Abstract
Radiologists are expected to expediently communicate critical and unexpected findings to referring clinicians to prevent delayed diagnosis and treatment of patients. However, competing demands such as heavy workload along with lack of administrative support resulted in communication failures that accounted for 7% of the malpractice payments made from 2004 to 2008 in the United States. To address this problem, we have developed a novel machine learning method that can automatically and accurately identify cases that require prompt communication to referring physicians based on analyzing the associated radiology reports. This semi-supervised learning approach requires a minimal amount of manual annotations and was trained on a large multi-institutional radiology report repository from three major external healthcare organizations. To test our approach, we created a corpus of 480 radiology reports from our own institution and double-annotated cases that required prompt communication by two radiologists. Our evaluation on the test corpus achieved an F-score of 74.5% and recall of 90.0% in identifying cases for prompt communication. The implementation of the proposed approach as part of an online decision support system can assist radiologists in identifying radiological cases for prompt communication to referring physicians to avoid or minimize potential harm to patients.
Keywords: Semi-supervised learning, Distributional semantics, Cluster analysis, Radiology report, Radiologist prompt communication
1. Introduction
A major challenge in current radiology practice is ensuring the communication of critical or unexpected findings in radiology exams to referring physicians [1]. Radiologists are expected to communicate critical and unexpected findings to referring clinicians in a timely manner to expedite patient diagnosis and treatment. Both the American College of Radiology (ACR) and The Joint Commission (TJC) have published practice guidelines and requirements that mandate both timely communication and documentation of such communications (e.g., typically within the radiology report). Still, communication breakdowns between radiologists and referring physicians remain problematic as a result of heavy workload and the lack of an automated system to communicate significant clinical findings. In fact, the World Alliance for Patient Safety, a World Health Organization (WHO) initiative to improve patient safety, has identified the poor communication of medical exam results as a serious problem affecting patient care and increasing the risk of missed or delayed diagnoses worldwide [2]. Data from medical malpractice insurance companies show that in the United States, failure to communicate critical and abnormal radiological findings in a timely fashion is estimated to be the second most common cause of litigation against radiologists [3].
The goal of the work presented in this paper is to build an effective and accurate system to automatically identify patients with radiological findings that need to be promptly communicated to their referring physicians based on their corresponding radiology reports. Our Natural Language Processing (NLP) methodology aims to classify whether a patient’s radiology report requires such prompt communication based on analyzing the free-text narratives in the report. Thus, our method could potentially be utilized as an online decision support system for radiologists to ensure appropriate communication with referring physicians to improve patient health outcomes.
Medical records and clinical notes, such as radiology reports, are recognized as a rich but difficult-to-analyze source of medical information [4]. Radiology reports contain a great amount of information that characterizes a patient’s medical condition and radiological findings. However, this information is mostly in an unstructured format, taking the form of free text, and is therefore difficult to search, sort, analyze, summarize, and present [5]. The free-text format and the ambiguities and variations of natural language hinder the extraction of reusable information from radiology reports for research and clinical decision support [6].
While the specifics of an urgent communication policy vary by institution, some examples of critical radiology findings that require prompt communication are as follows: pneumothorax, pulmonary embolism, and ectopic pregnancy [7,8]. Since institutional processes vary with regard to communicating critical findings, there is no definitive, complete list of specific findings that require prompt communication. The free-text nature of the radiology reports and the indeterminate nature of which findings necessitate prompt communication lend themselves toward an automatic document classification solution using machine learning – creating a computational model from the body of reports in a training set labeled for prompt communication that can then be used to classify whether new reports require prompt communication. The labeling of radiology reports as to whether or not they need to be promptly communicated is a standard example of a documentation classification problem. In text classification, various syntactic and semantic features can be extracted as inputs for a machine learning framework to train a prediction model for a target outcome variable based on the document contents [9].
In the domain of biomedical informatics, text-mining and machine learning methods [10] have been developed and utilized to help researchers identify important clinical information from a medical narrative in a high throughput manner [11]. There are many examples of previous efforts to automatically analyze radiology reports and other medical records. For instance, Hobbs developed a system for information extraction based on domain-dependent patterns, mapping unstructured biomedical text to predefined structured templates [12]. MetaMap has been utilized to map radiology reports and other clinical note text to concepts from the Unified Medical Language System (UMLS) Metathesaurus [13]. In another work, an unsupervised machine learning approach has been built to group radiology reports from a large multi-institutional repository based on their contents in free-text narrative [6]. The clinical Text Analysis and Knowledge Extraction System (cTAKES), commonly used for information extraction from radiology reports and other medical records, combines rule-based and supervised machine learning techniques to analyze clinical free text[14]. Medical Language Extraction and Encoding System (MEDLEE) was developed to extract information from Columbia-Presbyterian Medical Center’s chest radiology report repository by using a controlled vocabulary and grammatical rules to translate text into a structured database format [15]. Finally, a supervised text classification system was developed to annotate and extract clinically significant information from free-text radiology reports [16].
In addition to supervised machine learning approaches for text classification, which require a large amount of hand-labeled data for model development and training, semi-supervised approaches for text classification utilize small, labeled datasets along with large, unlabeled datasets. Banerjee et al. [17] proposed a semi-supervised model that combines a neural embedding method with a semantic dictionary mapping technique, creating a dense vector representation of unstructured radiology reports, to classify free-text reports of pulmonary embolisms. Gupta et al. [18] proposed an unsupervised model to extract relations and their associated named entities, using automated clustering of similar relations in narrative mammography radiology reports. Wang et al. [19] developed a semi-supervised set covering machine to detect ovarian cancer and coronary angiogram related results. Chai et al. [20] developed a semi-supervised statistical text classification model to automatically identify health information technology incidents in the USA Food and Drug Administration (FDA) Manufacturer and User Facility Device Experience (MAUDE) database.
Machine learning techniques have become increasingly common for biomedical information extraction and clinical decision support systems due to their scalability and accuracy. Recent efforts to develop automated systems for biomedical information extraction and text processing have been undertaken, but little work has been done to identify the characteristics in clinical notes and reports to determine the need for prompt communication in the clinical workflow, particularly with a limited amount of annotations. To tackle this problem, we present a semi-supervised learning approach that relies on a small amount of labeled data (i.e., seed data) in conjunction with a large amount of unlabeled data to develop a machine learning system that can use the free-text content in radiology reports to identify the radiological cases that require prompt communication to referring physicians. Of note, none of the model training steps in our approach, including the development of the seed-labeled dataset, require manual data review and labeling by domain experts. We also tested the generalizability of the proposed approach across different healthcare organizations in our evaluation and we expect our methodology can be utilized to identify other characteristics of clinical notes by using a small amount of labeled data. The details of the proposed approach and its evaluation are presented in the rest of the paper.
2. Materials and methods
We can formalize the identification of cases that need to be promptly communicated by radiologists to referring physicians as a text classification problem, in which we classify a patient’s radiology report as positive or negative for “prompt communication” based on the corresponding radiology report contents. Generally, the need for prompt communication arises from a critical or unexpected diagnosis in a radiology finding, and it is an accepted practice that this communication should be documented [7,8]. A common practice is for the radiologist to document these non-routine, prompt communications using a pre-defined, free-text template within the radiology report, such as, “I < radiologist name > discussed these critical results with < referring physician name > on < date > at < time > and verified that (s)he understood these results,” but there is no widely-accepted standard for this template, nor is there uniform guidance on when a finding should be communicated. Thus, in practice, cases with urgent findings may not be communicated promptly to referring physicians or the communications may not be properly documented.
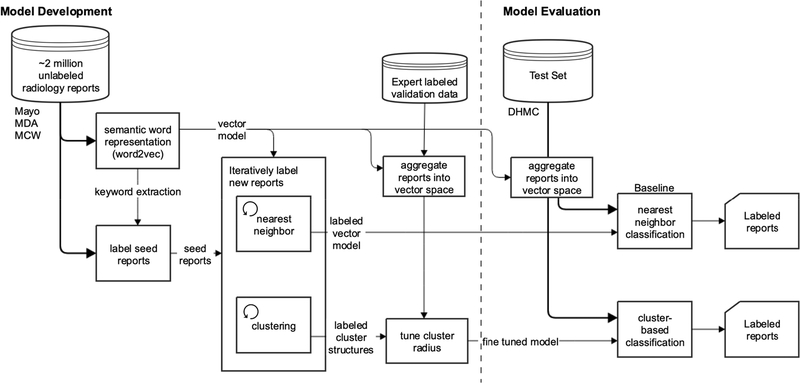
As mentioned in the Introduction section, generating labeled data to indicate whether or not findings in a radiology report need to be promptly communicated to a referring physician is the limiting factor in leveraging a supervised learning approach for this task. Therefore, in this paper, we propose a semi-supervised approach for solving this classification problem. Our approach uses unsupervised distributional representations [21] to extract reports we label as seed data, based initially on the common practice of radiologists documenting communication within a report. We then use the seed-labeled data and distributional representations to model a large dataset of free-text radiology reports. Two models are separately created using both nearest neighbor and k-means clustering [22] in iterative approaches to discover the structures around the seed-labeled data and these structures are then used to assign labels to unlabeled radiology reports. A smaller dataset (n = 180) of expert-labeled data is used to fine-tune the structure of our k-means cluster model. Both approaches (nearest neighbor and clustering-based methods) rely on an initial small set of labeled data (i.e., seed-labeled data), and both approaches iteratively expand the seed-labeled dataset to generate the final results. Therefore, both methods can be considered as semi-supervised approaches. The nearest neighbor model and cluster model structures were then each used to label an evaluation dataset. This retrospective study is approved by the Dartmouth Institutional Review Board (IRB). An overview of these approaches is shown in Fig. 1.
Fig. 1.
Overview of the proposed semi-supervised learning approach.
2.1. Datasets
Training Set:
We extracted free-text radiology reports from the RadCore radiology report repository [16] to train our semi-supervised model. RadCore, with about 2 million radiology reports, is a multi-institutional corpus of radiology reports aggregated in 2007 from three major health care organizations: Mayo Clinic (Mayo), MD Anderson Cancer Center (MDA), and Medical College of Wisconsin (MCW). All RadCore radiology reports have been de-identified by their source institutions. This dataset is described in full detail in our previous study [16].
Validation and Evaluation Sets:
We constructed two smaller, fully-labeled sets of radiology reports for fine-tuning our models (i.e., validation set) and for evaluation (i.e., test set). These radiology reports were extracted from our institution, Dartmouth-Hitchcock Medical Center (DHMC), a tertiary academic care center in Lebanon, New Hampshire. Two radiologists (YYC and RTS) manually annotated the reports to establish the ground truth labels for these reports. In this manual annotation, a binary label was assigned to an individual report to indicate whether a prompt communication was needed on the basis of critical or unexpected radiological findings in the report. The disagreements were resolved through further discussions between annotators in an adjudication process, and reports with commonly agreed labels from the two radiologists were used for validation and evaluation. In these datasets, we utilized a balanced mixture of three imaging modalities: (1) Computed Tomography (CT); (ii) Magnetic Resonance Imaging (MRI); and (iii) X-ray. Of note, for a rigorous validation and evaluation in this study, we programmatically removed the standard tags in reports in our dataset that were used at the DHMC Radiology Department to indicate unexpected findings and communications to referring physicians. In this process, we removed the word “unexpected” and the sentences for “discussed critical results” from the DHMC radiology reports to generalize our approach across reports that do not contain specific documentation of prompt communication. Through this process, 480 radiology reports that balanced across positive and negative cases for prompt communication, and that also balanced across all three modalities, were randomly selected as our final test set for evaluation. Additionally, a 180-report dataset was selected from the reports not chosen for the test set, balanced across positive and negative cases for prompt communication, and was used to validate and fine-tune our structures from k-means clustering.
2.2. Learning distributional semantics
To capture the semantics and variability of the textual information, we trained an unsupervised distributional semantics neural network on the entire corpus of radiology reports in RadCore. This neural network constructed a semantic vector representation for each existing word in the texts. This distributional semantics neural network, known as the word2vec model [21], relies on the linguistic principle that the meaning of a word (i.e., semantics) can be inferred based on surrounding words (i.e., context). Word2vec training takes a large corpus of text as its input and produces a vector space, typically consisting of several hundred dimensions, with each unique word in the corpus being assigned a corresponding vector in the space. The word2vec model considers word distribution in the surrounding context windows for each word in the corpus. Word vectors are computed in the vector space in such a way that words that share common contexts in the corpus are located in close proximity to one another in terms of vector space [21]. One of the most common measures of semantic similarity in NLP is the cosine similarity defined by the dot product between two vectors. Cosine similarity is an effective solution for measuring vector similarities in a high-dimensional space compared to other distance metrics because it does depend on the length of the vector. In particular, the word2vec neural network model is trained in an unsupervised fashion to maximize the cosine similarity between vector representations of words that have similar co-occurrence patterns. In this work, we tuned our parameters utilizing a grid search on the effect of different model architectures, such as continuous bag-of-words and skip-gram [21], semantic representation dimensions (from 200 to 500 with step of 100), and context window sizes (from 3 to 10), for training our model through cross-validation on the loss function to select the best model configuration for our application. The best word2vec model configuration in our application was the skip-gram architecture with a semantic representation dimension of 300 and a context window size of 8, which provides a tradeoff between the quality of the resulting word vectors and the computational complexity.
2.3. Extracting keywords to label seed data
To create a training set for our semi-supervised approaches, we label a small number of radiology reports as seed data by incorporating prior domain knowledge in regards to whether a report has been communicated. Through discussions with a domain expert (YYC), a board- certified academic radiologist with over 18 years of subspecialty practice, we recognized that if a radiology report explicitly contains the word “communicated”, the radiological findings were most likely communicated to a stakeholder, mainly to the referring physician and infrequently to the patient. Of note, the communication with the referring physician can happen either before or after the radiology report is finalized. If the communication occurs after the finalization of the report, the documentation of this communication usually is included in the report as an addendum. Therefore, radiology reports can contain the documentation of the radiologist’s communications regardless of the timing of the radiology reports.
Since these reports are free text, not all of the radiology reports that require prompt communication to referring physicians contain the word “communicated”, nor do all the reports that contain the word “communicated” require prompt communication. Due to variations in free-text reporting, there are many ways for radiologists to explicitly record their communication of critical or unexpected findings to referring physicians in radiology reports. In order to identify other possible keywords to build our seed-labeled dataset, we used distributional semantic representations to find other keywords highly similar to “communicated” in our training set corpus. The similarity to words in our corpus was calculated through cosine similarity [23]. Table 1 shows the top words in our corpus most similar to “communicated”. As shown in this table, the keywords most semantically similar to “communicated” according to our word2vec model are: (1) “relayed”, (2) “conveyed”, (3) “called”, (4) “phoned”, and (5) “discussed”. We discussed these words with our domain expert (YYC), who helped to verify the valid use of these words in radiology reports.
Table 1.
Words most similar to “communicated” according to our word2vec model.
| Word | Similarity score |
|---|---|
| “relayed” | 0.787 |
| “conveyed” | 0.775 |
| “called” | 0.670 |
| “phoned” | 0.616 |
| “discussed” | 0.596 |
To construct a seed-labeled dataset of cases requiring prompt communication to referring physicians, we extracted radiology reports from the RadCore repository that contained any of the six keywords identified for prompt communication under either the “findings” or “impression” section headings in these free-text reports. Focusing on these common sections in radiology reports narrows our attention to the current findings and filters out past or unrelated cases of communication.
In many cases, communications between radiologists and patients have nothing to do with critical findings. For example, the following report excerpt was typical communication between a radiologist and a patient: “The proposed procedure, comments, techniques, and possible complications were discussed in detail with the patient.” Also, radiologists may sometimes communicate their findings with other physicians on the patient’s care team, such as primary care providers, when the referring clinician is not available. Only communications between radiologists and referring physicians or other physicians on the patient’s care team are considered in our training. To remove likely false positive cases, where the keywords in radiology reports refer to communications with patients, we used a simple rule-based regular expression [24] approach to identify the co-occurrences of keywords communicated with “patient” in a sentence of the seed-labeled data. Through this approach, we excluded the reports with these “patient” co-occurrences to make sure the referred communications were performed with referring physicians instead of patients in our seed-labeled dataset.
This labeling of seed data resulted in 261 reports identified for prompt communication with referring physicians. Table 2 shows the distribution of the different keywords in the radiology reports of our labeled seed dataset. In this seed dataset, two cases contained two keywords each. Therefore, the total number of keyword occurrences in our dataset is 263. This seed-labeled dataset makes up only 0.013% of all the radiology reports in the repository. While the seed dataset contains cases in which the radiology reports contain explicit records of prompt communication with referring physicians, not all records with critical or unexpected findings, which require prompt communication, contain these keywords. Our proposed approaches aim at detecting cases in which the radiology reports do not explicitly include these keywords, but the communication is nonetheless necessary due to the nature of the radiological findings. As mentioned in the previous section, semi-supervised learning can be instrumental in this case where there is far more unlabeled data than labeled data for training a machine learning model [25]. In the rest of this section, we describe our approach that builds upon this seed-labeled data to identify the unlabeled cases that require the radiologist to have prompt communication with the referring physician.
Table 2.
Distribution of keywords in our extracted seed dataset.
| Keyword | # Keyword in the dataset | Percentage of seed dataset (261 reports) |
|---|---|---|
| “discussed” | 232 | 88.2% |
| “communicated” | 11 | 4.2% |
| “called” | 10 | 3.8% |
| “conveyed” | 5 | 1.9% |
| “relayed” | 4 | 1.5% |
| “phoned” | 1 | 0.4% |
| Total | 263 | 100% |
2.4. Radiology report representation
To develop our machine learning model, we require a mathematical representation of radiology reports. Among different sections of a radiology report, such as clinical history, indication, technique, findings, and impression [26], we focus on representing the text in the “impression” section. This is because the impression section contains information about diagnosis and follow-up recommendations. Therefore, the impression section is the most relevant part of a radiology report in identifying cases that require prompt communication to referring physicians. Of note, instead of only focusing on particular sentences in identifying critical recommendations [27], in this study, we aim to make use of all the text in the impression section.
To represent this text, we leverage the distributional semantics of the words to convert the free-text narrative to a vector. To do that, we retrieve the corresponding semantic vector representations for words in an impression section through our word2vec model. Subsequently, the word2vec semantic vector representations of these words are aggregated to reflect the appropriate semantic representation of the full impression section by using the following formula:
where Vimp is the semantic representation of the full impression section, w is each word in the impression section, Vw is the semantic representation of word w, Wimp is the set of words in the impression section, and ||Wimp|| is the number of words in the impression section. The effectiveness of this method for aggregating the vectors has been shown in previous work [18]. As discussed in Section 2.2, our best word2vec model was trained using the skip-gram architecture with a semantic representation dimension of 300 and a context window size of 8.
2.5. Nearest neighbor-based classification approach
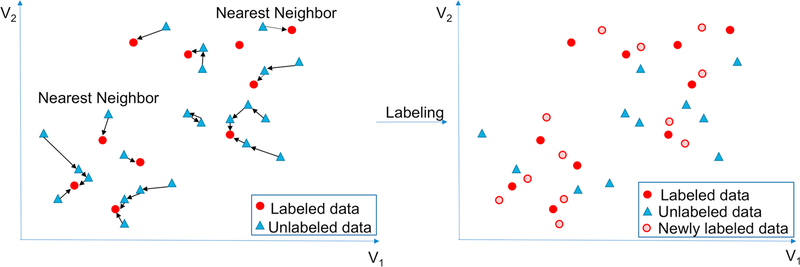
The nearest neighbor algorithm [28] is a simple but effective non- parametric method for classification and has been widely used in different applications [29]. The nearest neighbor algorithm uses Euclidean distance to calculate a similarity measure between data points; in our approach, this is the distance between the vectors representing the impression section of different reports. Hence, the algorithm classifies an unlabeled report by assigning it to the class of its most similar impression section vector. Fig. 2 shows a conceptual overview for the application of this baseline approach on a sample dataset. We used the nearest neighbor approach as a method on the vector representations of seed-labeled and unlabeled radiology reports to identify the new cases that require prompt communication with referring physicians. Newly labeled cases that require prompt communication are added back into the seed-labeled dataset. We iteratively labeled new cases based on the extended seed-labeled dataset until it stopped growing. The seed-labeled dataset became stable after the 7th iteration in our experiment. Including the 261 initial seed reports and 657 newly labeled reports, a total of 918 reports were labeled for prompt communication in the training set using the nearest neighbor baseline approach.
Fig. 2.
Conceptual overview of an iteration in the nearest neighbor approach in two-dimensional (2D) space (V1 and V2). For each unlabeled vector, we find the nearest neighboring vector (left); if the nearest neighbor is labeled (as a communicated report), we also label the unlabeled vector as communicated (right). Of note, the actual dimension of the vector space in our study is 300, and the 2D simplification is only for visualization purposes.
2.6. Clustering-based classification approach
2.6.1. Clustering seed-labeled data
Clustering algorithms are used in an unsupervised fashion to identify the underlying structures in a dataset. Clustering methods partition a dataset according to a similarity measure between data points. In this work, we iteratively apply k-means clustering [22] on the vector representations of the impression sections in radiology reports of the seed-labeled data. K-means clustering is a simple and efficient clustering algorithm that has been used in various applications [30].
To estimate the optimal number of clusters in the training set, we employed the gap statistic method [31], which compares the total intra-cluster variation for different values of k with their expected values under null reference distribution of the data. The gap statistic method aims to find an optimal number of clusters from a given range to relieve the bias caused by the initialization process of k-means clustering. We examined the potential range from ten to twenty for the number of clusters in our dataset based on a previous topic modeling study on the RadCore repository [6]. The optimal cluster was selected based on the largest gap statistic score. Similar to the nearest neighbor classification approach, we iteratively ran k-means clustering to label new cases. This iterative approach added newly labeled cases for prompt communication back into the seed-labeled dataset until no more cases were added. In our experiment, our method stopped adding more new cases after the 39th iteration. For each iteration, we examined the gap statistic, evaluated the number of clusters (k) from ten to twenty, and used the k value, which yielded the largest gap statistic as the optimal k value for that iteration.
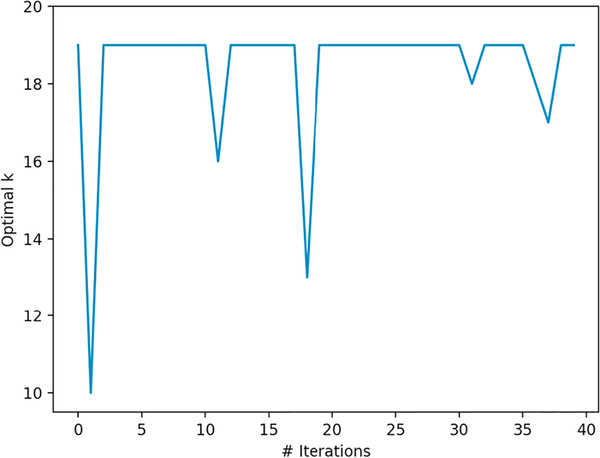
Fig. 3 shows the optimal number of clusters (k) generated by the gap statistic method for the seed data (iteration 0) and each of the 39 iterations. Nineteen was the most frequent optimal number of clusters generated by the gap statistic method, occurring 34 times. An optimal number of nineteen clusters was also verified by a domain expert radiologist in our previous topic modeling study performed on the RadCore dataset [6]. Including the 261 initial seed reports and 5855 newly labeled reports, a total of 6116 reports were labeled for prompt communication in the training set using the clustering approach.
Fig. 3.
The optimal number of clusters (k) by the gap statistic method for each iteration.
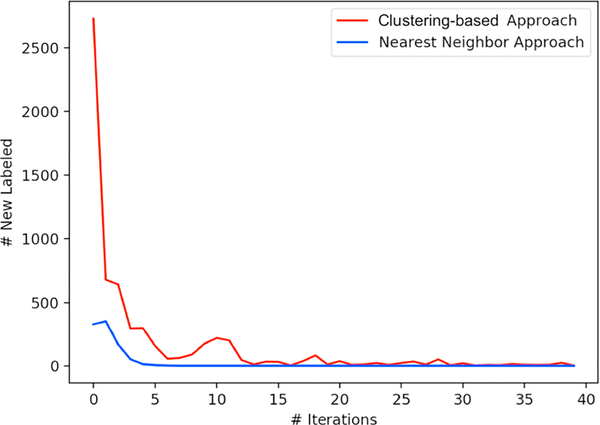
Fig. 4 shows a comparison between the nearest neighbor approach and the clustering-based approach with regard to the number of newly labeled cases for prompt communication in each iteration throughout training.
Fig. 4.
Newly labeled cases as promptly communicated by iteration. Nearest neighbor approach became stable after 7 iterations; clustering-based approach became stable after 39 iterations.
2.6.2. Cluster-based classification
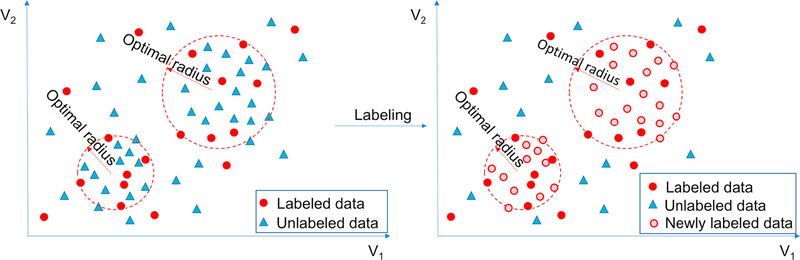
This approach uses the final nineteen k-means clusters of the training set, after iterating stopped adding new labels, as a basis to classify new, unlabeled radiology reports. Fig. 5 shows a conceptual overview of this cluster-based approach on a sample dataset.
Fig. 5.
Conceptual overview of an iteration in the clustering-based approach in two-dimensional (2D) space (V1 and V2). Clusters are created based on the seed-labeled dataset (left); if any unlabeled vector falls within the radius of any cluster, we label the unlabeled vector as communicated (right). Of note, the actual dimension of the vector space in our study is 300, and the 2D simplification is only for visualization purposes.
In order to select optimal radii of these clusters, we employed a grid search for the best F-score by labeling our validation dataset utilizing the clusters. We calculated the Euclidean distance between the impression section vector for reports labeled for prompt communication in a cluster from our training dataset and the cluster’s centroid to determine a radius for each report. For all of the nineteen clusters, we divided the difference between the shortest radius and the longest radius from the centroid in each cluster into twenty segments for grid search.
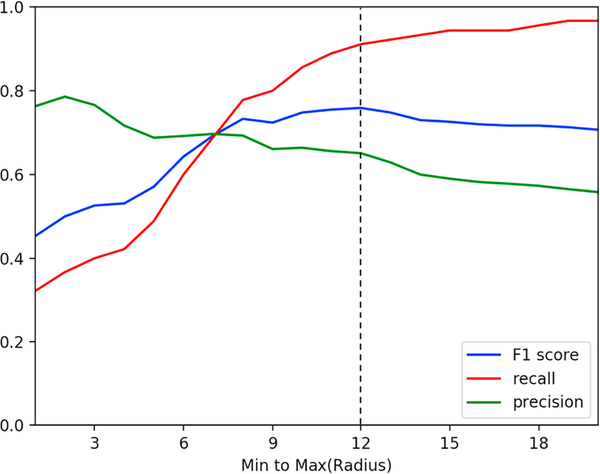
The grid search was performed on our validation dataset, and its overview is shown in Table 3 and is described in Section 2.1. The impression sections of these reports were aggregated through our existing word2vec model as described in Section 2.4. For each of the twenty cluster radius segment sets, we labeled each report in the validation dataset as positive for prompt communication if the vector representation for that report’s impression section fell within the radii for the clusters. The set of cluster radii that yielded the best F-score on this expert-labeled validation dataset was chosen as the “optimal radius” of the clusters; this validation result is shown in Table 4 and is graphed in Fig. 6. Subsequently, in our cluster-based classification approach, the unlabeled cases, for which the vector representations of their impression sections in radiology reports fall within the optimal radius of any cluster, are labeled as positive for prompt communication with referring physicians, as is shown in Fig. 5.
Table 3.
An overview of the validation set from DHMC.
| Expert Label | Modality | Total |
|---|---|---|
| Normal | CT: 22 | 90 |
| MRI: 36 | ||
| X-ray: 32 | ||
| Prompt Communication | CT: 22 | 90 |
| MRI: 36 | ||
| X-ray: 32 |
Table 4.
The performance of the clustering-based method on the validation dataset (N = P = 90). * mark indicates the optimal radius based on F-score.
| Search candidate | # True Positive | # False Positive | # True Negative | # False Negative | Recall (%) | Precision (%) | Specificity (%) | Negative predictive value (%) | F-score (%) |
|---|---|---|---|---|---|---|---|---|---|
| Radius 1 (shortest) | 29 | 9 | 81 | 61 | 32.2 | 76.3 | 90.0 | 57.0 | 45.3 |
| Radius 2 | 33 | 9 | 81 | 57 | 36.7 | 78.6 | 90.0 | 58.7 | 50.0 |
| Radius 3 | 36 | 11 | 79 | 54 | 40.0 | 76.6 | 87.8 | 59.4 | 52.6 |
| Radius 4 | 38 | 15 | 75 | 52 | 42.2 | 71.7 | 83.3 | 59.1 | 53.1 |
| Radius 5 | 44 | 20 | 70 | 46 | 48.9 | 68.8 | 77.8 | 60.6 | 57.1 |
| Radius 6 | 54 | 24 | 66 | 36 | 60.0 | 69.2 | 73.3 | 64.7 | 64.3 |
| Radius 7 | 62 | 27 | 63 | 28 | 68.9 | 69.7 | 70.0 | 69.2 | 69.3 |
| Radius 8 | 70 | 31 | 59 | 20 | 77.8 | 69.3 | 65.6 | 74.7 | 73.3 |
| Radius 9 | 72 | 37 | 53 | 18 | 80.0 | 66.1 | 58.9 | 74.6 | 72.4 |
| Radius 10 | 77 | 39 | 51 | 13 | 85.6 | 66.4 | 56.7 | 81.0 | 74.8 |
| Radius 11 | 80 | 42 | 48 | 10 | 88.9 | 65.6 | 53.3 | 82.8 | 75.5 |
| Radius 12* | 82 | 44 | 46 | 8 | 91.1 | 65.1 | 51.1 | 85.2 | 75.9 |
| Radius 13 | 83 | 49 | 41 | 7 | 92.2 | 62.9 | 45.6 | 85.4 | 74.8 |
| Radius 14 | 84 | 56 | 34 | 6 | 93.3 | 60.0 | 37.8 | 85.0 | 73.0 |
| Radius 15 | 85 | 59 | 31 | 5 | 94.4 | 59.0 | 34.4 | 86.1 | 72.6 |
| Radius 16 | 85 | 61 | 29 | 5 | 94.4 | 58.2 | 32.2 | 85.3 | 72.0 |
| Radius 17 | 85 | 62 | 28 | 5 | 94.4 | 57.8 | 31.1 | 84.8 | 71.7 |
| Radius 18 | 86 | 64 | 26 | 4 | 95.6 | 57.3 | 28.9 | 86.7 | 71.7 |
| Radius 19 | 87 | 67 | 23 | 3 | 96.7 | 56.5 | 25.6 | 88.5 | 71.3 |
| Radius 20 (longest) | 87 | 69 | 21 | 3 | 96.7 | 55.8 | 23.3 | 87.5 | 70.7 |
Fig. 6.
F-score, recall, and precision of our clustering-based method with different radii on the validation dataset. Radius 12 was selected as the optimal radius based on the highest F-score.
3. Evaluation and results
The evaluation was performed based on our DHMC test dataset to compare the performance of the clustering-based approach to the nearest neighbor classification approach as our baseline. As described in Section 2.1, we randomly selected 480 reports with ground truth labels that were annotated by two radiologists as positive (240 reports) and negative (240 reports) for prompt communication cases, balanced across the three imaging modalities of CT, MRI and X-ray (80 positive and 80 negative for each modality), as our test set for final evaluation (Table 5). The impression sections of these reports were aggregated through our word2vec model in this evaluation as described in Section2.4.
Table 5.
An overview of the test set from DHMC.
| Expert Label | Modality | Total |
|---|---|---|
| Normal | CT: 80 | 240 |
| MRI: 80 | ||
| X-ray: 80 | ||
| Prompt Communication | CT: 80 | 240 |
| MRI: 80 | ||
| X-ray: 80 |
For the nearest neighbor approach, each report in the test set was assigned a label based on the label of its nearest neighbor from the labeled training set of the nearest neighbor method. For the clustering- based approach, we calculated the distance between each report vector in the test set and the centroid of each cluster in the training set and compared this distance to the optimal radius of the cluster as computed in Section 2.6.2. Cases that fell within the optimal radius of any cluster centroids were labeled as positive for prompt communication. Table 6 shows the results along with precision, recall, and F-score calculated based on the expert-labeled test set for both approaches.
Table 6.
Statistical performance on test dataset (480 reports).
| Method | # True positive | # False positive | # True negative | # False negative | Recall (%) | Precision (%) | Specificity (%) | Negative predictive value (%) | F-score (%) |
|---|---|---|---|---|---|---|---|---|---|
| Nearest Neighbor | 114 | 69 | 171 | 126 | 47.5 | 62.3 | 71.0 | 58.0 | 53.5 |
| Clustering-Based | 216 | 124 | 116 | 24 | 90.0 | 63.5 | 48.0 | 83.0 | 74.5 |
4. Discussion
In this work, we presented a new machine learning pipeline as the first step toward a generalizable and accurate semi-supervised framework to assist radiologists in identifying cases that require prompt communication to referring physicians. This framework can potentially help with improving the communication between radiologists and referring physicians in the appropriate timeframe to avoid potential harm to patients. Our evaluation on an independent test set and the comparison of this clustering-based approach to a baseline nearest neighbor method showed our approach could achieve a considerably higher performance (F-score: 74.5%, recall: 90.0%, precision: 63.5%) in comparison to the nearest neighbor approach (F-score: 53.5%, recall47.5%, precision: 62.3%). Also, our evaluation on an internal dataset showed the generalizability of the proposed approach across different healthcare organizations. With a relatively low number of false negatives, our approach can potentially be utilized to help radiologists to identify critical cases that require prompt communication with referring physicians. Of note, our approach produced a considerable number of false positives in our evaluation, which may lead to alert fatigue in clinical settings. In future work, we plan to further improve the precision of our model by extending our labeled dataset and leveraging ensemble learning to address this limitation. Such an ensemble learning approach can combine our cluster-based and nearest neighbor methods to improve the results and their balance between precision and recall for clinical applications. In addition, advances in electronic health record systems can lead to real-time tracking of communications between radiologists and providers and will be instrumental in collecting labeled data to improve the proposed model in this study.
Our proposed approach in this paper analyzes free-text radiology reports to provide a decision support system to help radiologists identify cases requiring prompt communication with referring physicians. These are cases that would be otherwise be missed or overlooked due to a radiologist’s heavy workload, lack of administrative support, or complex team dynamics within the radiology department as well as with other care groups. This approach leverages various machine learning and NLP techniques, such as distributional semantics and cluster analysis, to build a semi-unsupervised learning method to identify cases requiring prompt communication. The evaluation of our method showed that using only unlabeled free text in a semi-un-supervised learning approach along with a minimally labeled set for fine tuning can achieve high performance, which is often achievable only through access to a large amount of hand-labeled data for model development and training in supervised learning approaches. Furthermore, because this semi-unsupervised learning method does not require extensive effort to collect labeled training data manually, it can be easily extended to other data sources and institutions. To identify reports needing prompt communication, institutions with a large radiology report dataset available to train a word2vec model can extend the proposed mechanisms to identify seed data and to fine-tune cluster parameters. In future steps, we also plan to generalize this approach to classify reports based on change, important findings, and urgency.
We randomly sampled 10% of the errors made by our best model (cluster-based classification) in our evaluation to conduct an error analysis. In this error analysis, our senior radiologist collaborator (YYC) manually reviewed the radiology reports associated with each case to assess the potential causes of the errors made by our approach. Through this analysis, we observed several patterns in our errors that once remedied can help to improve the performance of our methods in future work. For example, some false negatives were caused due to explicit requests from referring physicians to radiologists to contact them. Therefore, these cases needed to be promptly communicated regardless of the characteristics of radiological findings. In some other cases, false negatives were due to the size of the radiological findings. For example, small incidental pulmonary nodules (< 6mm) mostly do not require prompt communication for low-risk patients, however, larger incidental pulmonary nodules can be considered urgent. Of note, in most of the false positive cases, although the clinical findings, such as malignancy or brain aneurysm, were significant, prompt communication with referring physicians was not needed because the radiology exams were performed on in-patients and showed stable or unchanged conditions. In other false positive cases, radiologists included recommendations for further actions or follow-ups in radiology reports, which were mislabeled for prompt communications by our method. This error analysis showed that although the overall performance of our approach is promising, careful consideration of the context for radiological exams, such as patient clinical history and status, the nature of communication/follow-up requests, and change in radiological findings and their size, in our data modeling and classification can improve the performance of our methods.
In our cluster-based approach, we used the clusters from our seed-labeled data as a basis to classify unlabeled reports. Through cluster-based classification, unlabeled cases with radiology report vector representations that fell within the radius of our seed-labeled data cluster centroids were labeled as positive for prompt communication with referring physicians. These clusters help to identify different topics and substructures for the radiology report in our repository. Therefore, our approach could leverage different substructures in the dataset to increase the number of identified cases without compromising the accuracy. In future work, we plan to explore the use of other topic modeling approaches [32,33] to identify additional potentially helpful substructures in the dataset in an unsupervised fashion.
In this study, we used word2vec distributional semantic vectors to capture different findings in free text and to represent impression sections of radiology reports. However, the word2vec semantic vectors for the words in an impression section of a radiology report were aggregated in a bag-of-words model, without considering the dependency relationships among them and the sentence structures. In future work, we plan to leverage statistical parsers, such as Stanford Parser [34], and sentence representation models, such as Sent2vec [35], to include the grammatical structures of sentences and word dependencies in our text modeling. We expect that including these grammatical and dependency relationships in mathematical representations of radiology reports will improve the performance of our semi-unsupervised learning approach. The presented approach in this manuscript includes a systematic search (i.e., grid search) mechanism to identify the optimal parameters to train our model for a given dataset. However, investigating the effects of the training set size and other parameters on the performance and stability of our models requires additional datasets and experiments that we will pursue in future studies.
Of note, as a post-processing step, to improve the quality of our extracted seed-labeled data, we relied on a simple rule-based regular expression method to filter out communications with patients in our dataset. This simple, regular expression approach may not be generalizable enough to filter out all false positive cases of communication that occurred with persons other than referring physicians. As future work, we plan to leverage current state-of-the-art co-reference resolution methods [36,37] to more accurately identify and filter out from our seed-labeled data the conducted communications that were not with referring physicians. Finally, as future work, we plan to leverage the cluster-based topic modeling approaches to identify subcategories among cases that we identify as positive for prompt communication with regard to their follow-up plans to further assist radiologists in their communications with referring physicians.
5. Conclusion
In this paper, we described a semi-unsupervised machine learning approach to identify cases that require prompt communication between radiologists and referring physicians based on analyzing the corresponding radiology reports. Our method relies on an unsupervised distributional semantics neural network to model radiology report free-text narratives. In this work, we automatically identified 261 cases of prompt communication based on keywords generated through domain- expert knowledge and a distributional semantics neural network as seed-labeled data for our semi-unsupervised learning approach. We clustered this seed-labeled dataset and used the underlying structure of the clustered seed data to classify unlabeled reports. We compared the results of this clustering-based, semi-supervised approach to a baseline nearest neighbor classification method. The evaluation showed that our clustering-based, semi-supervised approach achieved an F-score of 74.5%, recall of 90%, and precision of 63.5% for identifying cases for prompt communication, outperforming the nearest neighbor approach. This clustering-based, semi-supervised approach could potentially be used as part of an online-decision support system in clinical settings to help radiologists identify cases for prompt communication with the referring physicians to avoid or minimize possible harm to patients.
Acknowledgments
The authors would like to thank Daniel Rubin, Chuck Kahn, Kevin McEnery and Brad Erickson for their help compiling the RadCore database; Daniel Rubin for providing access to the database; Curt Langlotz for helpful discussions; and Lamar Moss for their feedback on the manuscript. This research was supported in part by a U.S. National Institute of Health grant, R01LM012837.
Footnotes
Conflicts of interest
The authors have no competing interests to declare.
References
- [1].Hayes SA, Breen M, McLaughlin PD, Murphy KP, Henry MT, Maher MM, Ryan MF, Communication of unexpected and significant findings on chest radiographs with an automated PACS alert system, J. Am. Coll. Radiol 11 (2014) 791–795, 10.1016/j.jacr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- [2].Kitch B, Ferris T, Campbell E, Summary of the evidence on patient safety: implications for research, Summ. Evid. Patient Saf. Implic. Res (2008) 54–56 ISBN 978 92 4 159654 1. [Google Scholar]
- [3].Berlin L, Failure of radiologic communication: an increasing cause of malpractice litigation and harm to patients, Appl. Radiol 39 (2010) 17–23. [Google Scholar]
- [4].Zech J, Pain M, Titano J, Badgeley M, Su A, Costa A, Bederson J, Lehar J, Oermann EK, Natural language – based machine learning models for the annotation of clinical radiology 000 (2018) 1–11. [DOI] [PubMed] [Google Scholar]
- [5].Taira RK, Soderland SG, Jakobovits RM, Automatic structuring of radiology free-text reports 1, Radiographics 21 (2001) 237–245. [DOI] [PubMed] [Google Scholar]
- [6].Hassanpour S, Langlotz CP, Unsupervised topic modeling in a large free text radiology report repository, J. Digit. Imag 29 (2016) 59–62, 10.1007/s10278-015-9823-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hussain S, Communicating critical results in radiology, JACR 7 (2010) 148–151, 10.1016/j.jacr.2009.10.012. [DOI] [PubMed] [Google Scholar]
- [8].College A, ACR practice parameter for communication of diagnostic imaging findings, 1076 (2014) 1–9. [Google Scholar]
- [9].Sebastiani F, Machine learning in automated text categorization 34 (2002) 1–47. [Google Scholar]
- [10].J. D, Martin JH, Speech and Language Processing, 2009, doi: 10.1007/s00134-010-1760-5. [DOI] [Google Scholar]
- [11].Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, Gainer VS, Shaw SY, Xia Z, Szolovits P, Churchill S, Kohane I, Development of phenotype algorithms using electronic medical records and incorporating natural language processing, Bmj 350 (2015), 10.1136/bmj.h1885h1885-h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hobbs JR, Information extraction from biomedical text, J. Biomed. Inform 35 (2002) 260–264. [DOI] [PubMed] [Google Scholar]
- [13].Aronson AR, Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program, Proc. AMIA Symp 17 (2001) [pii] D010001275. [PMC free article] [PubMed] [Google Scholar]
- [14].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG, Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications, J. Am. Med. Inform. Assoc 17 (2010) 507–513, 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Friedman C, Hripcsak G, DuMouchel W, Johnson SB, Clayton PD, Natural language processing in an operational clinical information system, Nat. Lang. Eng 1 (1995) 83–108. [Google Scholar]
- [16].Hassanpour S, Langlotz CP, Information extraction from multi-institutional radiology reports, Artif. Intell. Med 66 (2016) 29–39, 10.1016/j.artmed.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Banerjee I, Chen MC, Lungren MP, Rubin DL, Radiology report annotation using intelligent word embeddings: applied to multi-institutional chest CT cohort, J. Biomed. Inform 77 (2018) 11–20, 10.1016/j.jbi.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gupta A, Banerjee I, Rubin DL, Automatic information extraction from unstructured mammography reports using distributed semantics, J. Biomed. Inform 78 (2018) 78–86, 10.1016/j.jbi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- [19].Wang Z, Shah AD, Tate AR, Denaxas S, Shawe-Taylor J, Hemingway H, Extracting diagnoses and investigation results from unstructured text in electronic health records by semi-supervised machine learning, PLoS One 7 (2012), 10.1371/journal.pone.0030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chai KEK, Anthony S, Coiera E, Magrabi F, Using statistical text classification to identify health information technology incidents, J. Am. Med. Informatics Assoc 20 (2013) 980–985, 10.1136/amiajnl-2012-001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mikolov T, Chen K, Corrado G, Dean J, Efficient estimation of word representations in vector space (2013) 1–12. doi: 10.1162/153244303322533223. [DOI] [Google Scholar]
- [22].Gentle JE, Kaufman L, Rousseuw PJ , Finding groups in data: an introduction to cluster analysis, Biometrics 47 (1991) 788, 10.2307/2532178. [DOI] [Google Scholar]
- [23].Singhal A, Modern INFORMATION RETRIEVAL: A BRIEF OVERView, Bull. Ieee Comput. Soc. Tech. Comm. Data Eng 24 (2001) 1–9 doi:10.1.1.117.7676. [Google Scholar]
- [24].Thompson K, Regular expression search algorithm, Commun. ACM 11 (1968) 419–422, 10.1145/363347.363387. [DOI] [Google Scholar]
- [25].Zhu X, Goldberg AB, Introduction to semi-supervised learning, Synth. Lect. Artif. Intell. Mach. Learn 3 (2009) 1–130, 10.2200/S00196ED1V01Y200906AIM006. [DOI] [Google Scholar]
- [26].Ganeshan D, Duong P-AT, Probyn L, Lenchik L, McArthur TA, Retrouvey M, Ghobadi EH, Desouches SL, Pastel D, Francis IR, Structured reporting in radiology, Acad. Radiol 25 (2018) 66–73, 10.1016/j.acra.2017.08.005. [DOI] [PubMed] [Google Scholar]
- [27].Yetisgen-Yildiz M, Gunn ML, Xia F, Payne TH, A text processing pipeline to extract recommendations from radiology reports, J. Biomed. Inform 46 (2013) 354–362, 10.1016/j.jbi.2012.12.005. [DOI] [PubMed] [Google Scholar]
- [28].Cover T, Hart P, Nearest neighbor pattern classification, IEEE Trans. Inf. Theory 13 (1967) 21–27, 10.1109/TIT.1967.1053964. [DOI] [Google Scholar]
- [29].Stuart Russell GI, Norvig Peter, Artificial Intelligence: A Modern Approach, third ed, 2010. [Google Scholar]
- [30].Schubert HKE, Zimek A, The (black) art of runtime evaluation: are we comparing algorithms or implementations? Knowl. Inf. Syst 52 (2017) 341–378, 10.1007/s10115-016-1004-2. [DOI] [Google Scholar]
- [31].Tibshirani R, Walther G, Hastie T, Estimating the number of clusters in a data set via the gap statistic, J. R. Stat. Soc. Ser. B Stat. Methodol 63 (2001) 411–423, 10.1111/1467-9868.00293. [DOI] [Google Scholar]
- [32].Blei DM, Ng AY, Jordan MI, Latent Dirichlet allocation, J. Mach. Learn. Res 3 (2003) 993–1022. [Google Scholar]
- [33].Cheng X, Yan X, Lan Y, Guo J, BTM: topic modeling over short texts, IEEE Trans. Knowl. Data Eng 26 (2014) 2928–2941, 10.1109/TKDE.2014.2313872. [DOI] [Google Scholar]
- [34].De Marneffe M-C, MacCartney B, Manning CD, Generating typed dependency parses from phrase structure parses, Proc. 5th Int. Conf. Lang. Resour. Eval. (LREC 2006) (2006) 449–454 doi:10.1.1.74.3875. [Google Scholar]
- [35].Pagliardini M, Gupta P, Jaggi M, Unsupervised learning of sentence embeddings using compositional n-gram features (2017), http://arxiv.org/abs/1703.02507. [Google Scholar]
- [36].Lee H, Peirsman Y, Chang A, Chambers N, Surdeanu M, Jurafsky D, Stanford’s multi-pass sieve coreference resolution system at the CoNLL-2011 shared task, Proc. Fifteenth Conf. Comput. Nat. Lang. Learn. Shar. Task (2011) 28–34. [Google Scholar]
- [37].Lee H, Chang A, Peirsman Y, Chambers N, Surdeanu M, Jurafsky D, Deterministic coreference resolution based on entity-centric, precision-ranked rules, Comput. Linguist 39 (2013) 885–916, 10.1162/COLI_a_00152. [DOI] [Google Scholar]