Abstract
Behavioral neuroendocrinology has benefited tremendously from the use of a wide range of model organisms that are ideally suited for particular questions. However, in recent years the ability to manipulate the genomes of laboratory strains of mice has led to rapid advances in our understanding of the role of specific genes, circuits and neural populations in regulating behavior. While genome manipulation in mice has been a boon for behavioral neuroscience, the intensive focus on the mouse restricts the diversity in behavioral questions that can be investigated using state-of-the-art techniques. The CRISPR/Cas9 system has great potential for efficiently generating mutants in non-traditional animal models and consequently to reinvigorate comparative behavioral neuroendocrinology. Here we describe the efficient generation of oxytocin receptor (Oxtr) mutant prairie voles (Microtus ochrogaster) using the CRISPR/Cas9 system, and describe initial behavioral phenotyping focusing on behaviors relevant to autism. Oxtr mutant male voles show no disruption in pup ultrasonic vocalization, anxiety as measured by the open field test, alloparental behavior, or sociability in the three chamber test. Mutants did however show a modest elevation in repetitive behavior in the marble burying test, and an impairment in preference for social novelty. The ability to efficiently generate targeted mutations in the prairie vole genome will greatly expand the utility of this model organism for discovering the genetic and circuit mechanisms underlying complex social behaviors, and serves as a proof of principle for expanding this approach to other non-traditional model organisms.
Keywords: CRISPR/Cas9, oxytocin receptor, prairie voles, autism spectrum disorder, social novelty preference, social behavior, genome editing, non-traditional models
Introduction
We are in an unprecedented time in behavioral neuroscience when the rapid advances in technology allows for the direct interrogation of the role of specific genes, circuits and neuronal populations in regulating complex behavior (Kohl et al., 2018). The greatest advances in this regard have been made possible by the ability to efficiently manipulate the genome in a targeted manner, and the laboratory mouse has become the vertebrate model of choice for this advancing technology. With this intensive focus on mouse models, however, we are limiting the diversity of behaviors and neural processes that can be explored (Brenowitz and Zakon, 2015; Yartsev, 2017), for example, laboratory mice do not show long-lasting familiarity to their conspecifics (Beery et al., 2018). Behavioral neuroendocrinology has thrived over the past several decades because investigators have utilized not only traditional laboratory models, such as mice and rats, but frequently select species that are most well-suited for addressing the experimental question of interest. However, in the past several years there has been a shift in attention from traditional neuroendocrine approaches, such as hormone manipulation and behavioral pharmacology, toward using genetically modified mouse lines to identify the roles of genes and circuits in regulating behavior with great precision. Often, with what we gain in precision, we lose in diversity and generalizability (Johnson and Young, 2018). We believe that it is imperative to develop state-of-the-art genome manipulation approaches in non-traditional model organisms so that we preserve the advantages of the comparative approach while benefiting from the remarkable power of genetic manipulation. The advance of viral vector mediated gene transfer, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated-proteins 9 (Cas9) system and other genome editing approaches are now making it possible to bring state-of-the-art technology to the field of comparative behavioral neuroendocrinology (Amadei et al., 2017; Keebaugh et al., 2015; Yokoi et al., 2015). Here we describe the development of the first targeted mutant of a model organism that has made a significant contribution to understanding the neuroendocrine mechanisms underlying social behavior, the prairie vole (Microtus ochrogaster).
The prairie vole is a socially monogamous rodent that forms enduring pair bonds between mates. The mates care for their pups bi-parentally and display selective aggression toward unfamiliar conspecifics, and the neural mechanisms underlying these behaviors are being rapidly elucidated (Aragona et al., 2006; Gobrogge et al., 2009; Johnson and Young, 2015; Walum and Young, 2018; Young et al., 2011; Young and Wang, 2004). Additionally, empathy-related consolation behavior toward familiar individuals that received mild stressors has been demonstrated in prairie voles (Burkett et al., 2016), as well as the consequences of loss of the partner (Bosch et al., 2016; Bosch et al., 2009; Bosch and Young, 2017; Pohl et al., 2018). These social behaviors are different from those of mice and rats, which have made the prairie vole a preferred model animal of social attachment and affiliation behaviors (McGraw and Young, 2010).
Oxytocin (OXT) and the oxytocin receptor (OXTR) play key roles in modulating social attachment behaviors in the prairie vole (Barrett et al., 2015; Donaldson and Young, 2008; Johnson et al., 2016; Johnson et al., 2017; Smith and Wang, 2014; Walum and Young, 2018). OXT is a nonapeptide produced in the paraventricular nucleus and supraoptic nucleus and projected throughout the brain to influence behavior (Johnson and Young, 2017). OXTR is a G-protein coupled receptor (GPCR) that mediates many of the behavioral effects of OXT. Studies in Oxt and Oxtr mutant mice have revealed that the OXT system plays a role in social recognition and the mutants display behavioral phenotypes relevant to autism spectrum disorder (ASD) (Ferguson et al., 2001; Pobbe et al., 2012; Sala et al., 2011). Indeed, the OXT system is being explored as a pharmacological target for treating ASD (Andari et al., 2010; Modi et al., 2015; Young and Barrett, 2015).
As the prairie vole is a suitable model for studying the role of OXTR signaling in social attachment, and Oxtr knockout mice have been suggested as a model relevant to ASD, we sought to create Oxtr mutant prairie voles using the CRISPR/Cas9 system.
The CRISPR/Cas system is derived from an RNA-based adaptive immune system of archaea and bacteria against bacteriophage or foreign nucleic acids (Barrangou et al., 2007; Wiedenheft et al., 2012). The CRISPR/Cas9 system from Streptococcus pyogenes belongs to the type II - A CRISPR/Cas system which requires a single nuclease protein and two short RNAs, a crRNA and a trans-activating crRNA (tracrRNA), to mutate foreign DNAs. The crRNA recognizes 20 nucleotide sequences of the foreign DNAs, through homology, adjacent to an NGG protospacer adjacent motif (PAM), and the tracrRNA is involved in the assembly of the RNA-protein complex. The crRNA-tracrRNA-Cas9 protein complex at the targeted DNA region induces targeted specific double strand breaks (DSB) (Deltcheva et al., 2011). DSB causes an endogenous error-prone DNA repair, such as non-homologous end joining or micro homology mediated end joining, resulting in mutation at the targeted DNA sequence, usually resulting in a deletion. If homologous DNA template, matching the targeted area, exists around the targeted site, homology directed repair (HDR) occurs. This provide a powerful means of inducing mutations or precise editing with insertion of any gene of interest in the genome. In the recent CRISPR/Cas9 approaches designed for gene engineering, the two short RNAs are replaced with one synthetic single-guide RNA (sgRNA) which is the fusion of a crRNA, with homology to the target sequence, and the tracrRNA (Cho et al., 2013; Cong et al., 2013; Mali et al., 2013), to guide the sgRNA-Cas9 complex and the mutation to the desired targeted site. Injection of Cas9 and sgRNA into the cytosol of zygotes can efficiently induce mutations at specific targeted site of the genome, which are passed on to subsequent generations (Hai et al., 2014; Kou et al., 2015; Li et al., 2013a; Li et al., 2013b; Wang et al., 2013; Zou et al., 2015).
We demonstrate that the CRISPR/Cas9 system is an efficient means of generating targeted mutations in the prairie vole and we successfully generated Oxtr mutants. We performed a preliminary screen of social or non-social behavioral tests, with an initial focus on behavioral phenotypes relevant to ASD in males. This initial report lays the foundation for further behavioral analyses and for the generation of an array of genetically modified prairie voles that will greatly enhance the utility of this model organism and for understanding of the neuroendocrine regulation of social behavior.
Materials and Methods
Animals
Prairie voles were maintained at Tohoku University and were derived from stock obtained from the colony maintained by Larry J. Young at Emory University in Atlanta, Georgia. These animals are derived from wild animals captured in Illinois, USA. Mating pairs were housed together to breed. After weaning at postnatal day (P) 21, the offspring were separated from their parents and housed in groups of two to four same sex siblings per cage. All animals were housed using alternating 12-h periods of light and dark at 25 °C and were allowed ad libitum access to food (Labo MR Stock, NOSAN, Japan) and water. All animal experiments were performed according to the Tohoku University guidelines for animal experimentation. All experimental procedures were approved by the Center for Laboratory Animal Research, Tohoku University.
Super-ovulation and embryo collection
Super-ovulation and embryo collection were performed as previously described (Horie et al., 2015). Briefly, 10 week-old female voles were injected (IP) with 60 IU pregnant mare serum gonadotropin in 100 μl saline and cohoused with a sexually experienced male separated by a mesh divider to prevent mating. After 48 h, females were injected (IP) with 60 IU human chorionic gonadotropin in 100 μl saline and the mesh divider was removed to allow mating. Twelve hours later the female voles were euthanized with isoflurane. Embryos were collected from oviducts in M-16 medium (Sigma-Aldrich) and cultured in G-l PLUS medium (Vitrolife, Gothenburg, Sweden) in a humidified 5% CO2 chamber at 37 °C until microinjection.
Prairie vole Oxtr sequence and design of sgRNAs
The prairie vole Oxtr sequence (Gene ID: 101979991) was obtained from GenBank (National Center for Biotechnology Information). Six sgRNAs next to 5′-NGG-3′ PAM sequences were designed around the coding site (CDS) 1 of Oxtr as illustrated in Figure 1.
Figure 1.
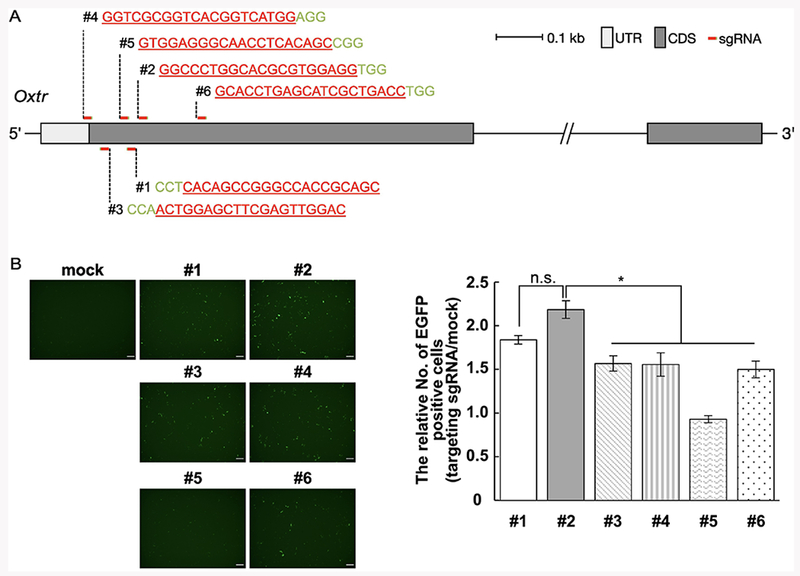
Construction of sgRNAs targeting the prairie vole oxytocin receptor gene (Oxtr). (A) The target regions of each sgRNA in Oxtr. All sgRNAs were targeted against the 5’ region of the coding sequence (CDS). UTR = untranslated region. (B) Evaluation of cutting efficiencies by SSA assay in HEK293T cells (n = 3). The greater the number of EGFP positive cells, the greater the cutting efficiency, n.s = not significant. * = p<0.05. Data are presented as mean +/− Standard Error of the Mean (SEM). Scale bar = 150 μm.
Validation of sgRNA targeting efficiencies by single-strand annealing assay (SSA)
Pairs of annealed oligo DNAs containing each sgRNA sequence were integrated into pX330 (Addgene Plasmid #42230) cut with Bbsl. Genomic sequences containing sgRNA targeted sequence were amplified by PCR and the amplicon was digested with EcoRI and BamHI. The digested amplicon was then ligated into pC AG EGxxFP (Addgene Plasmid #50716) cut with EcoRI and BamHI. For genomic PCR, the forward primer was 5′-CTAGGATCCACGCTTTAAAGAGCAGCAAG-3′ and the reverse primer was 5′-CTAGAATTCTCGAGCGACATAAGCAGCAG-3′. For the construction of pX330 vectors targeting the Oxtr locus, the sense oligo DNAs were sgRNAl, 5′-CACCGCTGCGGTGGCCCGGCTGTG-3′; sgRNA2, 5′-CACCGGCCCTGGCACGCGTGGAGG-3′; sgRNA3, 5′-CACCGTCCAACTCGAAGCTCCAGT-3′; sgRNA4, 5′-CACCGGTCGCGGTCACGGTCATGG-3′; sgRNA5, 5′-CACCGTGGAGGGCAACCTCACAGC-3′; and sgRNA6, 5′-CACCGCACCTGAGCATCGCTGACC; and the antisense oligo DNAs were sgRNAl, 5′-AAACCACAGCCGGGCCACCGCAGC-3′; sgRNA2, 5′-AAACCCTCCACGCGTGCCAGGGCC-3′; sgRNA3, 5′-AAACGTCCAACTCGAAGCTCCAGT-3′; sgRNA4, 5′-AAACCCATGACCGTGACCGCGACC-3′; sgRNA5, 5′-AAACGCTGTGAGGTTGCCCTCCAC-3′; and sgRNA6, 5′-AAACGCTGTGAGGTTGCCCTCCAC-3′.
The SSA assay in HEK293T cells was conducted as previously described (Mashiko et al., 2013). In this assay, generation of indels would give rise to in frame mutations of enhanced green fluorescent protein (EGFP) and mutations were detected by fluorescence microscopy using a model FSX100 microscope (Olympus). The number of EGFP positive cells, indicative of mutation efficiency, was counted using Image J software (NIH).
In vitro transcription (IVT) of sgRNA and Cas9 messenger RNA (mRNA)
The pGEM sgRNA vector for IVT of sgRNAs was constructed by the insertion of intact sgRNA sequence amplified by PCR from pX330 into the pGEM T easy vector (Promega). Each pair of oligo DNAs containing each sgRNA sequence targeting Oxtrwas integrated into the pGEM sgRNA vector digested with Bbsl. Individual vectors containing one of each sgRNA digested with Spel were used as the templates for IVT. IVT was performed using the MEGAshortscript T7 Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
For Cas9 mRNA IVT, the pBluescript Cas9 vector was constructed by inserting Cas9 cDNA from pX330 into a modified pBluescript SK digested with Agel and EcoRI. The pBluescript Cas9 vector digested with EcoRI was used as a template for IVT using mMESSAGE mMACHINE T7 ULTRA Transcription Kit (Thermo Fischer Scientific) according to the manufacturer’s protocol.
For the construction of the pGEM sgRNA vector, the forward primer was 5′-TAATACGACTCACTATAGGGGGGTCTTCGAGAAGACCTG-3′ and the reverse primer was 5′-AAAAGCACCGACTCGGTGCC-3′. For the integration of sgRNA targeting sequence, the sense oligo DNAs were sgRNAl, 5′-AGGGGCTGCGGTGGCCCGGCTGTG-3′ and sgRNA2, 5′-AGGGGGCCCTGGCACGCGTGGAGG-3′; and the antisense oligo DNAs were sgRNA1, 5′-AAACCACAGCCGGGCCACCGCAGC-3′ and sgRNA2, 5′-AAACCCTCCACGCGTGCCAGGGCC-3′.
Microinjection in the cytosol of zygotes
An sgRNA targeting Oxtr (50 ng/μl) and Cas9 mRNA (100 ng/μl) in ribonuclease free distilled deionized water were injected into the cytosol of zygotes (Wang et al., 2013). After 24 h of culture in G-l PLUS medium (Vitrolife), two to ten cells that had developed to the 2-cell stage were transferred into each oviduct of pseudopregnant female prairie voles (36 2-cells transferred to 3 pseudopregnant female voles).
T7 endonuclease I (T7E1) assay
The T7EI assay was performed as described previously (Guschin et al., 2010). Briefly, the genomic regions targeted by each sgRNA were amplified by nested PCR. The PCR amplicons were denatured for 3 min at 95 °C and then re-annealed at 85-30 °C (ramp rate −0.1 °C/sec). Hybridized PCR products were digested with T7EI (New England Biolabs) and then separated by 2% agarose gel. For PCR, the first forward primer was 5′-AGATCAGTGCCCGGGTGCCC-3′, the second forward primer was 5′-ACGCTTTAAAGAGCAGCAAG-3′, and the first and second reverse primer was 5′-TCGAGCGACATAAGCAGCAG-3′.
Oxtr genotyping and sequencing from PCR amplicons and sub-cloned PCR amplicons
The target region around Oxtr was PCR-amplified from genomic DNA using the primer pairs described in the T7E1 assay. PCR amplicons were purified by gel extraction. A portion of the amplicons was cloned into the pGEM T Easy Vector. PCR amplicons and sub-cloned amplicons were sequenced with each primer using an ABI PRISM Genetic Analyzer (Applied Biosystems).
Transforming growth factor-alpha (TGFα) shedding assay
The in vitro function of mutated OXTR was evaluated using the TGFα shedding assay as previously described (Inoue et al., 2012). Briefly, each mutant Oxtr and alkaline phosphatase fused TGFα (AP-TGFα) expression plasmids were transfected into HEK293 cells, and OXT was added to the culture medium. After a 1-h incubation, the AP substrate para-nitrophenylphosphate (p-NPP) was added to the medium and the optical density at 405 nm (OD405a) was measured using an absorption spectrometer. Absorbance was regarded as the relative activity of OXTR in vitro.
OXTR autoradiography
OXTR autoradiography was performed as previously described (Ross et al., 2009). Briefly, voles were euthanized by isoflurane. Brains were removed from the skull, and then quickly frozen on dry ice powder. Brains were shipped to Emory University for processing. Coronal 20 μm sections were cut in a cryostat, mounted on Superfrost plus slides (Fisher, Pittsburgh, PA), and stored at −80°C until used. Sections were dried and fixed with 0.1% paraformaldehyde in PBS for 2 min at room temperature and incubated in 50pM of selective 125I OXTR ligand, ornithine vasotocin analogue (125I-OVTA, 2200 Ci/mmol, PerkinElmer, Boston, MA) for 1 hour. After unbound 125I-OVTA was removed from the sections in Tris- MgCl2 buffer, sections were dried and exposed to BioMax MR film (Kodak, Rochester, NY, USA) for 72 hours. Previous studies demonstrated that 125I-OVTA is highly specific for the prairie vole OXTR and co-incubation with unlabeled OXTR ligand yields a uniformly low background in brain (Insel and Shapiro, 1992). Digital images were obtained at 1200dpi with 8-bit gray scale settings with QCAM CCD camera (Qimaging, Surrey, Canada). Contrast and brightness of the images were adjusted using Adobe photoshop CS6 (San Jose, CA).
Off-target analysis
Off-target sites were searched by CasOT (Xiao et al., 2014) with the default condition. If the seed sequence, which is a 12-bp sequence next to the PAM sequence, had a mismatch within 2 bp, it was regarded as the candidate off-target site. Each of the six potential off-target sites was amplified by PCR and the PCR amplicons were sequenced by each forward primer directly. The primer pairs were Off-T #1 (forward, 5′-CCAGAGATATTTCAGGGTTG-3′; reverse, 5′-TTACCATAGAGGGTTCATCC-3′); Off-T #2 (forward, 5′-AGACCTGGTCCCCCACTAGC-3′; reverse, 5′-AACCATCGGCCTCTGCCTGT-3′); Off-T #3 (forward, 5′-CCGCCCACCAGCCCGCGTGA-3′; reverse, 5′-GAGACCGCGGCGCAGTTCCC-3′); Off-T #4 (forward, 5′-GAAGGGCGTGCACACGCCTG-3′; reverse, 5′-GAAGGGCGTGCACACGCCTG-3′); Off-T #5 (forward, 5′-TCGTGGGTAAGGAGAGTGC-3′; reverse, 5′;AGATCTTGCTCCAAGGCGGC-3′), Off-T #6 (forward, 5′-AGCTGCCAAGCAAACAGGTA-3′; reverse, 5′-CATGTTTTGAAACTGGAAAC-3′).
Behavioral tests
Ultrasonic Vocalization (USV) recording:
USV recording was performed as previously described (Miyazaki et al., 2016). After 1 min isolation from their mothers at P5, the number and duration of USV calls (15 - 90 Hz) were recorded in a dark soundproof box for 5 min. All data were analyzed automatically by SONOTRACK (Metris).
Open Field Test (OFT):
OFT was performed as previously described (Hiroi and Neumaier, 2006). OFT was performed in a 50 × 50 × 40 cm opaque white chamber under 500 lux LED light. All subjects were >P70 male voles that previously performed the USV test at P5. After 10 min acclimation in the test arena, the 10 min tests were performed. All data were analyzed automatically by ANY-maze (Stoelting Co.). The chamber was divided into 16 squares by the software. The four squares located in the center of the chamber were regarded as the center zone and the twelve squares located around the wall were regarded as the corner zone. The total time in each zone, the number of entries to each zone, the number and duration of freezing in each zone and the total distance in each zone were analyzed.
Marble Burying Test (MBT):
The MBT was performed as previously described (Thomas et al., 2009). The MBT was performed in a 20 × 30 × 30 cm non-transparent white chamber with the woodchip bedding under 500 lux LED light. The male subjects, which were previously tested in the USV test and OFT, were acclimated for 20 min in the test chamber without marbles. After that, the subjects were taken out once to place 20 marbles on the surface of woodchips, and then placed back in the chamber to perform the 20 min test. All the assays were recorded by a web camera, and the number of completely buried marbles were manually counted.
Alloparental Behavior Test (ABT):
The ABT was performed as previously described (Olazábal and Young, 2005) with some modifications. ABT was performed in the 29 × 43 × 20 cm transparent cage with paper bedding and cotton for nest building under 150 lux LED light. The male subjects tested in the previous tests, were acclimated for 1 day in the test cage. On the test day, they were acclimated in the test room for 90 min. Two pups at P2-5 were located at the opposite side of each corner from the nest in the test cage, and the 15 min tests were performed and video recorded. The duration of crouching and licking; and the number of retrieved pups were manually analyzed.
Three Chamber Test (TCT):
The TCT was performed as previously described (Nadler et al., 2004). Briefly, the TCT was performed in a 41 × 21 × 35 cm opaque white chamber divided into three sections by two transparent partitions which had small entrances under 10 lux LED light. The male subjects tested in the previous tests were isolated from their siblings for a week. On the test day, subjects were acclimated for 10 min in the test cage without stimulus voles. Afterwards, an unfamiliar male stimulus vole was placed in a triangle mesh cup at the corner and subjects were allowed to freely investigate in the chamber for 10-min (sociability test). After the sociability test, a novel vole was located in another cup at the opposite corner, and then, subjects were allowed to freely investigate in the chamber for 10-min (social novelty). All tests were recorded and the investigation time, which was defined as the time within the 5 cm around each cup, was analyzed by ANY-maze.
Statistical analyses
Experimental data were analyzed for significance by R software. P < 0.05 was considered statistically significant. For all the tests, except for SSA assay and TCT, the Student’s t-test or Welch’s t-test were performed. SSA assay was analyzed with ANOVA and a post hoc Bonferroni test. TCT data was analyzed with ANOVA and post hoc Tukey-Kramer tests. Effect size (Cohen’s d) was calculated by dividing the mean difference by the pooled standard deviation as previously described (Johnson et al., 2016).
Results
For efficient genome editing in vivo, we initially screened a set of sgRNAs in vitro to select the most highly active sgRNAs. Six sgRNAs targeting around the start codon in Oxtr exon 1 were selected (Figure 1A). We performed the SSA assay to evaluate sgRNA activities in vitro. The number of EGFP positive cells was significantly greater in cells transfected with sgRNA #2 compared with the other sgRNAs, except for sgRNA# 1 (Figure 1B; F5, 12 = 20.9, p < 0.001). Thus, we selected sgRNA #1 and #2 for the generation of Oxtr knockout (KO) prairie voles in the following experiments.
Zygotes injected with Cas9 mRNA and each sgRNA were cultured to the 2-cell embryo stage followed by transfer into oviducts of pseudopregnant prairie voles. Forty of 92 injected zygotes developed to the 2-cell stage (the developmental rate of embryos, 43%). Thirty six 2-cells were transferred into the oviducts of 3 pseudopregnant female voles. Twenty to 22 days after embryo transfer, six newborns were obtained. Three representative voles (vl, v2, and v3) are shown in Figure 2A. Their physical health did not appear visibly abnormal. The vl, v3, v4 and v5 mutants were males and the v2 and v6 were females. Genomic DNA was extracted from the vole by ear punches and the T7E1 assay was performed. The results suggested that all the voles had mutations in the target region (Figure 2B). Cloning and sequencing from the PCR amplicon demonstrated that all the voles had a mutation in the sgRNA targeting region (Figure 2C; mutation efficiency = mutant voles/newboms, 100%). For example, vl vole had a 1-bp nucleotide insertion at the Cas9 cutting site, and v4 had 6 bp and 60 bp deletions.
Figure 2.
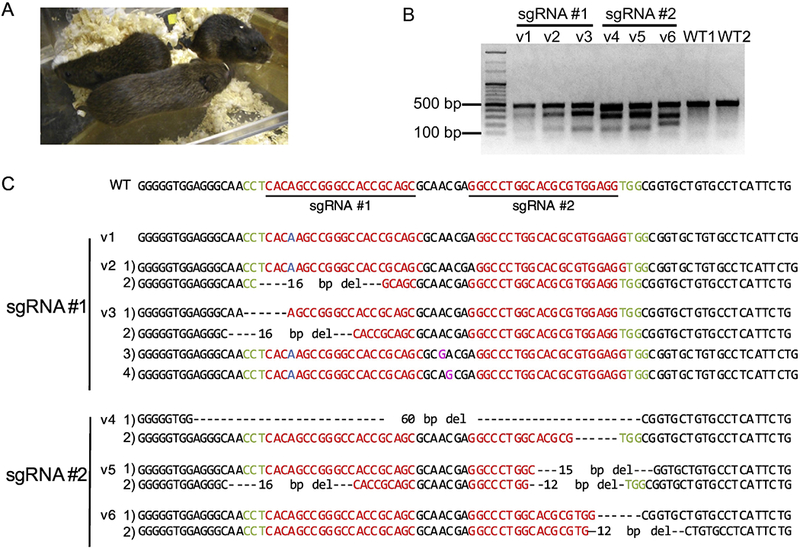
Generation of Oxtr gene modified prairie voles by CRISPR/Cas. (A) Picture of vl, v2 and v3 mutant voles demonstrating their healthy appearance. (B) T7E1 assay for detection of indels induced by CRISPR/Cas. The presence of multiple bands is indicative of the introduction of indels.(C) Sequences of each mutant vole. Note the most voles had indels in both alleles of the Oxtr. Four mutant alleles in the v3 vole indicated that CRISPR might work at the 2-cell stage. Red letters = protospacer sequence; green letters = PAM; blue letters = insertion; magenta letters = conversion; hyphen = deletion.
To analyze the function of mutated OXTR in vitro, the TGFα shedding assay was performed (Figure 3A; EC50: vl, N.D.; v4-l, N.D.; v4-2, 3.5×10−8 μM). This assay can measure the activity of GPCRs by measuring the activity of AP-TGFα. Upon the addition of OXT ligand to the culture medium, OXTR activates the TNF-α converting enzyme (TACE) in a dose-dependent manner. The activated TACE catalyzes the release of plasma membrane-tethered AP-TGFα to the culture medium. The activity of the released AP-TGFα as measured by absorbance indicated the activity of OXTR. The vl mutant-derived OXTR had no activity, possibly because the 1-bp insertion caused a frameshift mutation. The v4-2 mutant OXTR retained slight activity because the 6-bp nucleotide deletion resulted in the deletion of only two amino acid deletions. In contrast, the v4-l mutant OXTR, which featured a long (60 bp) in-frame deletion at the putative ligand binding domain, produced no activity in the TGFα shedding assay. Thus, we regarded the v4-l mutant Oxtr, hereafter referred to as OxtrΔ60/Δ60, as the best candidate Oxtr allele for generating an Oxtr KO prairie vole.
Figure 3.
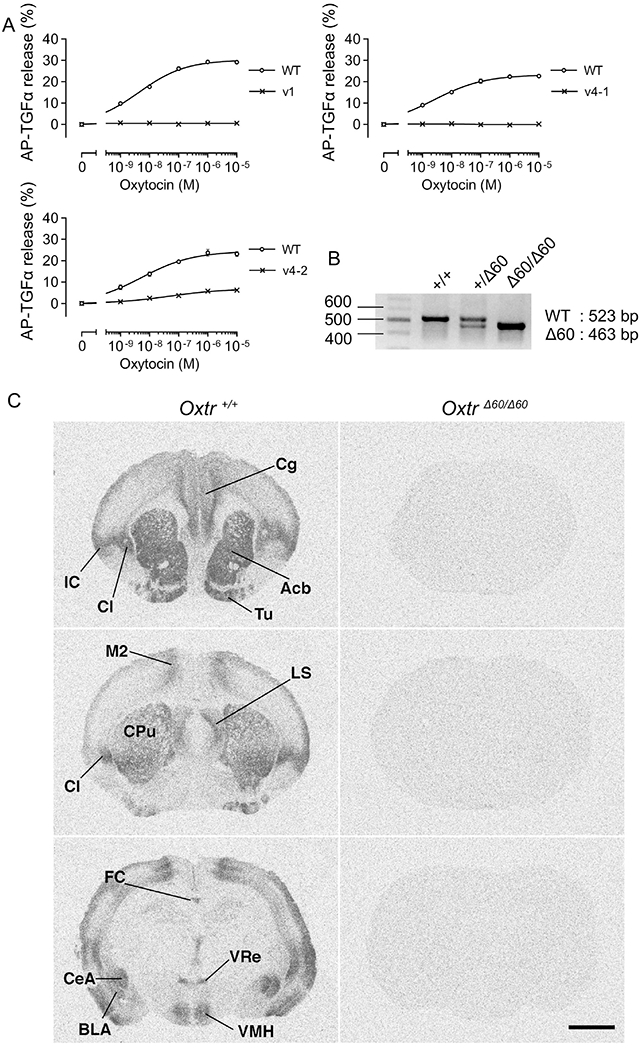
Evidence for functional abnormality of OXTR in OxtrΔ60/Δ60 voles. (A) Representative data of the function of mutated Oxtr by TGFα shedding assay in vitro for vl, v4-1 and v4-2 voles (n = 3). (B) Genotypes of Oxtr+/+, Oxtr+/Δ60 and OxtrΔ60/Δ60 detected by PCR and agarose gel electrophoresis. (C) OXTR receptor autoradiography in wildtype (Oxtr+/+) and OxtrΔ60/Δ60 Note the complete lack of signal in the OxtrΔ60/Δ60 brain. Cg, cingulate cortex; Acb, nucleus accumbens; Tu, tuberal region; Cl, claustrum; IC, insular cortex; M2, secondary motor cortex; LS, lateral septum; CPu, caudate putamen; FC, focus callosum; CeA, central amygdala; BLA, basolateral amygdala; VMH, ventromedial hypothalamus; VRe, ventral reuniens. Scale bar = 2mm.
We confirmed successful germline transmission of the v4-l genotype to subsequent generations (Figure 3B). Off-target effects were not detected by direct sequencing from the PCR amplicon of the six predicted off-target sites in the genome of the v4 mutant vole (Supplementary Figure 1). In the brain of OxtrΔ60/Δ60 voles, the OXTR protein was not detected by receptor autoradiography (Figure 3C), suggesting that the 60-bp deletion was sufficient to eliminate OXTR protein expression in vivo. Thus, we successfully generated Oxtr KO prairie voles.
Oxtr is known as the one of the key regulators of social and anxiety behaviors in prairie voles. Therefore, each behavioral test, USV, OFT, MBT, ABT and TCT, were sequentially performed to analyze the effect of OXTR deficiency in male prairie voles (Figure 4A). First, we measured USV calls induced by parental separation as an index of pup communication. OxtrΔ60/Δ60 voles showed normal USV duration and the number of calls during 5-min tests at P5 (Figure 4B: Oxtr+/+, n = 18; OxtrΔ60/Δ60, n = 12; duration of total calls: t(22) = 1.11; p = 0.28; d = 0.44; the number of total calls: t(23) = 1.19; p = 0.25; d = 0.46). Thus the Oxtr mutation did not affect the pup vocalization induced by maternal separation. Second, for measurement of anxiety behavior and locomotor activity in the adult male prairie voles, OFT was performed. OxtrΔ60/Δ60 voles showed normal center time and locomotor activity (Figure 4C: Oxtr+/+, n = 15; OxtrΔ60/Δ60, n = 15; center time: t(28) = 1.43; p = 0.16; d = 0.54; total distance: t(26) = 0.60, p = 0.55, d = 0.23). These results showed Oxtr does not influence anxiety and locomotor behaviors in male prairie voles in the OFT. Third, MBT was performed to detect compulsive-like or repetitive behaviors. The number of buried marbles were significantly higher in OxtrΔ60/Δ60 voles (Figure 4D; Oxtr+/+, n = 15; OxtrΔ60/Δ60, n = 15; t(25) = 2.12; p = 0.044; d = 0.80). This result suggests that anxiety or repetitive behaviors as measured by the MBT are regulated by Oxtr in male prairie voles. Then, alloparental behaviors, such as licking, crouching and grooming to pups, were measured in the ABT. All of the allopatemal behaviors were normal in male OxtrΔ60/Δ60 prairie voles (Figure 4E: ; duration of licking: Oxtr+/+, n = 10; OxtrΔ60/Δ60, n = 12; t(19) = 0.26; p = 0.80; d = 0.11; duration of crouching: Oxtr+/+, n = 10; OxtrΔ60/Δ60, n = 10; t(18) = 1.11; p = 0.28; d = 0.49; the relative number of retrieved pups: Oxtr+/+, n = 10; OxtrΔ60/Δ60, n = 12; t(18); p = 0.28; d = 0.39). Finally, three chamber tests were performed to assess sociability and social novelty (Figure 4F). The male Oxtr+/+ prairie voles spent more time with another vole than with empty cup and with a novel vole rather than with a familiar one (sociability: n = 10; F3, 16 = 6.55; p = 0.01; d = 1.22; social novelty: F3, 16 = 4.31; p = 0.01; d = 1.42). OxtrΔ60/Δ60 voles spent significantly more time with another vole than with empty cup in sociability test, whereas the time they spent with a novel conspecific and a familiar one was not significantly different in the social novelty test (n = 10; sociability: p = 0.03; d = 1.95; social novelty: p = 0.47; d = 0.70). These data indicate that while sociability is not influenced by Oxtr, social novelty appears impaired in Oxtr mutant male prairie voles.
Figure 4.
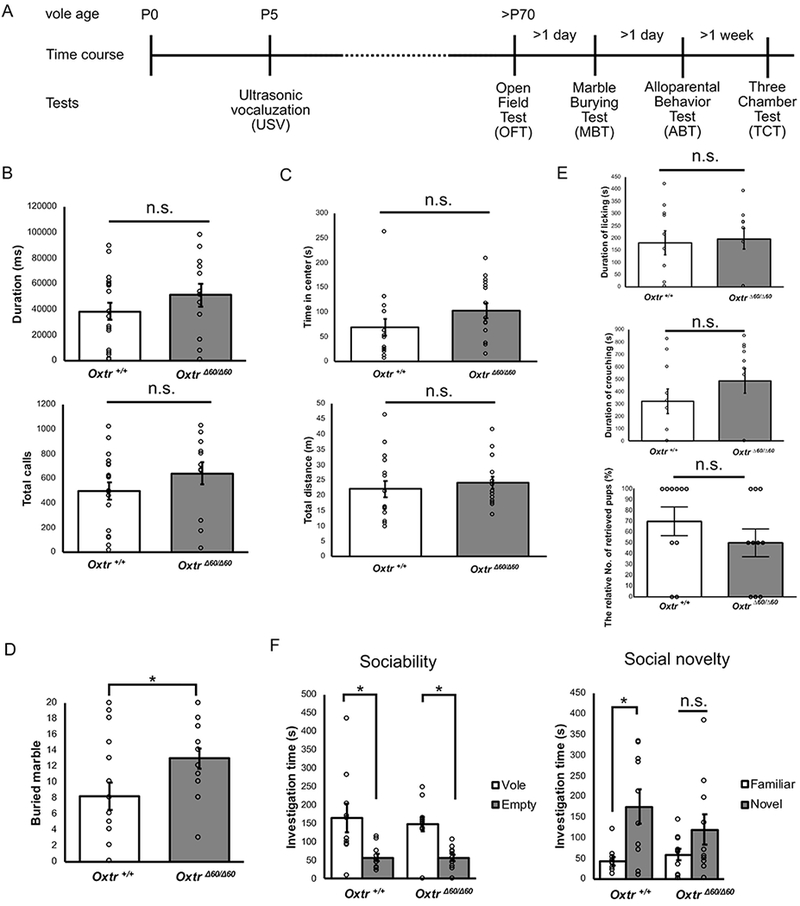
Behavioral phenotype of male OxtrΔ60/Δ60 prairie voles. (A) The time course of behavioral tests. Each animal was tested in each test. (B) The duration and number of total calls in USV tests. (C) The time in center and total distance in OFT. (D) The number of buried marbles in MBT (E) The duration of licking and crouching and the relative number of retrieved pups in ABT. (F) Sociability and social novelty in TCT. * = p<0.05. n.s. = not significant. Data were presented as mean +/− SEM.
Discussion
Here we report the successful generation of targeted Oxtr modified prairie voles by co-injecting Cas9 mRNAand an sgRNAinto the cytosol of zygotes. To our knowledge, this is the first published study to achieve targeted specific gene modification in prairie voles. We achieved efficient (100%) modification of the Oxtr gene in prairie voles by CRISPR/Cas9. This means that the sgRNAs which were chosen based on the SSA assay were highly effective. Each mutation was precisely induced around the 3-bp region next to the PAM sequence, which is the site at which Cas9 induces the double-strand break (Cong et al., 2013). These results suggested that the endogenous DNA repair pathway is conserved in prairie voles, as has been reported for mice and rats (Li et al., 2013b; Wang et al., 2013), making it possible to generate a wide array of mutants, including insertions of transgene sequences, such as fluorescent proteins, loxP sites, and Cre recombinase, in a targeted manner. While most mutant voles subjected to CRISPR/Cas9 editing had one or two mutated alleles, the v3 mutant vole had four different alleles in spite of the fact that the prairie vole is diploid. This suggests that genome editing occurred at the two cell stage in the v3 mutant vole resulting a chimera. Although the v5-2 allele had a 12 bp deletion around the DSB point, it had also another 16 bp deletion at the 5’ flanking region of the DSB point. It is possible that this allele was repaired by incomplete HDR using the other allele as the template. The functions of vl and v4-l mutant OXTRs were completely impaired in the TGFα shedding assay in vitro because the 1-bp deletion created a stop codon in the CDS1 and because the 60-bp deletion shortened the ligand binding domain, totally disrupting the function of OXTR. By contrast, the Oxtr v4-2 mutant with a 6-bp deletion retained slight activity, suggesting that this deletion was not sufficient to disrupt the function of OXTR in vitro. These data indicate that the full-length first extracellular domain of OXTR is important for transduction of the normal Gq/11 signaling.
The result of ligand binding assay with brain slices from OxtrΔ60/Δ60 voles confirmed that the Δ60 mutant OXTR is not functional in vivo. The in vivo activity of the Δ60 mutant OXTR was consistent with the in vitro function of the TGFα shedding assay, indicating that this assay has the potential to reflect the in vivo function of mutant GPCRs.
The induction of mutations by CRISPR/Cas9 in prairie voles was 100% efficient; all the voles obtained after zygote injection of Cas9 mRNA and an sgRNA had mutations at the targeted gene and no off-target effect was detected in our limited screen of likely off-target sites. The efficiency of genome editing is critical for the generation of targeted gene knock-in animals. Therefore, our procedure will enable the generation of gene knock-in prairie voles by CRISPR/Cas9. By using cloning-free knock-in methods, which is a highly efficient method involving the co-injection of Cas9 protein and two short RNAs, instead of an sgRNA, into pronuclear and cytosol of zygotes, we should soon be able to generate specific gene knock-in Oxtr prairie voles that will be of greater utility than the current KO vole (Aida et al., 2015).
We found that the male OxtrΔ60/Δ60 voles showed compulsive or repetitive-like behaviors in MBT and decreased social novelty in TCT, whereas they showed no deficits USV calls, locomotor activity, anxiety, alloparental behaviors and sociability.
OXT/OXTR signal is required for normal infant USV calls at P5 and P7 in male mice (Miyazaki et al., 2016; Takayanagi et al., 2005). In our current work, male OxtrΔ60/Δ60 prairie voles showed normal USV call frequency and duration after maternal separation at P5, suggesting that the regulatory mechanisms for mother calls may be different between mice and prairie voles at P5. The average of total mother calls for 5 min in wild type prairie voles were more than twice as much as that in wild type mice, consistent with differential regulation (Miyazaki et al., 2016). Infant USV calls are regulated not only by OXT, but also by vasopressin, dopamine and other factors in mice and rats (Curry et al., 2013; Fischer and Hammerschmidt, 2011; Scattoni et al., 2008). Our results are consistent with the hypothesis that USV calls induced by maternal separation in prairie voles are regulated by complex mechanisms including many neurotransmitters and their receptors.
Normal anxiety and locomotor activity of male OxtrΔ60/Δ60 prairie voles in the OFT suggests that Oxtr does not regulate non-social anxiety and locomotor activity in male prairie voles. Thus it is unlikely that behavioral deficits observed in future behavioral studies can be attributed to alterations in anxiety or general locomotion.
In mouse studies, abnormal repetitive marble burying behavior in MBT is regarded as a phenotype relevant to ASD (Angoa-Pérez et al., 2013; Silverman et al., 2010). Previous work showed that mice infected with lentivirus expressing Oxtr in nucleus accumbens, and intracerebroventricularly injected with OXT, reduced marble burying behaviors (Bahi et al., 2016; Sanathara et al., 2018). Additionally, lactating female mice intraperitoneally injected with OXTR antagonist buried a higher number of marbles than those injected with vehicle (Mitra et al., 2017). Although these results indicated that OXTR signaling modulates repetitive behavior in male and female mice, to our knowledge, there have been no study to analyze repetitive behavior in MBT with Oxtr gene modified mice, or prairie voles. The increase of buried marbles in the MBT in the male OxtrΔ60/Δ60 prairie voles suggested the role of Oxtr in prairie voles is also important for modulating repetitive behavior in the MBT, a function that is preserved across rodent species.
Robust paternal care, which consists of licking, huddling, and retrieving behaviors toward pups, is a characteristic of male prairie voles. In our current work, male OxtrΔ60/Δ60 prairie voles showed normal parental behaviors in the ABT. Previous work showed that co-administration of OXTR and vasopressin receptor la (AVPR1 A) antagonist decreased paternal behaviors, but each antagonist alone did not affect those behaviors (Bales et al., 2004), and also intraperitoneally administration of the OXTR antagonist, L-368,899, decreased alloparental behavior in male wild type prairie voles (Kenkel et al., 2017). These results are inconsistent with our results of male OxtrΔ60/Δ60 prairie voles. We hypothesize that OXTR antagonists like L-368,899, may not be highly specific for the prairie vole OXTR and may have some affinity for the AVPRla (Manning et al., 2008; Williams et al., 1994). Additionally, since OXTR and AVPR1A may both contribute to the regulation of paternal behaviors in male prairie voles, disruption of OXTR alone may not be sufficient to detect abnormal paternal behaviors. Previous studies have shown that a highly selective OXTR antagonist inhibits alloparental behavior in female prairie voles, but that study did not include males (Olazábal and Young, 2006).
Oxtr has also been reported to influence sociability and social novelty in the TCT in mice (Pobbe et al., 2012; Sala et al., 2011), but that effects were modest in some cases (Crawley et al., 2007). The sociability test detects the motivation to approach a novel conspecific relative to novel objects, and the social novelty test detects the preference for investigating an unfamiliar conspecific relative to a familiar conspecific, or the ability to distinguish familiar and unfamiliar social cues (e.g. social recognition). Reduction of these behaviors may be regarded as ASD-like behaviors in mice. The role of OXTR in regulation sociability and social novelty in prairie voles has not been investigated. Here, we found that OXTR was necessary for normal social novelty preference, but not sociability in male prairie voles in the TCT. Although we detected reduced social novelty, the change was modest and should be replicated. There was still some preference for the novel stimulus animal, although it was not significant.
We should note that the results provided herein represent our initial validation of the OxtrΔ60/Δ60 prairie vole as true null mutants and an initial screen of select behaviors relevant to ASD in males only. Although we restricted our study to males in this paper because we focused on autism related phenotypes and ASD is four times more prevalent in males, future studies can now use these OxtrΔ60/Δ60 voles to perform more comprehensive screens of both sexes to examine the effect of Oxtr knockout on a range of behaviors including maternal behavior, consolation and partner preference. Combined methods of immediate early gene detection in the vole brain after exposure to social stimuli and virus-mediated region-specific rescue of Oxtr will increase the understanding of the neural circuits in which Oxtr modulates social behaviors in voles.
General Conclusions
We successfully generated Oxtr knockout prairie voles by CRISPR/Cas9, which displayed some behavioral phenotypes that may be relevant to ASD. More importantly, we successfully demonstrated the high efficiency targeting of the prairie vole genome using the CRISPR/Cas9 system, opening the possibility of creating more mutant lines with greater utility, including enabling the expression of Cre recombinase, optogenetic proteins, and DREADDs in OXTR neurons. Additionally, future studies can target other genes such as Oxt, Avprla or dopamine receptor genes, greatly expanding the value of the prairie vole model for understanding the neural circuitry of social behavior. Finally, this work highlights the power of the CRISPR/Cas9 system for generating targeted mutants in non-traditional mammalian models that are useful behavioral neuroendocrine studies.
Supplementary Material
Highlights.
We successfully generated Oxtr gene modified prairie voles using CRISPR/Cas9.
OxtrΔ60/Δ60 prairie voles showed a complete absence of OXTR binding in brain.
Male OxtrΔ60/Δ60 prairie voles showed abnormal repetitive behavior in the marble burying test.
Male OxtrΔ60/Δ60 prairie voles showed impaired social novelty but normal sociability.
Genome editing will enhance the utility of prairie voles for studying mechanisms of behavior.
Acknowledgments
KH contributed all experiments. KI performed the OXTR autoradiography. SS performed TGFα shedding assay. LJY gave us prairie voles, consulted on experimental design and helped write the manuscript. TH, SY, SH supported management of the prairie voles. KN supervised our experiments. We thank Dr. Yuichi Hiraoka and Dr. Tomokazu Fukuda for comments and discussions. This research was supported by JSPS Grant-in-Aid for Scientific Research (A) [Grant Numbers 15H02442 (2015-2018)], JSPS Grant-in-Aid for challenging Exploratory Research [Grant Numbers 16K15698 (2016-2017)], JSPS Grant in Aid for JSPS Research Fellow [Grant Number 16J05070 (2016-2019)], MEXT Grant-in-Aid for Scientific Research on Innovative Areas “the evolutionary origin and neural basis of the empathetic systems” [Grant Numbers 16H01480 (2016-2017)], and Strategic Research Program for Brain Sciences from Japan Agency for Medical Research and development (AMED) [Grant Numbers 18dm0107076h0003 (2016-2020)]. KI and LJY’s contribution to this work was supported by NM grants R01MH096983, R01MH112788, R01MH112788 and P50MH100023 to LJY and P510D11132 to YNPRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors have no competing interests to declare.
References
- Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K, 2015. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 16, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadei EA, Johnson ZV, Jun Kwon Y, Shpiner AC, Saravanan V, Mays WD, Ryan SJ, Walum H, Rainnie DG, Young LJ, Liu RC, 2017. Dynamic corticostriatal activity biases social bonding in monogamous female prairie voles. Nature 546, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andari E, Duhamel JR, Zalla T, Herbrecht E, Leboyer M, Sirigu A, 2010. Promoting social behavior with oxytocin in high-functioning autism spectrum disorders. Proc Natl Acad Sci U S A 107, 4389–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angoa-Pérez M, Kane MJ, Briggs DI, Francescutti DM, Kuhn DM, 2013. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. J Vis Exp, 50978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona BJ, Liu Y, Yu YJ., Curtis JT, Detwiler JM, Insel TR, Wang Z, 2006. Nucleus accumbens dopamine differentially mediates the formation and maintenance of monogamous pair bonds. Nat Neurosci 9, 133–139. [DOI] [PubMed] [Google Scholar]
- Bahi A, Al Mansouri S, Al Maamari E, 2016. Nucleus accumbens lentiviral-mediated gain of function of the oxytocin receptor regulates anxiety- and ethanol-related behaviors in adult mice. Physiol Behav 164, 249–258. [DOI] [PubMed] [Google Scholar]
- Bales KL, Kim AJ, Lewis-Reese AD, Sue Carter C, 2004. Both oxytocin and vasopressin may influence alloparental behavior in male prairie voles. Horm Behav 45, 354–361. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P, 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Barrett CE, Arambula SE, Young LJ, 2015. The oxytocin system promotes resilience to the effects of neonatal isolation on adult social attachment in female prairie voles. Transl Psychiatry 5, e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beery AK, Christensen JD, Lee NS, Blandino KL, 2018. Specificity in Sociality: Mice and Prairie Voles Exhibit Different Patterns of Peer Affiliation. Front Behav Neurosci 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Dabrowska J, Modi ME, Johnson ZV, Keebaugh AC, Barrett CE, Ahern TH, Guo J, Grinevich V, Rainnie DG, Neumann ID, Young LJ, 2016. Oxytocin in the nucleus accumbens shell reverses CRFR2-evoked passive stress-coping after partner loss in monogamous male prairie voles. Psychoneuroendocrinology 64, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Nair HP, Ahern TH, Neumann ID, Young LJ, 2009. The CRF system mediates increased passive stress-coping behavior following the loss of a bonded partner in a monogamous rodent. Neuropsychopharmacology 34, 1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Young LJ, 2017. Oxytocin and Social Relationships: From Attachment to Bond Disruption. Curr Top Behav Neurosci. 35:97–117. DOI: 10.1007/7854_2017_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz EA, Zakon HH, 2015. Emerging from the bottleneck: benefits of the comparative approach to modern neuroscience. Trends Neurosci 38, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett JP, Andari E, Johnson ZV, Curry DC, de Waal FB, Young LJ, 2016. Oxytocin-dependent consolation behavior in rodents. Science 351, 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS, 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 230–232. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Chen T, Puri A, Washburn R, Sullivan TL, Hill JM, Young NB, Nadler JJ, Moy SS, Young LJ, Caldwell HK, Young WS, 2007. Social approach behaviors in oxytocin knockout mice: comparison of two independent lines tested in different laboratory environments. Neuropeptides 41, 145–163. [DOI] [PubMed] [Google Scholar]
- Curry T, Egeto P, Wang H, Podnos A, Wasserman D, Yeomans J, 2013. Dopamine receptor D2 deficiency reduces mouse pup ultrasonic vocalizations and maternal responsiveness. Genes Brain Behav 12, 397–404. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E, 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Young LJ, 2008. Oxytocin, vasopressin, and the neurogenetics of sociality. Science 322, 900–904. [DOI] [PubMed] [Google Scholar]
- Ferguson JN, Aldag JM, Insel TR, Young LJ, 2001. Oxytocin in the medial amygdala is essential for social recognition in the mouse. J Neurosci 21, 8278–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Hammerschmidt K, 2011. Ultrasonic vocalizations in mouse models for speech and socio-cognitive disorders: insights into the evolution of vocal communication. Genes Brain Behav 10, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobrogge KL, Liu Y, Young LJ, Wang Z, 2009. Anterior hypothalamic vasopressin regulates pair-bonding and drug-induced aggression in a monogamous rodent. Proc Natl Acad Sci U S A 106, 19144–19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ, 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649, 247–256. [DOI] [PubMed] [Google Scholar]
- Hai T, Teng E, Guo R, Li W, Zhou Q, 2014. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi R, Neumaier JF, 2006. Differential effects of ovarian steroids on anxiety versus fear as measured by open field test and fear-potentiated startle. Behav Brain Res 166, 93–100. [DOI] [PubMed] [Google Scholar]
- Horie K, Hidema S, Hirayama T, Nishimori K, 2015. In vitro culture and in vitro fertilization techniques for prairie voles (Microtus ochrogaster). Biochemical and biophysical research communications 463, 907–911. [DOI] [PubMed] [Google Scholar]
- Inoue A, Ishiguro J, Kitamura H, Arima N, Okutani M, Shuto A, Higashiyama S, Ohwada T, Arai H, Makide K, Aoki J, 2012. TGFα shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods 9, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Insel TR, Shapiro LE, 1992. Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Proc Natl Acad Sci U S A 89, 5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Walum H, Jamal YA, Xiao Y, Keebaugh AC, Inoue K, Young LJ, 2016. Central oxytocin receptors mediate mating-induced partner preferences and enhance correlated activation across forebrain nuclei in male prairie voles. Horm Behav 79, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Walum H, Xiao Y, Riefkohl PC, Young LJ, 2017. Oxytocin receptors modulate a social salience neural network in male prairie voles. Horm Behav 87, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Young LJ, 2015. Neurobiological mechanisms of social attachment and pair bonding. Curr Opin Behav Sci 3, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Young LJ, 2017. Oxytocin and vasopressin neural networks: Implications for social behavioral diversity and translational neuroscience. Neurosci Biobehav Rev 76, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, Young LJ, 2018. Evolutionary diversity as a catalyst for biological discovery. Integr Zool. doi: 10.1111/1749-4877.12339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, Young LJ, 2015. RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibits social attachment and parental care in monogamous female prairie voles. Soc Neurosci 10, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel WM, Perkeybile AM, Carter CS, 2017. The neurobiological causes and effects of alloparenting. Dev Neurobiol 77, 214–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Babayan BM, Rubinstein ND, Autry AE, Marin-Rodriguez B, Kapoor V, Miyamishi K, Zweifel LS, Luo L, Uchida N, Dulac C, 2018. Functional circuit architecture underlying parental behaviour. Nature 556, 326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Z, Wu Q, Kou X, Yin C, Wang H, Zuo Z, Zhuo Y, Chen A, Gao S, Wang X, 2015. CRISPR/Cas9-mediated genome engineering of the ferret. Cell Res 25, 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, 2013a. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31, 681–683. [DOI] [PubMed] [Google Scholar]
- Li W, Teng F, Li T, Zhou Q, 2013b. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31, 684–686. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM, 2013. RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning M, Stoev S, Chini B, Durroux T, Mouillac B, Guillon G, 2008. Peptide and non-peptide agonists and antagonists for the vasopressin and oxytocin V1a, V1b, V2 and OT receptors: research tools and potential therapeutic agents. Prog Brain Res 170, 473–512. [DOI] [PubMed] [Google Scholar]
- Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M, 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3, 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Young LJ, 2010. The prairie vole: an emerging model organism for understanding the social brain. Trends Neurosci 33, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Mucha M, Owen S, Bult-Ito A, 2017. Postpartum Lactation-Mediated Behavioral Outcomes and Drug Responses in a Spontaneous Mouse Model of Obsessive-Compulsive Disorder. ACS Chem Neurosci 8, 2683–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S, Hiraoka Y, Hidema S, Nishimori K, 2016. Prenatal minocycline treatment alters synaptic protein expression, and rescues reduced mother call rate in oxytocin receptor-knockout mice. Biochem Biophys Res Commun 472, 319–323. [DOI] [PubMed] [Google Scholar]
- Modi ME, Inoue K, Barrett CE, Kittelberger KA, Smith DG, Landgraf R, Young LJ, 2015. Melanocortin Receptor Agonists Facilitate Oxytocin-Dependent Partner Preference Formation in the Prairie Vole. Neuropsychopharmacology. 40, 1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR, Crawley JN, 2004. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav 3, 303–314. [DOI] [PubMed] [Google Scholar]
- Olazábal DE, Young LJ, 2005. Variability in “spontaneous” maternal behavior is associated with anxiety-like behavior and affiliation in naive juvenile and adult female prairie voles (Microtus ochrogaster). Dev Psychobiol 47, 166–178. [DOI] [PubMed] [Google Scholar]
- Olazábal DE, Young LJ, 2006. Oxytocin receptors in the nucleus accumbens facilitate “spontaneous” maternal behavior in adult female prairie voles. Neuroscience 141, 559–568. [DOI] [PubMed] [Google Scholar]
- Pobbe RL, Pearson BL, Defensor EB, Bolivar VJ, Young WS, Lee HJ, Blanchard DC, Blanchard RJ, 2012. Oxytocin receptor knockout mice display deficits in the expression of autism-related behaviors. Horm Behav 61, 436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl TT, Young LJ, Bosch OJ, 2018. Lost connections: Oxytocin and the neural, physiological, and behavioral consequences of disrupted relationships. Int J Psychophysiol. DOI: 10.1016/j.ijpsycho.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HE, Freeman SM, Spiegel LL, Ren X, Terwilliger EF, Young LJ, 2009. Variation in oxytocin receptor density in the nucleus accumbens has differential effects on affiliative behaviors in monogamous and polygamous voles. J Neurosci 29, 1312–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala M, Braida D, Lentini D, Busnelli M, Bulgheroni E, Capurro V, Finardi A, Donzelli A, Pattini L, Rubino T, Parolaro D, Nishimori K, Parenti M, Chini B, 2011. Pharmacologic rescue of impaired cognitive flexibility, social deficits, increased aggression, and seizure susceptibility in oxytocin receptor null mice: a neurobehavioral model of autism. Biol Psychiatry 69, 875–882. [DOI] [PubMed] [Google Scholar]
- Sanathara NM, Garau C, Alachkar A, Wang L, Wang Z, Nishimori K, Xu X, Civelli O, 2018. Melanin concentrating hormone modulates oxytocin-mediated marble burying. Neuropharmacology 128, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, McFarlane HG, Zhodzishsky V, Caldwell HK, Young WS, Ricceri L, Crawley JN, 2008. Reduced ultrasonic vocalizations in vasopressin 1b knockout mice. Behav Brain Res 187, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN, 2010. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci 11, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AS, Wang Z, 2014. Hypothalamic oxytocin mediates social buffering of the stress response. Biol Psychiatry 76, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, Yanagisawa T, Kimura T, Matzuk MM, Young LJ, Nishimori K, 2005. Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proc Natl Acad Sci U S A 102, 16096–16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R, 2009. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl) 204, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walum H, Young LJ, 2018. The neural mechanisms and circuitry of the pair bond. Nature Rev Neurosci doi: 10.1038/s41583-018-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R, 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA, 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338. [DOI] [PubMed] [Google Scholar]
- Williams PD, Anderson PS, Ball RG, Bock MG, Carroll L, Chiu SH, Clineschmidt BV, Culberson JC, Erb JM, Evans BE, 1994. 1-((7,7-Dimethyl-2(S)-(2(S)-amino-4-(methylsulfonyl)butyramido)bicyclo [2.2.1]-heptan-1(S)-yl)methyl)sulfonyl)-4-(2-methylphenyl)piperaz ine (L-368,899): an orally bioavailable, non-peptide oxytocin antagonist with potential utility for managing preterm labor. J Med Chem 37, 565–571. [DOI] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B, 2014. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182. [DOI] [PubMed] [Google Scholar]
- Yartsev MM, 2017. The emperor’s new wardrobe: Rebalancing diversity of animal models in neuroscience research. Science 358, 466–469. [DOI] [PubMed] [Google Scholar]
- Yokoi S, Okuyama T, Kamei Y, Naruse K, Taniguchi Y, Ansai S, Kinoshita M, Young LJ, Takemori N, Kubo T, Takeuchi H, 2015. An essential role of the arginine vasotocin system in mate-guarding behaviors in triadic relationships of medaka fish (Oryzias latipes). PLoS Genet 11, e1005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KA, Gobrogge KL, Liu Y, Wang Z, 2011. The neurobiology of pair bonding: insights from a socially monogamous rodent. Front Neuroendocrinol 32, 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LJ, Barrett CE, 2015. Neuroscience. Can oxytocin treat autism? Science 347, 825–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LJ, Wang Z, 2004. The neurobiology of pair bonding. Nat Neurosci 7, 1048–1054. [DOI] [PubMed] [Google Scholar]
- Zou Q, Wang X, Liu Y, Ouyang Z, Long H, Wei S, Xin J, Zhao B, Lai S, Shen J, Ni Q, Yang H, Zhong H, Li L, Hu M, Zhang Q, Zhou Z, He J, Yan Q, Fan N, Zhao Y, Liu Z, Guo L, Huang J, Zhang G, Ying J, Lai L, Gao X, 2015. Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol 7, 580–583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


