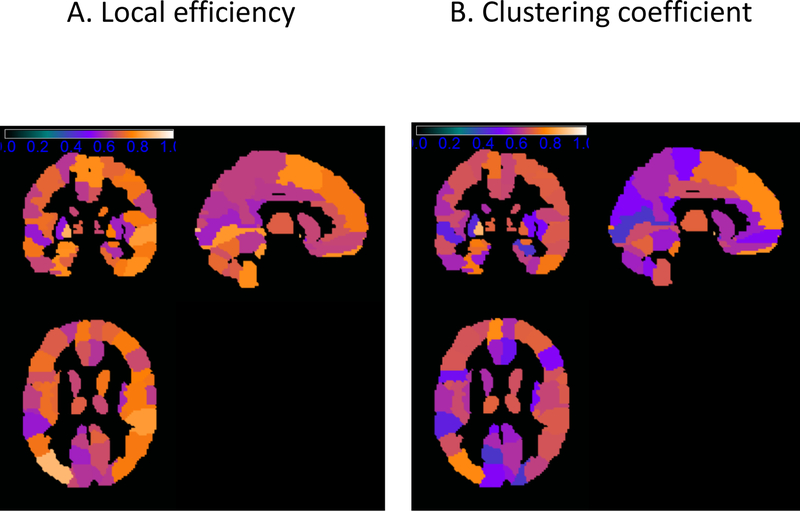
To provide more detailed information on the variability of these measures, Figure 1 present the scatterplot of the estimated global network measures for each of the nine subjects using the three scanners. For variability of local network measures, Table 2 presents the top 20 regions in the AAL atlas with the highest ICC for local efficiency and cluster coefficient. Figure 2 presents the ICC maps of the local efficiency and clustering coefficients for the regions in AAL atlas.
Figure 1:
Scatterplot for global graph theoretical metrics (a) global efficiency, (b) transitivity, and (c) characteristic path length on three scanners. Subjects 1 and 3 did not complete scan on the Skyra.
Table 2:
Top 20 regions in the Automated Anatomic Labeling atlas with highest intraclass correlation for the local network measures.
| Local efficiency | Clustering coefficient | ||
|---|---|---|---|
| Region | ICC | Region | ICC |
| ‘Occipital Mid L’ | 0.86 | ‘Pallidum L’ | 0.86 |
| ‘Frontal Mid Orb L’ | 0.85 | ‘Frontal Mid Orb L’ | 0.83 |
| ‘Pallidum L’ | 0.85 | ‘Parietal Inf L’ | 0.80 |
| ‘Temporal Pole Sup L’ | 0.81 | ‘Frontal Sup Orb L’ | 0.79 |
| ‘Frontal Inf Orb R’ | 0.81 | ‘Olfactory R’ | 0.77 |
| ‘Temporal Sup R’ | 0.80 | ‘Frontal Sup Med L’ | 0.77 |
| ‘Temporal Pole Sup R’ | 0.80 | ‘Occipital Mid L’ | 0.77 |
| ‘Cerebelum 3 L’ | 0.80 | ‘Cerebelum 3 R’ | 0.76 |
| ‘Lingual L’ | 0.79 | ‘Frontal Inf Orb R’ | 0.76 |
| ‘Frontal Mid Orb R’ | 0.79 | ‘Vermis 3’ | 0.74 |
| ‘Hippocampus L’ | 0.78 | ‘SupraMarginal R’ | 0.74 |
| ‘Frontal Sup Orb L’ | 0.78 | ‘Amygdala L’ | 0.74 |
| ‘Temporal Inf R’ | 0.78 | ‘Fusiform L’ | 0.74 |
| ‘Fusiform L’ | 0.78 | ‘Temporal Inf R’ | 0.73 |
| ‘Cerebelum 3 R’ | 0.78 | ‘Cerebelum 3 L’ | 0.74 |
| ‘Amygdala L’ | 0.78 | ‘Frontal Med Orb R’ | 0.73 |
| ‘Supp Motor Area L’ | 0.78 | ‘Cerebelum Crus1 R’ | 0.73 |
| ‘Cerebelum 9 L’ | 0.77 | ‘Temporal Pole Sup R’ | 0.72 |
| ‘Supp Motor Area R’ | 0.77 | ‘Temporal Pole Sup L’ | 0.72 |
| ‘Cerebelum Crus1 R’ | 0.76 | ‘Supp Motor Area L’ | 0.71 |
ICC, intraclass correlation; L, left; R, right; Sup, superior; Mid, middle; Inf, inferior; Orb, orbital; Supp, supplmental.
Figure 2:
ICC maps of local graph theoretical metrics (a) local efficiency (b) and clustering coefficient for regions in the automated anatomic labeling atlas. The color of the regions correspond to the ICC values as indicated by the color bar.
Discussion
Graph theoretical analysis of functional imaging is a promising tool for studying normal cognition and disease, and its variability both within and across scanner platforms is of paramount importance to its translation to clinical use. A recent review of 23 studies on the reproducibility of both functional and structural connectivity graph theory metrics found intersession agreement mostly reached good (ICC 0.60–0.74) and excellent (ICC>0.74) range.10 The implication of many methodological considerations on reproducibility have been evaluated by past studies, including scan interval and length, band-pass filter frequency range, global signal regression, correlation method, atlas choice, weighted or unweighted network, thresholding method for graph construction.13,20–24 While it is well known that these and other methodological factors will have significant impacts on rsfMRI results, a recent meta-analysis found that only frequency band selection and length of acquisition had a significant impact on test-retest reliability.9
Our study is the first to study the variability of graph theoretical metrics in rsfMRI across different scanner platforms. To this end, our results show good to excellent test-retest reliability for commonly used global and local measures, in a similar range to those report by same-scanner studies. Our study has several limitations, one of which is the limited types of scanners used. We used only Siemens 3T scanners in this study due to availability and wish to maintain consistency in acquisition parameters, and this can affect our results and limit its generalizability to a mix of other scanner vendors, types and strengths. The design of our study did not allow us to isolate the across-scanner variability from inter-session variability. A future study that would address this question would require repeated scans on each scanner in different sessions. Our results are nevertheless important in that such high consistency was obtained with both potential sources of variability. Although direct comparison with prior studies that evaluated inter-session variability cannot be done due to various methodologic differences, the variability in our results tended to trend lower than previously reported. This is encouraging and adds another data point to the existing literature that supports the same-scanner reproducibility of graph theory metrics. We strived to maintain of the scanning parameters across three scanners, although subjects 1–7 scanned on scanner 1 (Prisma) were scanned at a higher spatial resolution and shorter time than the rest due to unintended protocol deviation. As our analysis was performed on ROI-level and not voxel-wise data, and the acquisition time differences were small, these deviations did not seem to affect the pair-wise variability between this and other scanners (Table 1). It was also not our goal to evaluate the effect of various pre- and post-processing methods, so we used commonly used public atlas (AAL) and post-processing pipelines (Conn and Brain Connectivity Toolbox).
From our results, we can conclude that by using fairly consistent scanning parameters and standard processing, good to excellent consistency of graph theory metrics can be achieved across three different Siemens 3T scanner platforms. Future studies including scanners from a broader range of vendors and of different magnet strengths, perhaps at multiple sites, would provide additional information about the generalizability of these findings.
Acknowledgements and Disclosure:
Ying Guo: Research reported in this publication was supported by the National Institute of Mental Health of the National Institutes of Health under Award Number R01 MH105561 and R01MH079448. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Footnotes
No competing financial interests exist for any of the authors.
References
- 1.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 1995;34:537–41. [DOI] [PubMed] [Google Scholar]
- 2.Smitha KA, Arun KM, Rajesh PG, Thomas B, Kesavadas C. Resting-state seed-based analysis: an alternative to task-based language fMRI and its laterality index. AJNR Am J Neuroradiol 2017;38:1187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Zuo X, He Y. Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci 2010;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calhoun VD, de Lacy N. Ten key observations on the analysis of resting-state functional MR imaging data using independent component analysis. Neuroimaging Clin N Am 2017;27:561–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 2010;52:1059–69. [DOI] [PubMed] [Google Scholar]
- 6.Messé A, Caplain S, Pélégrini-Issac M, et al. Specific and evolving resting-state network alterations in post-concussion syndrome following mild traumatic brain injury. PLoS One 2013;8:e65470–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Zhong S, Jia Y, et al. Disrupted resting-state functional connectivity in nonmedicated bipolar disorder. Radiology 2016;280:529–36. [DOI] [PubMed] [Google Scholar]
- 8.Wang YF, Ji XM, Lu G-M, Zhang L-J. Resting-state functional MR imaging shed insights into the brain of diabetes. Metab Brain Dis 2016;31:993–1002. [DOI] [PubMed] [Google Scholar]
- 9.Andellini M, Cannatà V, Gazzellini S, Bernardi B, Napolitano A. Test-retest reliability of graph metrics of resting state MRI functional brain networks: a review. J. Neurosci Methods 2015;253:183–92. [DOI] [PubMed] [Google Scholar]
- 10.Welton T, Kent DA, Auer DP, Dineen RA. Reproducibility of graph-theoretic brain network metrics: a systematic review. Brain Connect 2015;5:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo CC, Kurth F, Zhou J, et al. One-year test-retest reliability of intrinsic connectivity network fMRI in older adults. Neuroimage 2012;61:1471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun U, Plichta MM, Esslinger C, et al. Test–retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage 2012;59:1404–12. [DOI] [PubMed] [Google Scholar]
- 13.Liang X, Wang J, Yan C, et al. Effects of different correlation metrics and preprocessing factors on small-world brain functional networks: a resting-state functional MRI study. PLoS One 2012;7:e32766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect 2012;2:125–41. [DOI] [PubMed] [Google Scholar]
- 15.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15:273–89. [DOI] [PubMed] [Google Scholar]
- 16.Smith SM, Miller KL, Salimi-Khorshidi G, et al. Network modelling methods for FMRI. Neuroimage 2011;54:875–91. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Kang J, Kemmer PB, Guo Y. An efficient and reliable statistical method for estimating functional connectivity in large scale brain networks using partial correlation. Front Neurosci 2016;10:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420–28. [DOI] [PubMed] [Google Scholar]
- 19.McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological Methods 1996;1:30–46. [Google Scholar]
- 20.Wang J-H, Zuo X-N, Gohel S, Milham MP, Biswal BB, He Y. Graph theoretical analysis of functional brain networks: test-retest evaluation on short- and long-term resting-state functional MRI data. PLoS One 2011;6:e21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao X-H, Xia M-R, Xu T, et al. Functional brain hubs and their test-retest reliability: a multiband resting-state functional MRI study. Neuroimage 2013;83:969–82. [DOI] [PubMed] [Google Scholar]
- 22.Braun U, Plichta MM, Esslinger C, et al. Test-retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage 2012;59:1404–12. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz AJ, McGonigle J. Negative edges and soft thresholding in complex network analysis of resting state functional connectivity data. Neuroimage 2011;55:1132–46. [DOI] [PubMed] [Google Scholar]
- 24.Cao H, Plichta MM, Schäfer A, et al. Test-retest reliability of fMRI-based graph theoretical properties during working memory, emotion processing, and resting state. Neuroimage 2014;84:888–900. [DOI] [PubMed] [Google Scholar]