Abstract
Budding yeast is an excellent model organism for studying the dynamics of the Golgi apparatus. To characterize Golgi function, it is important to visualize a secretory cargo as it traverses the secretory pathway. We describe a recently developed approach that generates fluorescent protein aggregates in the lumen of the yeast ER, and allows the fluorescent cargo to be solubilized for transport through the Golgi by addition of a small molecule ligand. We further describe how to generate a yeast strain expressing the regulatable secretory cargo, and we provide basic protocols for visualizing the cargo by 4D confocal microscopy and immunoblotting.
Keywords: yeast, Golgi, cisternal maturation, fluorescence microscopy, cargo
INTRODUCTION
The budding yeast Saccharomyces cerevisiae is ideally suited to analyzing the dynamic operation of the secretory pathway, for two reasons. First, the versatility of this experimental organism has generated a wide range of strains, plasmids, and methods for studying membrane traffic (Duden et al., 1997). Second, the S. cerevisiae Golgi is not stacked, so individual cisternae can be tracked in live cells by fluorescence microscopy (Wooding et al., 1998). These properties allowed for a test of two competing models for Golgi function. The stable compartments model predicted that Golgi compartments would be long-lived, whereas the cisternal maturation model predicted that early Golgi cisternae would undergo a biochemical transformation into late Golgi cisternae. 4D confocal microscopy of yeast Golgi cisternae provided definitive support for cisternal maturation (Losev et al., 2006; Matsuura-Tokita et al., 2006).
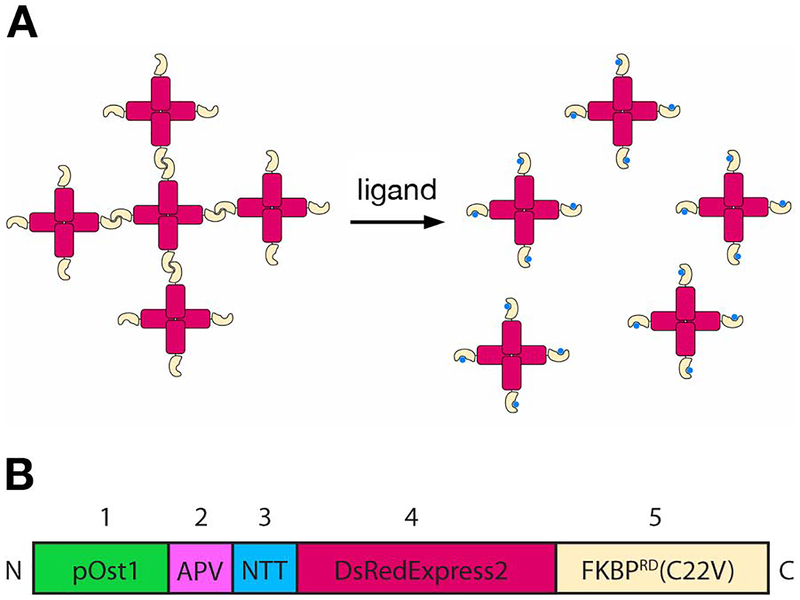
A deficient component of the S. cerevisiae toolbox has been secretory cargo proteins that could be visualized by microscopy during transport. Ideally, a cohort of fluorescent secretory cargo molecules would be trapped in the ER until a stimulus released them for synchronous transport through the secretory pathway. For mammalian cells, several methods have been developed to trap secretory cargo proteins reversibly in the ER (Boncompain et al., 2013). We adapted one such method for yeast. In the original approach, a GFP-tagged monomeric secretory cargo protein was fused to four copies of a dimerizing mutant of the FK506-binding protein FKBP to generate aggregates in the ER lumen (Rivera et al., 2000). Addition of a ligand dissolved the aggregates by blocking FKBP dimerization, thereby allowing the solubilized monomers to exit the ER. As shown in Figure 1, we modified this approach by fusing the tetrameric red fluorescent protein DsRed-Express2 (Strack et al., 2008) to a single copy of a reversibly dimerizing mutant of FKBP. Our construct forms ER-localized aggregates, which can be dissolved by addition of a ligand to create soluble tetramers that proceed through the secretory pathway. Although this concept is simple, the implementation required significant engineering of the cargo and the yeast strain (Barrero et al., 2016; Casler et al., 2018).
Figure 1. A reversibly aggregating fluorescent secretory cargo.
(A) Strategy for generating and dissolving fluorescent aggregates. DsRed-Express2 tetramers (red) fused to a dimerizing variant of FKBP (gold) associate with one another to form aggregates. Addition of the FKBP ligand SLF (blue) blocks dimerization, thereby dissolving the aggregates into soluble tetramers. (B) Schematic of the functional segments of the regulatable secretory cargo polypeptide. (1) pOst1 (green): ER signal sequence that directs cotranslational translocation. (2) APV (pink): ER export signal tripeptide. (3) NTT (blue): N-linked glycosylation signal tripeptide. (4) DsRed-Express2 (red): tetrameric fluorescent protein. (5) FKBPRD(C22V) (gold): reversibly dimerizing variant of FKBP. The lengths of the different segments are not to scale.
Here we describe protocols for generating a drug-sensitive yeast strain that expresses the regulatable secretory cargo (Basic Protocol 1), for visualizing the secretory cargo in yeast cells by microscopy (Basic Protocol 2), and for detecting the secreted cargo by immunoblotting (Basic Protocol 3).
Strategic Planning
The red fluorescent aggregates generated by our construct can be dissolved by addition of a synthetic ligand of FKBP (SLF) (Holt et al., 1993) to create soluble tetramers. Optimization of the FKBP mutant was needed because the original reversibly dimerizing FKBP generated aggregates that dissolved slowly and required high concentrations of SLF. Therefore, we rationally engineered a reversibly dimerizing FKBP mutant to form aggregates that dissolve faster and at lower SLF concentrations (Barrero et al., 2016). A further concern was that position 22 of FKBP was a cysteine, which could cause unwanted disulfide formation in the ER lumen. The corresponding residue in some FKBP homologs is valine (Galat, 2008), so we introduced a C22V mutation that does not alter the formation or dissolution of aggregates (Casler et al., 2018). We term the final reversibly dimerizing FKBP mutant FKBPRD(C22V), and the fusion construct DsRed-Express2-FKBPRD(C22V).
While optimizing the FKBP mutant, we discovered that yeast are adept at removing SLF via the action of pleiotropic drug transporters (Rogers et al., 2001). Removal of SLF causes reaggregation of the cargo. Deletion of the single drug transporter Pdr5 reduced but did not abrogate the removal of SLF. A much more successful approach was to delete Pdr1 and Pdr3, which are transcription factors that control the expression of many pleiotropic drug transporters (Coorey et al., 2015; Schüller et al., 2007). In a pdr1Δ pdr3Δ background, addition of SLF allows for sustained dissolution of the aggregates (Barrero et al., 2016).
The next challenge was to optimize entry into and exit from the ER. Many soluble yeast secretory cargo proteins undergo posttranslational translocation (Ast et al., 2013; Ng et al., 1996), but it was essential to direct DsRed-Express2-FKBPRD(C22V) for cotranslational translocation to ensure its passage across the ER membrane. We appended the Ost1 signal sequence, which efficiently directs cotranslational translocation (Fitzgerald et al., 2014). Addition of this signal sequence generates ER-localized DsRed-Express2-FKBPRD(C22V) tetramers that form aggregates (see the red channel in Movie 1 and Figure 2). Upon addition of SLF, the aggregates quickly dissolve, and the solubilized tetramers fill the ER lumen (Casler et al., 2018).
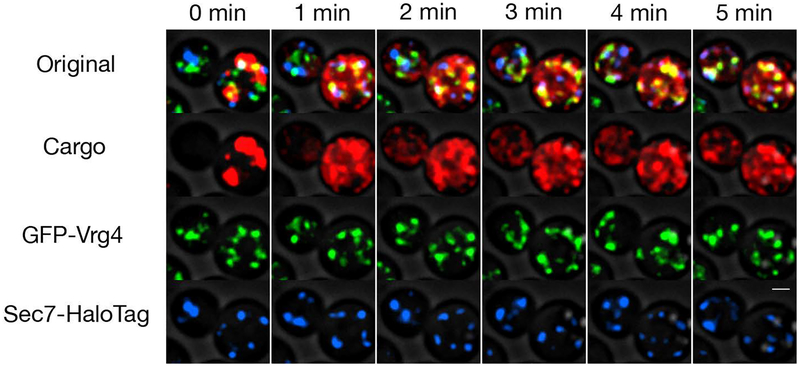
Figure 2. Visualizing the secretory cargo together with Golgi markers.
A strain expressing pOst1-APVNTT-DsRed-Express2-FKBPRD(C22V), GFP-Vrg4 (early Golgi marker), and Sec7-HaloTag (late Golgi marker) was grown to mid-log phase in NSD, labeled with the HaloTag-JF646 dye (Grimm et al., 2015), washed, mounted in a flow chamber, and imaged by 4D confocal microscopy while SLF was flowed over the cells. Images, taken from Movie 1, are average projected Z-stacks at the indicated time points. Scale bar, 2 μm.
Unfortunately, the solubilized tetramers exit the ER at the slow rate of bulk flow (Barlowe et al., 2016). To circumvent this problem, we inserted the first six residues of the yeast α-factor pro region after the Ost1 signal sequence. That hexapeptide sequence is APVNTT, and it contains both the evolutionarily conserved ER export signal APV (Yun et al., 2018), which interacts with the cargo receptor Erv29 (Barlowe et al., 2013), and the N-linked glycosylation site NTT (Kornfeld et al., 1985). Addition of the hexapeptide to generate APVNTT-DsRed-Express2-FKBPRD(C22V) tetramers (Figure 1B) preserves the formation of aggregates in the ER lumen and allows rapid signal-mediated ER export upon addition of SLF. As shown in Figure 1B, the complete construct consists of the Ost1 signal sequence, the APVNTT hexapeptide, DsRed-Express2, and FKBPRD(C22V).
Another concern was that a significant fraction of the cargo molecules were diverted to the vacuole. This problem can be alleviated by introducing the vps10–104 allele, which encodes a Vps10 protein that lacks one of the two sortilin homology domains (Jørgensen et al., 1999). The Vps10–104 mutant receptor continues to recognize endogenous vacuolar proteases but no longer recognizes foreign secretory cargo proteins (Fitzgerald et al., 2014; Jørgensen et al., 1999). Our results also indicate that for unknown reasons, glycosylation of the secretory cargo further suppresses diversion to the vacuole (Casler et al., 2018). Thus, in a vps10–104 strain, the glycosylated APVNTT-DsRed-Express2-FKBPRD(C22V) tetramers efficiently traverse the secretory pathway to the plasma membrane.
BASIC PROTOCOL 1
GENERATING A DRUG-SENSITIVE YEAST STRAIN THAT EXPRESSES THE REGULATABLE SECRETORY CARGO
In this protocol we describe the construction of a pdr1Δ pdr3Δ vps10–104 yeast strain suitable for tracking the regulatable fluorescent secretory cargo. The two gene deletions can be generated by standard PCR-based recombination techniques using G418-resistance and nourseothricin-resistance cassettes (Goldstein et al., 1999; Wach et al., 1994), and suitable primers are listed in Table 1. The vps10–104 allele can be generated using pop-in/pop-out gene replacement with the plasmid vps10–104-YIplac211. Expression of the secretory cargo is driven by the strong constitutive TPI1 promoter and the CYC1 terminator, and is achieved by integrating an expression vector at either the TRP1 or LEU2 locus using the plasmid YIplac204-pOst1-APVNTT-DsRed-Express2-FKBP(LV,C22V) or YIplac128-pOst1-APVNTT-DsRed-Express2-FKBP(LV,C22V), respectively (Table 1). Single-copy integrants are verified by PCR.
Table 1.
Primers, plasmids, and yeast strains.
| Purpose | Primers |
|---|---|
| PDR1 deletion | 5’-CAGCCAAGAATATACAGAAAAGAATCCAAGAAACTGGAAGCGTACGCTGCAGGTCGAC-3’ 5’-GGAAGTTTTTGAGAACTTTTATCTATACAAACGTATACGTATCGATGAATTCGAGCTCG-3’ |
| PDR3 deletion | 5’-ATCAGCAGTTTTATTAATTTTTTCTTATTGCGTGACCGCACGTACGCTGCAGGTCGAC-3’ 5’-TACTATGGTTATGCTCTGCTTCCCTATTTTCTTTGCGTTTATCGATGAATTCGAGCTCG-3’ |
| pdr1Δ verification | 5’-CCTAATGAGTGGCAATAAGAGGCGC-3’ 5’-CACTCGCATCAACCAAACCGTTATTC −3’ |
| pdr3Δ verification | 5’-CCGTGCTCTGCTGCTTTCAG-3’ 5’-GTGAACCCCATCCGCCGGTA-3’ |
| vps10–104 verification | 5’-CTCCCGTGGGCTTATGCGTTGCAG-3’ 5’-GGAGGGATTCGTCGGTGACTGTGTC-3’ |
| TRP1 integration | 5’-GTGTACTTTGCAGTTATGACGCCAGATGG-3’ 5’-AGTCAACCCCCTGCGATGTATATTTTCCTG-3’ |
| LEU2 integration | 5’-GGAGGTCGACTACGTCGTTAAGGC-3’ 5’-GTCCTGTACTTCCTTGTTCATGTGTG-3’ |
| Plasmid | Marker | Addgene # | Notes |
|---|---|---|---|
| YIplac204-pOst1-APVNTT-DsRed-Express2-FKBP(LV,C22V) | TRP1 | 115420 | Express cargo from a cassette integrated at TRP1 |
| YIplac128-pOst1-APVNTT-DsRed-Express2-FKBP(LV,C22V) | LEU2 | 115421 | Express cargo from a cassette integrated at LEU2 |
| vps10–104-YIplac211 | URA3 | 115426 | Create vps10–104 allele by gene replacement |
| Yeast Strain | Genotype |
|---|---|
| yAG56 | pdr1Δ pdr3Δ vps10–104 |
| yAK7 | pdr1Δ pdr3Δ vps10–104 trp1::PTPI1-pOst1-DsRed-Express2-FKBPRD(C22V) |
| yAL45 | pdr1Δ pdr3Δ vps10–104 leu2::PTPI1-pOst1-DsRed-Express2-FKBPRD(C22V) |
Materials
SD selection plates (see recipe)
YPD selection plates (see recipe)
Reagents and materials for yeast PEG/lithium acetate transformation (Gietz et al., 2002)
Thermal cycler
Herculase or Herculase II polyermase (Agilent, product numbers 600264, 600675)
Genomic DNA purification kit (Epicentre, product number MPY80200)
Generating a pdr1Δ pdr3Δ Strain
-
To delete PDR1, amplify a G418-resistance cassette from plasmid pFA6a-kanMX6 by PCR using the primers listed in Table 1 and the protocols listed in Table 2. Isolate the amplified 1583-bp fragment from an agarose gel, and transform it into a yeast strain of choice using PEG/lithium acetate.
The pFA6a-kanMX6 plasmid is available from Addgene (number 39296). When transforming yeast cells to obtain antibiotic resistance, spread the cells evenly on the plate because otherwise clumps of cells will generate pseudo-colonies. -
Select for transformants on YPD + G418 plates at 30°C for 2–3 days. Restreak colonies on fresh YPD + G418 plates to ensure that the clones are truly drug-resistant.
To allow expression of the drug resistance gene, we recommend incubating the transformed yeast cells in YPD for at least 4 hours prior to spreading on the selective plates. Confirm deletion of PDR1 by colony PCR (Amberg et al., 2006) using the primers listed in Table 1. The wild-type PDR1 allele will not yield an amplified fragment, whereas the pdr1Δ allele will yield an amplified fragment of 1155 bp.
-
Use a similar procedure to delete PDR3. In this case, amplification of the nourseothricin-resistance cassette from pAG25 will generate a 1349-bp fragment, and transformants will be selected on YPD + nourseothricin plates. After colony PCR, the wild-type PDR3 allele will not yield an amplified fragment, whereas the pdr3Δ allele will yield an amplified fragment of 1240 bp.
The pAG25 plasmid is available from Addgene (number 35121). Both of the drug resistance cassettes use the same promoter and terminator. Therefore, it is crucial when generating the second deletion to select on YPD plates that contain both nourseothricin and G418, to avoid selecting clones that have recombined at the previously deleted gene.
Table 2.
PCR reaction conditions.
| Herculase Reaction | |
|---|---|
| Water | 37.5 μL |
| Buffer | 5 μL |
| DNA | 1.5 μL (~150 ng) |
| Primer Forward | 1 μL (10 μM stock) |
| Primer Reverse | 1 μL (10 μM stock) |
| dNTPS | 2 μL (10 mM stock) |
| Polymerase | 2 μL |
| Herculase Thermocycler Conditions | ||
|---|---|---|
| Temperature | Time | Repeats |
| 92°C | 2 min | 1 |
| 92°C | 30 s | 30 |
| 55°C | 30 s | |
| 68°C | 10 min | |
| 68°C | 15 min | 1 |
| 4°C | Indefinite | Indefinite |
| Herculase II Reaction | |
|---|---|
| Water | 30.5 μL |
| Buffer | 10 μL |
| DNA | 1.5 μL (~150 ng) |
| Primer Forward | 2.5 μL (10 μM stock) |
| Primer Reverse | 2.5 μL (10 μM stock) |
| dNTPS | 2 μL (10 mM stock) |
| Polymerase | 1 μL |
| Herculase II Thermocycler Conditions | ||
|---|---|---|
| Temperature | Time | Repeats |
| 92°C | 2 min | 1 |
| 92°C | 30 s | 10 |
| 55°C | 30 s | |
| 68°C | 11 min | |
| 92°C | 30 s | 20 |
| 55°C | 30 s | |
| 68°C | 11 min + 20 s/cycle | |
| 68°C | 12 min | 1 |
| 4°C | Indefinite | Indefinite |
Introducing the vps10–104 Allele by Pop-in/Pop-out Gene Replacement
-
Linearize plasmid vps10–104-YIplac211 with HpaI to direct integration at the VPS10 locus. To linearize the plasmid, digest it thoroughly with HpaI, then clean the DNA with a commercial kit.
The vps10–104-YIplac211 plasmid, which contains the URA3 selectable marker, is available from Addgene (number 115426). This plasmid is suitable for pop-in/pop-out gene replacement (Rothstein, 1991). In this procedure, integration at the VPS10 locus generates adjacent wild-type and mutant alleles. Then counterselection with 5-fluoroorotic acid (5-FOA) selects for recombinants that have excised the URA3 gene. Recombination will either regenerate the wild-type VPS10 allele or cause a clean replacement of the wild-type allele with the mutant vps10–104 allele. Multiple pop-out clones need to be screened to ensure identification of a vps10–104 strain. Using PEG/lithium acetate, transform the linearized DNA (~250 ng total) into a ura3 auxotrophic yeast strain carrying the pdr1Δ and pdr3Δ alleles.
-
Select for transformants on SD – Ura dropout plates at 30°C for 2–3 days. Restreak the colonies on fresh SD – Ura plates, and then work with single colonies from those plates.
With dropout plates, only cells that have integrated the URA3-containing linearized plasmid will be able to grow and generate colonies. -
Confirm successful pop-in integration by colony PCR (Amberg et al., 2006) with the primers listed in Table 1.
Wild-type band = 3.2 kb
Mutant band = 1.1 kb
-
Grow a culture of a confirmed pop-in strain in YPD overnight. We recommend working with two or three separate pop-in clones to increase the chance of obtaining successful pop-out clones.
In theory, the pop-in clones should all be genetically identical. In practice, yeast transformation sometimes causes unexpected genomic rearrangements that elude detection by PCR, so it is good practice to work with several candidate clones. -
Spread an aliquot from each pop-in clone culture on an SD + 5-FOA plate. Incubate at 30°C for 2–3 days until colonies appear.
Plating ~50 μL of a saturated culture tends to give a good colony density, but this amount may need to be determined empirically.
Restreak the pop-out clones on fresh YPD plates, and then work with single colonies from those plates.
-
Confirm the presence of the mutant vps10–104 allele by the same colony PCR protocol described in Step 4, using primers listed in Table 1.
In our hands, out of 24 pop-out clones, 6 had the mutant allele.
Generating and Confirming Single-Copy Integrants of a Cargo Expression Construct
-
Linearize plasmid YIplac204-pOst1-APVNTT-DsRed-Express2-FKBP(LV,C22V) with EcoRV or plasmid YIplac128-pOst1-APVNTT-DsRed-Express2-FKBP(LV,C22V) with ClaI, to direct integration at the TRP1 or LEU2 locus, respectively.
These plasmids are available from Addgene (numbers 115420 and 115421). To linearize a plasmid, digest it extensively with the appropriate enzyme, then clean the DNA with a commercial kit. -
Transform the linearized DNA into a pdr1Δ pdr3Δ vps10–104 yeast strain using PEG/lithium acetate.
We recommend transforming with ~100–150 ng of linearized DNA. Higher amounts of DNA increase the chance of multiple integrations. -
Select for transformants on SD – Trp or SD – Leu dropout plates at 30°C for 2–3 days.
With dropout plates, only cells that have integrated the TRP1- or LEU2-containing linearized plasmid will be able to grow and generate colonies. Restreak 8–16 clones on fresh YPD plates.
Work with individual colonies to confirm expression of the construct by fluorescence microscopy. For this purpose, use a sterile toothpick to suspend a small amount of cells from a colony in 1–2 μL of water on a glass slide. Then spread the cells by overlaying a 22×22 mm No. 1.5 glass coverslip, and examine the cells by widefield microscopy for the presence of red fluorescent aggregates.
-
For fluorescent clones, isolate high purity genomic DNA.
We typically use the MasterPure Yeast DNA Purification Kit (Epicentre, product number MPY80200).
-
Verify single-copy integrants by PCR using the primers listed in Table 1 and the protocols listed in Table 2.
Some of the transformants will likely have experienced multiple tandem integrations of the linearized plasmid. Single-copy integrants are more genetically stable, and they will generate reliable results that allow for direct comparisons between different strains.
BASIC PROTOCOL 2
VISUALIZING THE SECRETORY CARGO IN YEAST CELLS BY MICROSCOPY
This protocol describes a method to track the fluorescent secretory cargo by 4D confocal microscopy. Cells are immobilized on a coverslip dish that contains liquid medium, and the dish is viewed with an inverted microscope. An example of cargo solubilization and transport to Golgi compartments is shown in Figure 2 and Movie 1. In addition, we describe a fluorescence recovery after photobleaching (FRAP) procedure that detects secretory cargo transfer between cisternae during Golgi maturation (Casler et al., 2018).
When cells are grown in minimal medium to facilitate fluorescence microscopy, the secreted cargo becomes trapped between the plasma membrane and the cell wall (Casler et al., 2018). The resulting periplasmic fluorescence can complicate image analysis. To avoid this effect, imaging can be performed in minimal medium at a pH of approximately 4 to quench the fluorescence of any extracellular DsRed-Express2 molecules, which have a chromophore pKa of ~4.5 (Strack et al., 2009).
Materials
50-mL baffled flask, sterilized by autoclaving
Shaker
NSD minimal medium (Bevis et al., 2002), pH approximately 4
Concanavalin A (Sigma-Aldrich, product number C2010)
MatTek P35G-1.5–14-C coverslip dishes (Thermo Fisher, product number NC0625009)
SLF solution, 100 mM in ethanol (Cayman Chemical, product number 10007974)
Leica SP8 or comparable confocal microscope with inverted optics
Nocodazole (Thermo Fisher, product number AC358240500)
Preparing Concanavalin A-Coated Coverslip Dishes
-
Add 250 μL of a 2 mg/mL solution of concanavalin A in deionized water to a coverslip dish.
a. Aliquots of the concanavalin A solution can be frozen in liquid nitrogen, stored at −80°C, and thawed just before use.
Incubate at ~23°C for 10–15 minutes to allow the concanavalin A to bind to the glass.
-
Wash gently with deionized water, and allow the glass surface to dry prior to adhering cells. It is best to use coverslip dishes that have been coated with concanavalin A the same day.
Concanavalin A will bind carbohydrates in the yeast cell wall, thereby gently immobilizing the cells on the coverslip dish.
Preparing Cells for 4D Confocal Microscopy
-
Inoculate a fresh culture of the yeast strain in 5 mL NSD in a 50-mL baffled flask. Allow the culture to grow overnight with shaking at 225 rpm at ~23°C.
A saturated preculture of the yeast strain can be stored in a 15-mL culture tube at 4°C for up to a month. To initiate a fresh culture, dilute a small aliquot of the preculture in NSD. A typical dilution for overnight growth is 1:1000. -
Verify that the OD600 is between 0.5–0.8. If it is too high, dilute the culture in fresh NSD to an OD600 of ~0.2, and let the culture grow for at least 2 more hours.
The OD600 is the optical density at 600 nm. It can be measured with a standard spectrophotometer using a disposable plastic cuvette. Add 250 μL of the culture to a concanavalin A-coated coverslip dish. Allow the cells to settle and adhere for 10 minutes, then gently wash with NSD. Add 1 mL of NSD to cover the cells.
Prepare NSD supplemented with 200 μM SLF by adding 2 μL of the 100 mM SLF stock solution to 1 mL NSD and mixing.
Place the dish on the microscope stage. Set the microscope parameters as described below, and adjust the objective position so that cells immobilized on the coverslip are in focus.
-
Remove the dish from the microscope stage. Add 1 mL of the NSD + SLF medium to the dish to yield a final concentration of 100 μM SLF, and mix gently by swirling. Immediately replace the dish on the microscope stage, and identify the cells that will be imaged.
Cargo aggregates will begin to dissolve immediately and will be fully dissolved within ~2 minutes.
Appearance of the solubilized cargo in early and late Golgi compartments should be maximal at ~2 minutes and ~4 minutes after SLF addition, respectively.
4D Confocal Microscopy
The following parameters are suitable for a Leica SP8 with a 1.4-NA oil objective. Other confocal microscopes should have similar settings.
-
Choose a frame size of 256×128 pixels.
The scan time is strongly influenced by the frame size. A 256×128 image should be sufficient to capture several cells. -
Zoom to a pixel size of ~70–80 nm.
This pixel size results in imaging at the Nyquist limit, thereby minimizing light exposure while theoretically avoiding information loss. Choose the maximum scan speed. Enable bidirectional scanning for rapid imaging.
-
Adjust the pinhole to 1.2 Airy units.
The standard recommendation for confocal microscopy is to use 1.0 Airy unit. But empirically, when imaging live cells, a setting of 1.2 Airy units yields significantly stronger signals with no discernible loss in image quality. Use line accumulation of at least 2 (preferably 3 or 4 if the total scan time is under 3 seconds).
-
Set the Z-stack to ~20–30 steps at 0.25–0.35 μm/step, to cover the entire cell with extra optical sections above and below.
This step size results in imaging at the Nyquist limit, thereby minimizing light exposure while theoretically avoiding information loss. To avoid excessive scan times and photobleaching, keep the number of optical slices above and below the cells to the minimum needed to ensure full capture of the fluorescence signals. Aim for the total time to capture a Z-stack of between 1–3 seconds.
-
With the laser set to 561 nm, a range of 4–7% laser intensity typically provides an adequate, non-saturated signal.
Even though the regulatable fluorescent secretory cargo is expressed from an integrated construct that does not vary in copy number, heterogeneity between cells in the sizes of aggregates may require adjustment of the laser intensity on a cell-by-cell basis.
-
Perform deconvolution and bleach correction to compensate for the inherently noisy nature of 4D confocal microcopy data (Day et al., 2016).
During imaging, individual pixels may accumulate only one or a few photons, so structures will be difficult to see in the raw optical sections. Capturing data in this manner can be disconcerting. However, after deconvolution and projection of the Z-stacks, structures should be readily visible and trackable through time.
FRAP Experiments with Nocodazole-Treated Cells
Inoculate a fresh culture of the yeast strain in 5 mL NSD in a 50-mL baffled flask. Allow the culture to grow overnight with shaking at 225 rpm at ~23°C.
Verify that the OD600 is between 0.5–0.8. If it is too high, dilute the culture in fresh NSD to an OD600 of ~0.2, and and let it grow for at least 2 more hours.
-
Add nocodazole to 8 μg/mL from a 1000× stock solution in DMSO, and incubate with shaking for 2 hours.
Nocodazole depolymerizes microtubules, causing a yeast cell to arrest in mitosis with a large budded daughter that lacks a nucleus. After 2 hours, nearly every cell in the culture is arrested at this point in the cell cycle, but cellular function is not yet significantly compromised. The available evidence indicates that operation of the yeast secretory pathway is normal under these conditions (Casler et al., 2018). Prepare cells for 4D confocal microscopy as described above, except that the cells should be washed and covered with NSD supplemented with 8 μg/mL nocodazole.
Add 1 mL NSD + 200 μM SLF + 8 μg/mL nocodazole to the coverslip dish to yield a final concentration of 100 μM SLF, and mix gently.
-
Identify budded daughters by lack of perinuclear ER fluorescence in the cargo channel. This identification can be performed within ~2 minutes of adding SLF.
The nuclear envelope comprises a large fraction of the ER in yeast cells. Because a budded daughter that lacks a nucleus has much less ER than an unperturbed yeast cell, imaging of Golgi dynamics in a budded daughter is relatively easy. Draw a region of interest (ROI) around the entire budded daughter.
Increase the laser power to 100% (with a background of 5% outside the ROI), and bleach by taking about 20 Z-stacks over a total period of 40 seconds.
Drop the laser power to 4–7% and begin imaging normally. Look for recovery of cargo fluorescence in Golgi cisternae in the bleached daughter. Any recovered fluorescence is due to fluorescent cargo molecules that were originally present in the unbleached mother.
BASIC PROTOCOL 3
DETECTING THE SECRETED CARGO BY IMMUNOBLOTTING
This protocol describes a method to track secretion of the cargo by immunoblotting. To enable release of the cargo from the periplasm, the cells are grown in rich YPD medium to yield a more permeable cell wall (Casler et al., 2018; de Nobel et al., 1990). The secreted cargo is hyperglycosylated, and is therefore treated with endoglycosidase H to remove the oligosaccharide and generate a sharp band when analyzed by SDS-PAGE (Casler et al., 2018).
Materials
Protein Precipitation Kit (National Diagnostics, product number EC-888)
Endoglycosidase H (New England Biolabs, product number P0702S)
0.5-mm diameter glass beads (BioSpec Products)
Rich glucose medium (YPD)
SLF solution, 100 mM in ethanol (Cayman Chemical, product number 10007974)
Phosphate-buffered saline (PBS)
Acetone
70% ethanol
SDS-PAGE sample buffer containing β-mercaptoethanol
Polyclonal rabbit anti-FKBP12 antibody (Abcam, product number ab2918)
Goat anti-rabbit secondary antibody conjugated to Alexa Fluor 647 (Thermo Fisher, product number A21245)
Cargo Secretion and Cell Lysis
-
Inoculate a fresh culture of the yeast strain in 5 mL YPD in a 50-mL baffled flask. Allow the culture to grow overnight with shaking at 225 rpm at ~23°C.
To detect secreted cargo, it is essential to grow the cells in rich medium. When cells are grown in minimal medium, most of the cargo becomes trapped in the periplasm.
Verify that the OD600 is between 0.5–0.8. If it is too high, dilute the culture in fresh YPD to an OD600 of ~0.2, and let it grow for at least 2 more hours.
Spin the sample at 3000 rpm for 5 minutes, and wash twice with fresh YPD.
-
Resuspend the cell pellet in the initial volume of YPD.
At this point the cargo is still in the form of ER-localized aggregates. Transfer of the cells to fresh medium has removed any cargo molecules that were secreted during growth of the culture.
Add SLF from the 100 mM stock solution to a final concentration of 100 μM. Continue to incubate the culture with shaking.
-
At each of the desired time points, transfer 1.6 mL of the culture to a snap-cap tube.
Secreted cargo molecules should begin to appear in the medium within about 10 minutes after SLF addition.
Spin at 5000 rpm for 2 minutes in a microcentrifuge, and separate the pellet (cells) and supernatant (secreted protein) fractions. Keep both samples on ice. The supernatant sample is ready for protein precipitation, and the pellet sample is further processed as described below.
Wash the cell pellet once with deionized water by resuspending the cells and then spinning at 5000 rpm for 2 minutes in a microcentrifuge.
Resuspend the cell pellet in 100 μL PBS, and add 100 μL of 0.5-mm diameter glass beads.
-
Vortex this sample for 1 minute followed by 1 minute on ice. Repeat twice.
This procedure mechanically disrupts the cells, thereby ensuring that the intracellular cargo molecules will be efficiently solubilized when the sample is eventually boiled in SDS-PAGE sample buffer.
Add 800 μL PBS to the lysed cell sample, mix briefly, and then transfer 800 μL of the liquid to a fresh snap-cap tube.
Protein Precipitation
-
Precipitate proteins using the National Diagnostics Protein Precipitation Kit, as follows.
In our hands, this kit is more effective than standard protein precipitation methods that employ trichloroacetic acid. Add 80 μL or 40 μL of Reagent A to the secreted protein fraction or the lysed cell fraction, respectively.
Add 160 μL or 80 μL of Reagent B to the secreted protein fraction or the lysed cell fraction, respectively.
Mix by inversion, and incubate at ~23°C for 15 minutes.
Collect the precipitated proteins by spinning at 12,000 rpm in a microcentrifuge for 10 minutes.
Discard the supernatants, and resuspend each of the pellets in 1 mL acetone. For resuspension, vortexing typically works, but pipetting may be necessary.
Spin at top speed in a microcentrifuge for 15 minutes. Discard the supernatant.
Wash each pellet twice with 70% ethanol.
Resuspend each final pellet in 50 μL SDS-PAGE sample buffer. Boil for 5–10 minutes.
Endoglycosidase H Treatment and Immunoblotting
The enzyme and buffers are from New England Biolabs.
-
Add glycoprotein denaturing buffer to 1× to the protein samples in SDS-PAGE sample buffer.
In our hands, 9 μL of the lysed cell fraction and 14 μL of the secreted protein fraction provide good signals.
Boil for 5–10 minutes to denature the proteins.
Add Glycobuffer 3 to 1×, together with 2 μL of endoglycosidase H and water to 20 μL.
Incubate at 37°C for at least 1 hour.
Subject the samples to standard SDS-PAGE and immunoblotting.
-
Detect the cargo using an anti-FKBP primary antibody together with a suitable secondary antibody. The protein band will have an apparent MW of approximately 38–39 kDa.
For unknown reasons, we were unable to detect the cargo using anti-DsRed antibodies.
REAGENTS AND SOLUTIONS
Complete Supplement Mixture (CSM)
CSM (Sunrise Science, product number 1001–100)
CSM – Ura (Sunrise Science, product number 1004–100)
CSM – Leu (Sunrise Science, product number 1005–100)
CSM – Trp (Sunrise Science, product number 1007–100)
Antibiotics
G418, Disulfate Salt (Teknova, product number G5001)
Nourseothricin Sulfate (Research Products International, product number N51200–1.0)
5-Fluoroorotic Acid (5-FOA) (Oakwood Products, product number 50204161)
Growth Media
SD Plates for Selection
-
For 1 liter:
Glucose: 20 g
Ammonium Sulfate: 5 g
CSM lacking a nutrient: 0.79 g
Yeast Nitrogen Base: 1.7 g
Agar: 20 g
Adjust the pH to 5.5 with NaOH. Sterilize by autoclaving. Let cool for ~30 minutes, and pour the plates.
SD + 5-FOA Plates for Counterselection
-
For 700 mL:
Glucose: 20 g
Ammonium Sulfate: 5 g
CSM: 0.79 g
Yeast Nitrogen Base: 1.7 g
Agar: 20 g
Adjust the pH to 5.5 with NaOH. Sterilize by autoclaving. Let cool for ~30 minutes to ~60°C. Meanwhile, dissolve 1 g 5-FOA in 300 mL H2O with stirring and gentle heating. Filter sterilize the 5-FOA solution, and add it slowly with stirring to the autoclaved and cooled SD, avoiding bubbles. Pour the plates.
YPD Plates for Growth and Selection
-
For 1 liter:
Glucose: 20 g
Peptone: 20 g
Yeast Extract: 10 g
Adenine Sulfate: 20 mg
Uracil: 20 mg
Sterilize by autoclaving. Let cool for ~30 minutes. If desired, add G418 to 200 μg/mL or nourseothricin to 100 μg/mL. Pour the plates.
NSD
-
For 1 liter:
Glucose: 20 g
Ammonium Sulfate: 5 g
CSM, or CSM lacking a nutrient: 0.79 g
Yeast Nitrogen Base: 1.7 g
500× Nonfluorescent Vitamin Mix: 2 mL
0.2 g/L Biotin: 2 mL
0.1 g/L CoCl2·6H2O: 2 mL
Adenine Sulfate: 20 mg
Adjust the pH with NaOH if desired. Without adjustment the pH is typically ~4. Filter sterilize, and store at room temperature.
Nonfluorescent Vitamin Mix (500×)
-
For 500 mL:
Calcium Pantothenate (stored at 4°C): 500 mg
Myo-inositol: 2.5 g
Niacin (Nicotinic Acid): 100 mg
p-Aminobenzoic Acid (stored at 4°C): 50 mg
Pyridoxine Hydrochloride: 100 mg
Thiamine Hydrochloride: 100 mg
Filter sterilize, and store at 4°C.
COMMENTARY
Background Information
Use of the regulatable secretory cargo requires a particular genetic background: the pdr1Δ pdr3Δ mutations ensure that SLF will be effective, and the vps10–104 mutation ensures that the cargo will avoid being diverted to the vacuole. With such a strain, cargo transport through the secretory pathway is reproducibly efficient, and this process can be readily tracked by fluorescence microscopy and immunoblotting (Casler et al., 2018).
After SLF addition, the ER-localized aggregates dissolve rapidly and the fluorescent tetramers begin to exit the ER. Cargo molecules almost immediately become concentrated in ER exit sites and nascent early Golgi cisternae, so the punctate pattern of ER-localized aggregates is replaced by a different punctate pattern (Figure 2). The ER export signal present in the cargo protein leads to a “cargo wave” as defined by the sequential appearance of red fluorescence in early and late Golgi cisternae. Golgi labeling persists until the cargo drains from the ER and is fully secreted.
Our method for tracking yeast secretion has limitations but also opens additional possibilities. The main limitation is that the cargo is artificial, and cannot easily be adapted to track natural or transmembrane cargoes. On the other hand, modification of the cargo with a vacuolar targeting signal should be feasible (Rothman et al., 1989), thereby enabling visualization of a fluorescent cargo as it transits through the Golgi and prevacuolar endosome to the vacuole. The use of fluorescent color variants of DsRed-Express2 (Strack et al., 2011) may allow plasma membrane- and vacuole-targeted cargoes to be tracked simultaneously in different color channels. We are actively exploring these options.
Below are summarized some of the critical parameters for this method as well as time considerations. Many of these points are also stated at the appropriate places in the protocols.
Critical Parameters
Because the secreted cargo becomes trapped in the periplasm when cells are grown in minimal medium, it is best to perform imaging in NSD medium at pH 4. This pH is below the chromophore pKa of DsRed-Express2 (~4.5), so the fluorescence signal in the periplasm is quenched while the intracellular signal is unaffected.
Although deletion of the Pdr1 and Pdr3 transcription factors dramatically inhibits the extrusion of SLF, a very small percentage of the cells may show some reaggregation of the cargo. This effect could be due to degradation or incomplete permeation of SLF. Any cells that show reaggregation should be excluded from the analysis.
Even with an integrated single-copy expression construct, there is always considerable heterogeneity between cells with regard to the sizes of the ER-localized aggregates. Thus, it may be necessary to adjust the 561-nm laser power for a given cell to obtain a stronger signal or to avoid saturating the detector.
Aggregation efficiency is temperature-sensitive (Casler et al., 2018). Significant aggregation in the ER is seen at room temperature, but the extent of aggregation progressively declines at higher temperature and is negligible at 37°C. Therefore, it may be difficult to use this regulatable secretory cargo in certain experiments that employ thermosensitive mutants.
An SLF concentration of 100 μM should be well above the threshold needed for complete dissolution of the aggregates (Barrero et al., 2016). This concentration of the drug does not seem to perturb yeast cells.
To avoid precipitation of SLF, we recommend allowing the SLF stock solution to warm to room temperature, and diluting it into room-temperature medium prior to adding the drug to cells. SLF can also be added directly to cell cultures.
To detect the secreted cargo by immunoblotting, the cells must be grown in rich medium. This requirement likely reflects higher permeability of the cell wall in rich medium than in minimal medium (de Nobel et al., 1990).
The secreted cargo is hyperglycosylated during transit through the Golgi. Therefore, to detect a clean band by immunoblotting, N-linked glycans must be removed enzymatically prior to SDS-PAGE. Endoglycosidase H works well for this purpose.
For unknown reasons, we have been unable to detect the cargo construct with commercially available anti-DsRed antibodies from TaKaRa/Clontech. However, an anti-FKBP antibody works well.
Time Considerations
Generation of a cargo-expressing strain with the appropriate genetic background typically takes about 4–6 weeks.
After addition of SLF at room temperature, complete dissolution of the cargo aggregates in a cell takes about 1–2 minutes, depending on the size of the aggregates. The solubilized cargo fully populates early Golgi compartments within 2 minutes and late Golgi compartments within 3–4 minutes. Strong Golgi labeling persists for about 20 minutes, and secretion of the original pool of cargo is nearly complete in 30 minutes.
When cells are grown in rich medium at room temperature and then treated with SLF, secreted cargo in the medium can be detected by immunoblotting within 10 minutes.
Supplementary Material
Movie 1. Visualizing the solubilization and Golgi traffic of the secretory cargo. A strain expressing pOst1-APVNTT-DsRed-Express2-FKBPRD(C22V), GFP-Vrg4 (early Golgi marker), and Sec7-HaloTag (late Golgi marker) was grown to mid-log phase in NSD, labeled with the HaloTag-JF646 dye, washed, mounted in a flow chamber, and imaged by 4D confocal microscopy while SLF was flowed over the cells. Full Z-stacks were taken every 3 seconds for 10 minutes, and average projected to create the movie frames. The top panel shows the complete movie, while the bottom three panels show the individual fluorescence channels. The cargo aggregates dissolve, and then the solubilized cargo appears in early and then in late Golgi compartments. Scale bar, 2 μm.
Key References.
Casler et al., 2018. See above.
Barrero et al., 2016. See above.
These are key supporting references for this unit.
The first reference provides details about the construction, characterization, and usage of the regulatable secretory cargo and suitable yeast strains. The second reference describes the development of an improved reversibly dimerizing FKBP, and the characterization of yeast strains engineered to allow the use of SLF.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 GM104010. J.C.C. was supported by NIH training grant T32 GM007183. Thanks for assistance with fluorescence microscopy to Vytas Bindokas and Christine Labno at the Integrated Microscopy Core Facility, which is supported by the NIH-funded Cancer Center Support Grant P30 CA014599. Additional thanks to Luke Lavis for providing the JF646 dye.
REFERENCES
- Amberg DC, Burke DJ, and Strathern JN (2006). Yeast colony PCR. CSH Protoc. doi: 10.1101/pdb.prot4170 [DOI] [PubMed] [Google Scholar]
- Ast T, Cohen G, and Schuldiner M (2013). A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell, 152, 1134–1145. [DOI] [PubMed] [Google Scholar]
- Barlowe C, and Helenius A (2016). Cargo capture and bulk flow in the early secretory pathway. Annu. Rev. Cell Dev. Biol, 32, 197–222. [DOI] [PubMed] [Google Scholar]
- Barlowe CK, and Miller EA (2013). Secretory protein biogenesis and traffic in the early secretory pathway. Genetics, 193, 383–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JJ, Papanikou E, Casler JC, Day KJ, and Glick BS (2016). An improved reversibly dimerizing mutant of the FK506-binding protein FKBP. Cell. Logist, 6, e1204848. doi: 10.1080/21592799.2016.1204848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis BJ, Hammond AT, Reinke CA, and Glick BS (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol, 4(10), 750–756. [DOI] [PubMed] [Google Scholar]
- Boncompain G, and Perez F (2013). Fluorescence-based analysis of trafficking in mammalian cells. Methods Cell Biol, 118, 179–194. [DOI] [PubMed] [Google Scholar]
- Casler JC, Papanikou E, Barrero JJ, and Glick BS (2018). Visualization of secretory cargo transport in a maturing Golgi apparatus. Submitted. [Google Scholar]
- Coorey NVC, Matthews JH, Bellows DS, and Atkinson PH (2015). Pleiotropic drug-resistance attenuated genomic library improves elucidation of drug mechanisms. Mol. Biosyst, 11, 3129–3136. [DOI] [PubMed] [Google Scholar]
- Day KJ, Papanikou E, and Glick BS (2016). 4D confocal imaging of yeast organelles. Methods Mol. Biol, 1496, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel JG, Klis FM, Priem J, Munnik T, and Van Den Ende H (1990). An assay of relative cell wall porosity in Saccharomyces cerevisiae, Kluyveromyces lactis and Schizosaccharomyces pombe. Yeast, 6, 483–490. [DOI] [PubMed] [Google Scholar]
- Duden R, and Schekman R (1997). Insights into Golgi function through mutants in yeast and animal cells In Berger EG & Roth J (Eds.), The Golgi Apparatus (pp. 219–246). Basel: Birkhäuser Verlag. [Google Scholar]
- Fitzgerald I, and Glick BS (2014). Secretion of a foreign protein from budding yeasts is enhanced by cotranslational translocation and by suppression of vacuolar targeting. Microb. Cell Fact, 13, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A (2008). Functional drift of sequence attributes in the FK506-binding proteins (FKBPs). J. Chem. Inf. Model, 48, 1118–1130. [DOI] [PubMed] [Google Scholar]
- Gietz RD, and Woods RA (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol, 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, and McCusker JH (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast, 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, and Lavis LD (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods, 12(3), 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt DA, Luengo JI, Yamashita DS, Oh HJ, Konialian AL, Yen HK, Rozamus LW, Brandt M, Bossard MJ, Levy MA, Eggleston DS, Liang J, Schultz LW, Stout TJ, and Clardy J (1993). Design, synthesis, and kinetic evaluation of high-affinity FKBP ligands and the X-ray crystal structures of their complexes with FKBP12. J. Am. Chem. Soc, 115, 9925–9938. [Google Scholar]
- Jørgensen MU, Emr SD, and Winther JR (1999). Ligand recognition and domain structure of Vps10p, a vacuolar protein sorting receptor in Saccharomyces cerevisiae. Eur. J. Biochem, 260, 461–469. [DOI] [PubMed] [Google Scholar]
- Kornfeld R, and Kornfeld S (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem, 54, 631–664. [DOI] [PubMed] [Google Scholar]
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, and Glick BS (2006). Golgi maturation visualized in living yeast. Nature, 441(22 June), 1002–1006. [DOI] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, and Nakano A (2006). Live imaging of yeast Golgi cisternal maturation. Nature, 441(22 June), 1007–1010. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, and Walter P (1996). Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol, 134, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Wang X, Wardwell S, Courage NL, Volchuk A, Keenan T, Holt DA, Gilman M, Orci L, Cerasoli FJ, Rothman JE, and Clackson T (2000). Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science, 287, 826–830. [DOI] [PubMed] [Google Scholar]
- Rogers B, Decottignies A, Kolaczkowski M, Carvajal E, Balzi E, and Goffeau A (2001). The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol, 3, 207–214. [PubMed] [Google Scholar]
- Rothman JH, Yamashiro CT, Kane PM, and Stevens TH (1989). Protein targeting to the yeast vacuole. Trends Biochem. Sci, 14(8), 347–350. [DOI] [PubMed] [Google Scholar]
- Rothstein R (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol, 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Schüller C, Mamnun YM, Wolfger H, Rockwell N, Thorner J, and Kuchler K (2007). Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in Saccharomyces cerevisiae. Mol. Biol. Cell, 18, 4932–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack RL, Bhattacharyya D, Glick BS, and Keenan RJ (2009). Noncytotoxic orange and red/green derivatives of DsRed-Express2 for whole-cell labeling. BMC Biotechnol, 9, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack RL, Keenan RJ, and Glick BS (2011). Noncytotoxic DsRed derivatives for whole-cell labeling. Methods Mol. Biol, 699, 355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, and Glick BS (2008). A noncytotoxic DsRed variant for whole-cell labeling. Nat. Methods, 5, 955–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, and Philippsen P (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wooding S, and Pelham HRB (1998). The dynamics of Golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell, 9, 2667–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y, Garcia MR, Novak AJ, Saunders AM, Ank RS, Nam AS, and Fisher LW (2018). Surf4 (Erv29p) binds amino-terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol, 16(8), e2005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Visualizing the solubilization and Golgi traffic of the secretory cargo. A strain expressing pOst1-APVNTT-DsRed-Express2-FKBPRD(C22V), GFP-Vrg4 (early Golgi marker), and Sec7-HaloTag (late Golgi marker) was grown to mid-log phase in NSD, labeled with the HaloTag-JF646 dye, washed, mounted in a flow chamber, and imaged by 4D confocal microscopy while SLF was flowed over the cells. Full Z-stacks were taken every 3 seconds for 10 minutes, and average projected to create the movie frames. The top panel shows the complete movie, while the bottom three panels show the individual fluorescence channels. The cargo aggregates dissolve, and then the solubilized cargo appears in early and then in late Golgi compartments. Scale bar, 2 μm.