Abstract
We found that a newly developed method named LITPOMS (ligand titration, fast photochemical oxidation of proteins and mass spectrometry) can characterize section-by-section of a protein the conformational changes induced by metal-ion binding. Peptide-level LITPOMS applied to Ca2+ binding to calmodulin reveals binding order and site-specific affinity, providing new insights on the behavior of proteins upon binding Ca2+. We established that EF hand-4 (EF-4) binds calcium first, followed by EF-3, EF-2 and EF-1 and determined the four affinity constants by modeling the extent-of-modification curves. We also found positive cooperativity between EF-4, EF-3 and EF-2, EF-1 and allostery involving the four EF-hands. LITPOMS recapitulates via one approach the calcium-calmodulin binding that required decades of sophisticated development to afford versatility, comprehensiveness, and outstanding spatial resolution.
Graphical Abstract:
Our goal is a single approach to understand the complexities of protein-metal binding, especially multiple binding.1–2 To do this, we chose calmodulin (CaM), a small calcium (Ca2+)-binding protein in all eukaryotic cells, that plays essential roles in Ca2+ signaling,3–4 regulating cell motility, growth, proliferation, and apoptosis.5–7 The protein has two homologous globular domains (N- and C-terminal) connected by a flexible linker.8–11 Each domain contains a pair of Ca2+-binding motifs called EF-hands and binds two Ca2+ ions cooperatively (CaM:Ca2+ stoichiometry is 1:4).12–13 Subsequent protease susceptibility studies revealed cooperativity between and N- and C-terminal domains.14 Each EF-hand binds with μM affinity, making the protein sensitive to intracellular Ca2+-concentration changes and responsive as a signaling protein. Each canonical EF-hand has a ‘helix-loop-helix’ configuration, where a calcium-loop sits between the two helices. Ca2+ binding alters interhelical angles in the EF-hands, moving them from ‘closed’ to ‘open’,12 exposing hydrophobic sites that bind, and activating many target proteins.7
Understanding these interactions has required decades of research, during which various approaches were developed2, 13–26 and applied.13–14, 19–23, 25 Fluorescence-based methods reveal overall conformational changes, Ca2+ binding affinities,13, 23 and, in combination with microfluidic mixers, the kinetics of conformational change upon binding.21 NMR shows the EF-hand local conformational change induced by binding.19 Isothermal titration calorimetry examines the system thermodynamically, providing valuable binding constants.20, 22 On the other hand, the needs for specific fluorophore labeling, special sample preparation, and high sample quantity limit applications. Thus, a new approach that overcomes these problems and delivers improved spatial resolutions will be a significant improvement in tool set for protein - metal-ion binding.
To date, NMR resolves protein solution-state structure with the highest spatial resolution. A titration-type affinity measurement for metal-ion binding, however, requires low micromolar concentrations, which are not accessible today to NMR. A mass spectrometry (MS) structural proteomics approach should significantly lower sample amount and eliminates the need for specific isotopic labeling as for NMR. Moreover, a structural proteomics approach may also greatly elevate the spatial resolution and sensitivity when compared with fluorescence.24–25 Indeed, spatial resolution is achievable by protease digestion and MS.27
Recently, we developed LITPOMS26 that combines ligand titration with footprinting by fast photochemical oxidation of proteins (FPOP).28–30 We demonstrated its efficacy for 1:1 binding, where the results are similar to those of HDX.25 That is, LITPOMS distinguishs binding from non-bonding, accurately reports the binding affinity, and reveals some of the critical binding residues.26
For more complicated signalling proteins, such as CaM, HDX in a titration format also shows binding and provides hints of remote conformational changes and allostery. But these proteins undergo complex conformational changes encompassing the whole protein where some regions tighten and bind, others open, and still provide cooperativity and allostery. To meet the challenges of tracking these changes, we extended FPOP with its high footprinting speed and irreversible labeling to capture, section-by-section this complexity and thereby reveal features not seen by HDX or specific amino-acid labeling.25, 31–32
Here, we demonstrate that application of LITPOMS can meet the challenge and determine binding sites, binding affinities, binding orders, site-specific affinities, and importantly the complex, composite behavior of the protein upon metal-ion binding. Moreover, it can reveal cooperativity and allostery. Information from LITPOMS recapitulates the long-term cumulative efforts to understand calcium-calmodulin binding.
In the current experiment, 1 μM calcium-free calmodulin (Apo-CaM) was first equilibrated with calcium at [Ca2+]-to-[Apo-CaM] of 0 to 60, and the proteins were footprinted by FPOP.26, 34 Global response was recorded by MS under denaturing conditions. For peptide-level responses, the complexes were digested with trypsin/Lys-C, and the digested peptides were submitted to LC/MS2 analysis.34 Integration of extracted ion chromatograms taken by LC-MS2 allowed calculation of the modification fraction.
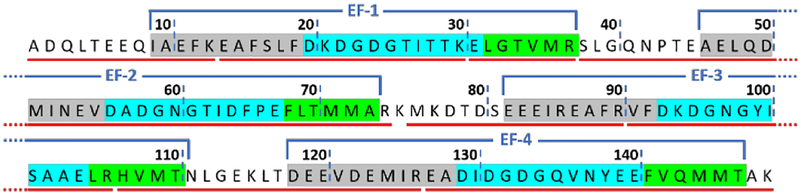
To assure good coverage and high S/N, an MS2 method with a precursor-ion inclusion list was used for signal acquisition. The inclusion list was developed based on several test runs. To meet standards of resolvable extracted ion chromatograms, accurate precursor/fragment masses, and confident MS2, eight distinct tryptic peptides were chosen to afford 99% sequence coverage (Figure 1, Figure S3, and detailed description in SI). Like many Ca2+-binding proteins, CaM contains four EF-hands consisting of an E helix, calcium loop, and F helix (peptides representing these regions are in grey, cyan and green in Figure 1, respectively).
Figure 1.
A 99% sequence coverage map of calmodulin showing regions of conformational change.11 Red lines under the sequence indicate the observed peptides. The four EF-hands are super-scored by blue brackets. Residues shaded with grey, cyan and green represents the E helix, calcium loop and F helix of each EF-hand, respectively.
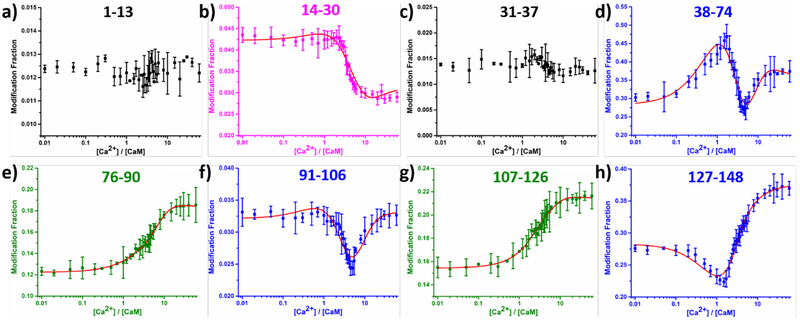
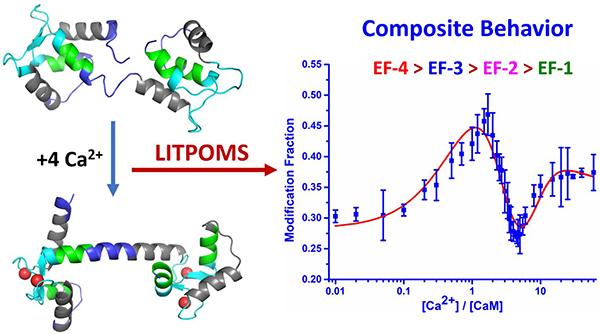
When titrated with Ca2+ by using the LITPOMS protocol, CaM shows four remarkable and distinct classes of behavior upon titration with Ca2+ (Figure 2), each successfully fit for binding. These curves are sensitive representations of opposing conformational changes that impart a composite of protection and deprotection (binding with tightening of structure vs. loosening or opening in allostery).
Figure 2.
LITPOMS response at peptide level, where modification fractions were plotted as a function of calcium:calmodulin ratio. Four different classes of behaviors are shown in black (a and c), magenta (b), blue (d, f and h) and olive (e and g). Red solid lines in (b), (d), (e), (f), (g) and (h) are from fitting using an algorithm reported previously.25, 33 Data points represent average of two runs, and error bars are standard deviations. Modification fractions reported here accounts for all resolvable oxidations, for which a maximum of +32 were observed at peptide level.
The first class of behavior is the loss of protection in the linker region where the well-known transformation from compact to ‘dumbbell-like’ occurs.7–8, 10, 34 The experimental observations recapitulate the loosening of structure for region 76–90 (Class I behavior), representing the central linker between the two lobes together with part of helix E in EF-3, and for the region 107–126 connecting EF-3 and EF-4.
Class II behavior is for peptides 1–13 and 31–37 (black in Figure 2a and 2c). Modification extents for these peptides remain relatively constant as Ca2+ binds, indicating no involvement in binding and no remote conformational changes. Indeed, peptide 1–13 is part of the N-terminal of calmodulin whereas peptide 31–37 is helix F in EF-1.
Class III behavior shows classical binding behavior, which is illustrated by peptide 14–30 that covers the helix E and calcium loop of EF-1 (magenta in Figure 2b). The modification fraction stays constant early in the titration, but when [Ca2+]:[CaM] ≅ 2, this region becomes protected and shows a decrease in modification. Once saturated with calcium, peptide 14–30 remains protected at a modification fraction of ~ 0.028.
The three other binding sites exhibit different behavior, termed Class IV. This behavior is composite, showing a combination of binding and conformational changes induced remotely (allostery).14, 21 Peptides 38–74, 91–106 and 127–148 join this category (blue panels in Figure 2d, 2f and 2h). Peptide 127–148 contains the calcium loop and F helix in EF-4, whose response shows a gain then a loss in protection when adding Ca2+. The initial gain is due to calcium binding in EF-4, whereby residues in the calcium loop chelate Ca2+, leading to increasing compactness and a decrease in solvent accessibility. Upon passing the critical point of [Ca2+]/[CaM] ≅ 1.5, the calcium loop in EF-4 becomes saturated with calcium whereas the F loop becomes more exposed owing to binding-induced conformational change. Loss of protection of the F loop contributes to an increase of the modification fraction for peptide 127–148 at the later stage of the calcium titration. The modification fraction of peptide 91–106 stays relatively stable until [Ca2+]/[CaM] ≅ 0.7, after which a decrease in modification occurs. Most of the calcium loop in EF-3 (residue 93–104) resides in this peptide, explaining the decrease of the modification fraction. Like peptide 127–148, an increase in modification fraction occurs at the later stage of the titration, owing to a binding-induced conformational change that facilitates calcium binding of other EF hands.
While falling in the class IV behavior, peptide 38–74 differs from its peers. Upon adding calcium, peptide 38–74 becomes less protected, seen as an increase in modification fraction. This increase peaks at [Ca2+]/[CaM] ≅ 1.7, where EF-4 finishes binding with calcium and EF-3 is on its way. Peptide 38–74 covers the linker between EF-1 and EF-2 as well as the whole EF-2 (helix E, calcium loop and helix F). The observed loss in protection at the early stage of the titration follows by a binding-induced conformational change. To be specific, binding of calcium by EF-3 and EF-4 facilitates a conformational change in EF-2, preparing it to take its own calcium. After [Ca2+]/[CaM] ≅ 1.7, peptide 38–74 becomes protected, indicating that EF-2 starts to bind calcium. This discloses the allosteric behavior of calcium binding, where EF-2 is prepared to take calcium upon calcium binding at remote sites. Similarly, after adding 5 equivalents of calcium, peptide 38–74 becomes less protected, along with structural transitions for most of the protein (peptide 38–74, 76–90, 91–106, 107–126 and 127–148) to more extended conformations. During the later stage of the titration (i.e., [Ca2+]/[CaM] ≥ 5), EF-1 is taking up calcium and transitioning into a calcium-bound state while most of the protein is opening. The conformation of the holo-calmodulin with four bound calcium ions stabilizes when [Ca2+] > 15 equivalents of the protein as seen by a nearly invariant modification extent.
Allostery upon Ca2+ binding has been difficult to assess. For example, proteolytic footprinting shows that, upon titration with Ca2+, the susceptibility profiles of residues E31 and R37 of an truncated CaM (N-domain) differ from that of the N-terminus of the full protein.14 This early insight into allosteric behavior of CaM upon Ca2+ binding was followed by protein grafting that reveals the intrinsic Ca2+ affinities of isolated calcium loops in each individual EF-hand differ from those of the EF-hands in intact CaM. Although the differences reflect structural effects of adjacent residues and allostery,35 neither study shows the actual behavior. LITPOMS greatly advances the understanding by revealing the overall calcium binding dynamics of CaM as a function of calcium concentration, which, until now, has not been understood with such spatial detail.
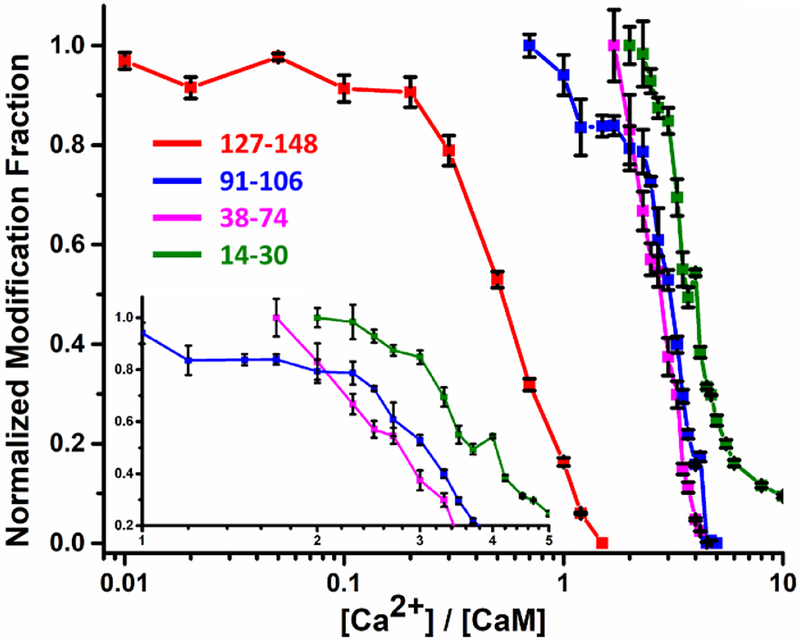
In addition to following the complex dynamics upon Ca2+ binding, the results permit a determination of the order of Ca2+ binding (Figure 3). Given that bond formation increases protection (decreases reactivity) at a binding site, the onsets for decreases in modification should reveal the starting points for Ca2+ binding at each EF-hand. The onsets clearly show that the first binding occurs at EF-4 (peptide 127–148) and the last at EF-1 (peptide 14–30). The order of binding for EF-3 (peptide 91–106) and EF-2 (peptide 38–74) is ambiguous. Most of the decrease in modification fraction of EF-3 lags that of EF-2, indicating that the two binding events are highly competitive. Considering the earlier onset for EF-3, however, we conclude that this is the second site. Thus, the binding order is assigned as EF-4 > EF-3 > EF-2 > EF-1, which not only agrees with some previous conclusions that the C-terminal lobe binds first,13 but also advances that picture by revealing the order within each lobe.
Figure 3.
Normalized (to 1) LITPOMS responses for selected peptides showing increases in protection (decreases in modification) for four peptides representing the four EF hands of CaM. Insert shows expanded decreases in modification for peptides 91–106, 38–74 and 14–30. Data points are averages of two independent runs, and error bars are standard deviations.
Following an assignment of binding order, we obtained four binding affinities for each EF hand (summary in Table 1) by fitting the various binding curves in Figure 2 by using a previously reported fitting algorithm,25, 33 where EF-1 and EF-2 were grouped into N-terminal lobe and EF-3 and EF-4 in the C-terminal lobe. Positive cooperativity was assumed in each lobe whereas the two lobes bind calcium independently.13 Each state (Apo-CaM, CaM-1-Ca2+, CaM-2-Ca2+ …) has its distinct modification fraction. A search utilizes the experimental data to determine the system composition at different calcium concentrations, after which binding affinities and a peptide-specific fitting curve are obtained.
Table 1.
Summary of binding order and site-specific binding affinities for calcium-calmodulin system
| Binding Order | EF-hand | LITPOMS Ki (M−1) | Literature Ki (M−1)13 |
|---|---|---|---|
| 1 | EF-4 | 1.4 × 106 | 8.0 × 104 |
| 2 | EF-3 | 6.2 × 106 | 4.0 × 106 |
| C-Term Lobe | 8.6 × 1012 | 3.2 × 1011 | |
| 3 | EF-2 | 4.1 × 104 | 2.5 × 104 |
| 4 | EF-1 | 2.9 × 106 | 4.0 × 105 |
| N-Term Lobe | 1.2 × 1011 | 1.0 × 1010 |
The results (Table 1) show that LITPOMS successfully reveals the binding order and the site-specific binding affinities for four EF-hands in calmodulin upon binding with calcium. The binding affinities for EF-2 and EF-3 agree reasonably well with those in the literature (within a factor of 1.6) whereas for EF-1 and EF-4 are within 20-fold.13 Although an advantage of LITPOMS to affinity measurements may be the peptide-level modeling is of at least four binding curves rather than a single global-level fluorescence response,13 we view the affinities as estimates that need further validation. Nevertheless, the affinity constants show that the binding in each lobe is positively cooperative (EF-3 > EF-4 and EF-1 > EF-2).
In summary, we find that LITPOMS reveals the composite behavior for regions covering all of calmodulin upon binding with calcium and shows clearly the allosteric changes in, for example, region 38–74 caused by the binding. The composite behavior at peptide level is due, in part, to the convolution from the long peptide (e.g., 38–74), which can potentially be dissected through enzymatic digestion by a different protease or by tracking specific residues in that peptide. Further, the outcome illustrates the cooperativity within N-terminal and C-terminal lobes, but also between two lobes as demonstrated by the behavior of peptide 38–74 at early stage of titration. In addition to this detail, we can also determine the binding sites, the binding order, and site-specific binding affinities, which agree reasonably well with some but not all previous determinations.13
The application of LITPOMS not only recapitulates, via a single approach, the understanding of the calcium-calmodulin binding system gained over many decades but also provides new insights that cannot be easily accessed through other approaches. The outstanding spatial resolution of FPOP, as an example of MS-based structural proteomics, affords insights that previously required combinations of different approaches. The improving sensitivity of modern mass spectrometers allows these advances to be made with high picomoles of samples whereas the irreversible labeling of FPOP broadens the application of LITPOMS by overcoming the major disadvantages of HD exchange,25, 36 making it possible to extend it to the amino-acid residue level. With proper selection of labeling reagents,37–39 it may be possible to characterize the affinity of a specific binding residue even at the nM level for many different ligands.26 This bodes well for studies of signaling proteins,3–4, 40 where conformational changes are widespread and concomitant changes in solvent accessibility are readily followed by reactions of free radicals and other reactive species.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institute of Health NIGMS Grant 5P41GM103422 and 1S10OD016298–01A1 (to MLG.). Authors are grateful to Dr. Jagat Adhikari for helpful discussions and Protein Metrics for software support.
Footnotes
Supporting Information
Detailed experimental conditions, global level LITPOMS response, details about peptide selection, and an example of peptide level data processing can be found in supporting information.
The authors declare no competing financial interests.
REFERENCES
- 1.Schellman JA, Macromolecular binding. Biopolymers 1975, 14 (5), 999–1018. [Google Scholar]
- 2.Williams M, Daviter Tina, Protein-Ligand Interactions Methods and Applications. Humana Press: New York, 2013. [Google Scholar]
- 3.Clapham DE, Calcium signaling. Cell 1995, 80 (2), 259–268. [DOI] [PubMed] [Google Scholar]
- 4.Clapham DE, Calcium Signaling. Cell 2007, 131 (6), 1047–1058. [DOI] [PubMed] [Google Scholar]
- 5.Cheung W, Calmodulin plays a pivotal role in cellular regulation. Science 1980, 207 (4426), 19–27. [DOI] [PubMed] [Google Scholar]
- 6.Means AR; Dedman JR, Calmodulin—an intracellular calcium receptor. Nature 1980, 285, 73. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M; Tanaka T; Ikura M, Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nature Structural Biology 1995, 2, 758. [DOI] [PubMed] [Google Scholar]
- 8.Babu YS; Bugg CE; Cook WJ, Structure of calmodulin refined at 2.2 Å resolution. Journal of Molecular Biology 1988, 204 (1), 191–204. [DOI] [PubMed] [Google Scholar]
- 9.Chattopadhyayal Rajagopal, M. WE, Means Anthony R., Quiocho Florante A., Calmodulin Structure Refined at l.7 A Resolution. J. Mol. Biol 1992, 228, 1177–1192. [DOI] [PubMed] [Google Scholar]
- 10.Kuboniwa Hitoshi, N. T, Grzesiek Stephan, Ren Hao, Klee Claude B., Bax Ad, Solution Structure of Calcium-free Calmodulin. Nat. Chem. Biol 1995, 2 (9), 768–776. [DOI] [PubMed] [Google Scholar]
- 11.Watterson DM; Sharief F; Vanaman TC, The complete amino acid sequence of the Ca2+-dependent modulator protein (calmodulin) of bovine brain. Journal of Biological Chemistry 1980, 255 (3), 962–975. [PubMed] [Google Scholar]
- 12.Lewit-Bentley A; Réty S, EF-hand calcium-binding proteins. Current Opinion in Structural Biology 2000, 10 (6), 637–643. [DOI] [PubMed] [Google Scholar]
- 13.Linse S; Helmersson A; Forsén S, Calcium binding to calmodulin and its globular domains. Journal of Biological Chemistry 1991, 266 (13), 8050–8054. [PubMed] [Google Scholar]
- 14.Sorensen BR; Shea MA, Interactions between Domains of Apo Calmodulin Alter Calcium Binding and Stability. Biochemistry 1998, 37 (12), 4244–4253. [DOI] [PubMed] [Google Scholar]
- 15.Rossi AM; Taylor CW, Analysis of protein-ligand interactions by fluorescence polarization. Nat Protoc 2011, 6 (3), 365–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer B; Peters T, NMR Spectroscopy Techniques for Screening and Identifying Ligand Binding to Protein Receptors. Angewandte Chemie International Edition 2003, 42 (8), 864–890. [DOI] [PubMed] [Google Scholar]
- 17.Johnsson B; Löfås S; Lindquist G, Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Analytical Biochemistry 1991, 198 (2), 268–277. [DOI] [PubMed] [Google Scholar]
- 18.Wiseman T; Williston S; Brandts JF; Lin L-N, Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical Biochemistry 1989, 179 (1), 131–137. [DOI] [PubMed] [Google Scholar]
- 19.Biekofsky RR; Turjanski AG; Estrin DA; Feeney J; Pastore A, Ab Initio Study of NMR 15N Chemical Shift Differences Induced by Ca2+ Binding to EF-Hand Proteins. Biochemistry 2004, 43 (21), 6554–6564. [DOI] [PubMed] [Google Scholar]
- 20.Henzl MT; Larson JD; Agah S, Estimation of parvalbumin Ca2+- and Mg2+-binding constants by global least-squares analysis of isothermal titration calorimetry data. Analytical Biochemistry 2003, 319 (2), 216–233. [DOI] [PubMed] [Google Scholar]
- 21.Park HY; Kim SA; Korlach J; Rhoades E; Kwok LW; Zipfel WR; Waxham MN; Webb WW; Pollack L, Conformational changes of calmodulin upon Ca2+ binding studied with a microfluidic mixer. Proceedings of the National Academy of Sciences 2008, 105 (2), 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilli R; Lafitte D; Lopez C; Kilhoffer MC; Makarov A; Briand C; Haiech J, Thermodynamic Analysis of Calcium and Magnesium Binding to Calmodulin. Biochemistry 1998, 37 (16), 5450–5456. [DOI] [PubMed] [Google Scholar]
- 23.Miyawaki A; Llopis J; Heim R; McCaffery JM; Adams JA; Ikura M; Tsien RY, Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882. [DOI] [PubMed] [Google Scholar]
- 24.Powell KD; Ghaemmaghami S; Wang MZ; Ma L; Oas TG; Fitzgerald MC, A General Mass Spectrometry-Based Assay for the Quantitation of Protein−Ligand Binding Interactions in Solution. Journal of the American Chemical Society 2002, 124 (35), 10256–10257. [DOI] [PubMed] [Google Scholar]
- 25.Zhu MM; Rempel DL; Du Z; Gross ML, Quantification of Protein−Ligand Interactions by Mass Spectrometry, Titration, and H/D Exchange: PLIMSTEX. Journal of the American Chemical Society 2003, 125 (18), 5252–5253. [DOI] [PubMed] [Google Scholar]
- 26.Liu XR; Zhang MM; Rempel DL; Gross ML, Protein-Ligand Interaction by Ligand Titration, Fast Photochemical Oxidation of Proteins and Mass Spectrometry: LITPOMS. Journal of The American Society for Mass Spectrometry 2019, 30 (2), 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H; Rempel DL; Giblin D; Frieden C; Gross ML, Peptide-Level Interactions between Proteins and Small-Molecule Drug Candidates by Two Hydrogen−Deuterium Exchange MS-Based Methods: The Example of Apolipoprotein E3. Analytical Chemistry 2017, 89 (20), 10687–10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hambly DM; Gross ML, Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom 2005, 16 (12), 2057–63. [DOI] [PubMed] [Google Scholar]
- 29.Guozhong XMRC, Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev 2007, 107, 3514–3543. [DOI] [PubMed] [Google Scholar]
- 30.Li KS; Shi L; Gross ML, Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization. Accounts of Chemical Research 2018, 51 (3), 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Mei M., R. DL, Zhao Jiang, Giblin Daryl E., and Gross Michael L., Probing Ca2+-Induced Conformational Changes in Porcine Calmodulin by H/D Exchange and ESI-MS: Effect of Cations and Ionic Strength. Biochemistry 2003, 42, 15388–15397. [DOI] [PubMed] [Google Scholar]
- 32.Guo C; Cheng M; Gross ML, Protein-Metal-Ion Interactions Studied by Mass Spectrometry-Based Footprinting with Isotope-Encoded Benzhydrazide. Analytical Chemistry 2019, 91 (2), 1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu MM; Rempel DL; Gross ML, Modeling data from titration, amide H/D exchange, and mass spectrometry to obtain protein-ligand binding constants. J Am Soc Mass Spectrom 2004, 15 (3), 388–97. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Hao, G. BC, Jones Lisa M., Vidavsky Ilan, and Gross Michael L., Fast Photochemical Oxidation of Proteins for Comparing Structures of Protein-Ligand Complexes: The Calmodulin-Peptide Model System. Anal. Chem 2011, 83, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Y; Lee H-W; Yang W; Shealy S; Yang JJ, Probing Site-Specific Calmodulin Calcium and Lanthanide Affinity by Grafting. Journal of the American Chemical Society 2005, 127 (11), 3743–3750. [DOI] [PubMed] [Google Scholar]
- 36.Weis DD, Hydrogen Exchange Mass Spectrometry of Proteins: Fundamentals, Methods, and Applications. Wiley: 2016. [Google Scholar]
- 37.Zhang B; Rempel DL; Gross ML, Protein Footprinting by Carbenes on a Fast Photochemical Oxidation of Proteins (FPOP) Platform. Journal of The American Society for Mass Spectrometry 2016, 27 (3), 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng M; Zhang B; Cui W; Gross ML, Laser-Initiated Radical Trifluoromethylation of Peptides and Proteins: Application to Mass-Spectrometry-Based Protein Footprinting. Angewandte Chemie International Edition 2017, 56 (45), 14007–14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang MM; Rempel DL; Gross ML, A Fast Photochemical Oxidation of Proteins (FPOP) platform for free-radical reactions: the carbonate radical anion with peptides and proteins. Free Radical Biology and Medicine 2019, 131, 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkman BF; Lipson D; Wemmer DE; Kern D, Two-State Allosteric Behavior in a Single-Domain Signaling Protein. Science 2001, 291 (5512), 2429–2433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.