Abstract
Graphene oxide (GO) has attracted enormous interests due to its extraordinary properties. Recent studies have confirmed the cytotoxicity of GO, we further investigate its mutagenic potential in this study. The results showed that GO interfered with DNA replication and induced mutagenesis at molecular level. GO treatments at concentrations of 10 and 100 μg/mL altered gene expression patterns at cellular level, and 101 differentially expressed genes mediated DNA-damage control, cell apoptosis, cell cycle, and metabolism. Intravenous injection of GO at 4 mg/kg for 5 consecutive days clearly induced formation of micronucleated polychromic erythrocytes in mice, and its mutagenesis potential appeared to be comparable to cyclophosphamide, a classic mutagen. In conclusion, GO can induce mutagenesis both in vitro and in vivo, thus extra consideration is required for its biomedical applications.
Graphene, firstly isolated from graphite in 20041, is a flat monolayer of carbon atoms tightly packed into a two-dimensional (2D) honeycomb lattice. Due to the unique physicochemical properties, high surface area, excellent thermal conductivity, electric conductivity, and strong mechanical strength, graphene and graphene oxide (GO) have shown great promise in many applications, such as electronics, energy storage and conversion, mechanics, and biotechnologies2,3,4,5,6. Recently, many studies reported that GO has outstanding potentials in the field of biomedicine. GO and PEGylated GO exhibit certain advantages in vitro and in vivo drug delivery, such as high drug loading efficiency, controlled drug release, tumor-targeting drug delivery, and reversal effect against cancer drug resistance7,8,9,10. In addition, GO has strong optical absorbance in the near-infrared (NIR) region, thus is suitable for the photothermal therapy11,12,13.
Now, it is possible to manufacture high-quality GO in large scale quantities14,15, and its industry production is increasing exponentially. Together with its potential applications in the biomedical field, the biosafety of GO is of critical importance. Many investigations have paid attentions to its biocompatibilty16,17,18,19. At a concentration approximate to 50 μg/mL or higher, GO begins to show the toxicity against erythrocytes, fibroblasts, and PC12 cells. It can induce cell apoptosis, hemolysis, and oxidative stress16,18,19. Surface chemical modification, such as PEGylation, is likely to improve the biocompatibility of GO20,21. However, the chemical bonds linking GO with modified polymer can be broken down in vivo, thus surface-modified GO can also induce in vivo toxicity.
Several investigations have reported that treatments with carbon nanomaterials, such as nanodiamonds and multiwalled carbon nanotubes, can elevate the expression of p53, MOGG-1, and Rad51, which reflect the chromosomal DNA damage22,23. However, it is not clear whether this DNA damage induced by carbon nanomaterials can cause mutagenesis. GO, due to its unique nanosheet structure, can interact with DNA. The interactions include DNA-intercalation and cleavage24. We therefore hypothesize that GO can perhaps induce mutagenesis and interfere with the flow of genetic information. Such mutagenic effects can be accumulated upon long term use and be passed on to next generation by germline mutation, thus is very dangerous. In this study, we firstly investigated the potential mutagenic effect of GO at molecular, cellular, and animal levels.
Results
Preparation and characterization of GO, and its anti-proliferative effect against cells
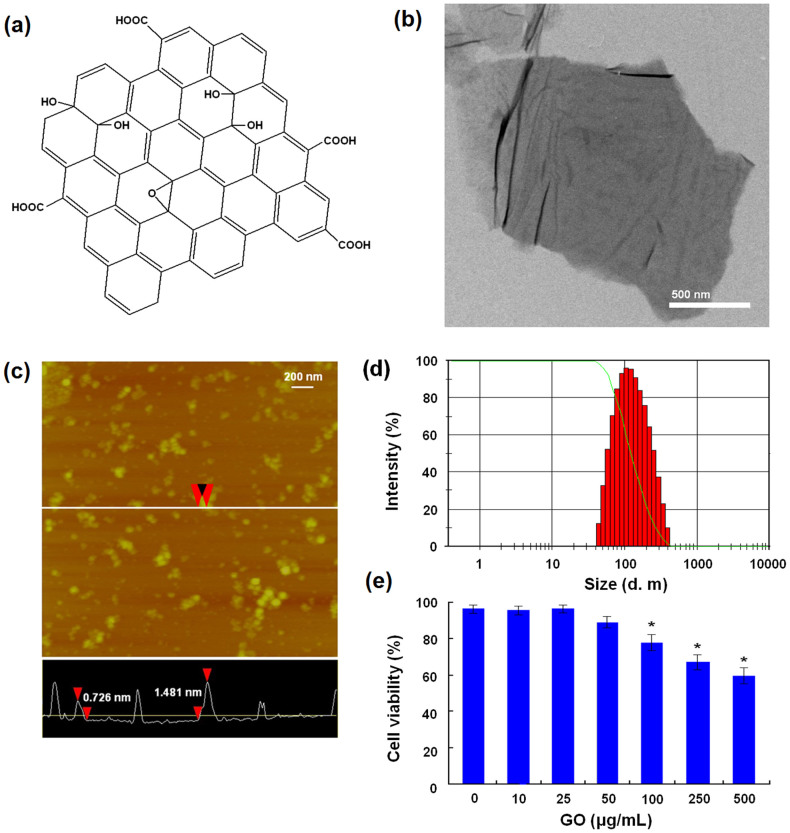
GO with carboxylic acid group content of 8.1% was prepared from purified natural graphite25, and its chemical structure was shown in Fig. 1a. The synthesized GO showed the irregular morphology with micro-scale under the TEM observation (Fig. 1b). Then, GO was dispersed in water and small sized by the sonication, according to our previous protocol26 (Supplementary information). The morphology and size of GO were characterized by atomic force microscope (AFM) and laser light scattering. Small sized GO had sheet structures and their thickness ranged from 0.7 to 1.5 nm, suggesting a single- or double-layered structure (Fig. 1c). Mean diameter of GO sheets was 156.4 nm and the polydispersity index was 0.386 (Fig. 1d). In addition, GO was also characterized by means of fourier transform infrared (FTIR), Ultra-visible (UV-vis), Raman, X-ray diffraction (XRD) and thermal gravimetric analysis (TG), and all results (Supplementary information) were basically consistent with the previous studies27,28,29. Next, we evaluated the cytotoxicity of GO by Cell Counting Kit-8 (CCK8) assay. After co-incubation for 48 h, GO exhibited dose-dependent cytotoxicity against MDA-MB-231 human breast cancer cells at concentrations ranging from 100 μg/mL to 500 μg/mL (Fig. 1e).
Figure 1. Characterization of GO and its impairment of cell viability.
(a) Chemical structure of GO. (b) TEM image of GO. (c)AFM image of small sized GO. (d) Size distribution of small sized GO. (e) Cytotoxicity of GO against MDA-MB-231 cells. * indicates P < 0.05 for comparison with the control at GO concentration of 0 μg/mL.
GO interfered with DNA replication and induced mutagenesis at molecular level
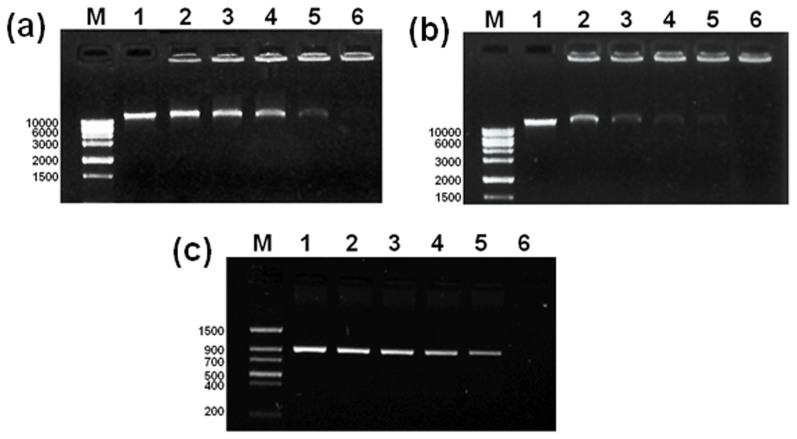
It has been reported that GO nanosheets can intercalate efficiently into DNA molecules24. We therefore hypothesize that GO may interact with genomic DNA and interfere with gene replication. To test this hypothesis, we examined the interaction between GO and genomic DNA. The genomic DNA was extracted, treated with GO at different concentrations for 2 h, and subjected to electrophoresis (Fig. 2a). Intensity of genomic DNA gradually decreased as GO concentration increasing and completely disappeared when GO concentration reached to 600 μg/mL, indicating that GO entirely interacted with genomic DNA. This interaction proceeded at a rapid speed, reaching the maximum effect at 2 h upon GO treatment (Fig. 2b). Next, the effect of GO in DNA replication was examined by polymerase chain reaction (PCR) using 50 ng genomic DNA as a template (Fig. 2c). GO inhibited replication of human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) gene in a dose-dependent manner, and completely blocked the replication at 1 μg/mL (total 25 ng GO in each reaction), a dose much lower than that was required to interact with genomic DNA.
Figure 2. The interaction of GO with genomic DNA and its interference with DNA replication.
(a) GO interacted with genomic DNA. Genomic DNA was incubated with GO at different concentrations for 2 h and then subjected to gel electrophoresis (lanes 1–6: the control and GO treatments at 200, 300, 400, 500, and 600 μg/mL). Full-length gels are presented in Supplementary Figure 2a. (b) Kinetic analysis of the interaction between GO and genomic DNA. Genomic DNA was incubated with GO at 600 μg/mL for 0–2 h (lane 1–6: the control and GO treatments at 10, 40, 80 min, and 2 h). Full-length gels are presented in Supplementary Figure 2b. (c) Impairment of GO on the gene replication by using PCR assay. 50 ng of genomic GNA was used as templates and the reaction mixture was 25 μL. (lane 1–6: the control and GO treatments at 200, 400, 600, 800, and 1000 ng/mL). Full-length gels are presented in Supplementary Figure 2c.
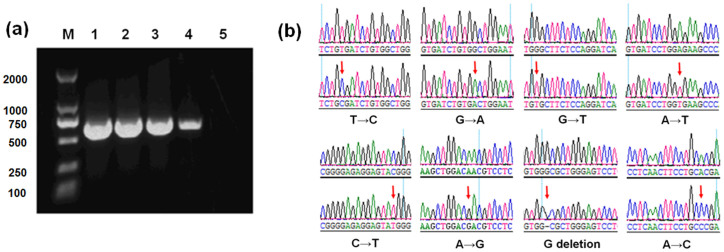
We next examined the mutagenic effect of GO on gene replication by PCR assays. 5 ng plasmid DNA containing human gene protein kinase Cζ (PKCζ) was used as a template for PCR in the presence of GO. As shown in Fig. 3a, band intensity of PCR product reduced when GO concentration increased and could not be detected at GO concentration of 100 ng/mL (total 2.5 ng GO in each reaction). Thus, GO blocked replication when the mass ratio of GO to template DNA was higher than 0.5. PCR product was cloned into pMD19T vectors, and transformed into Escherichia coli (E. coli) DH5α. The clones of the control and GO-treatment groups were then sequenced to examine the fidelity of gene replication. Mean mutation rate of the control group was 0.38%, probably due to the intrinsic properties of Taq DNA polymerase and the reaction condition (Table 1). GO treatment at 10 ng/mL, insufficient concentration to block replication, induced a 37% increase in mutation rate, reaching 0.52% (Table 1). Mutation types included T → C, C → T, G → A, A → G, G → T, A → T, A → C transitions, and G deletion (Fig. 3b). The above results confirmed mutagenic effect of GO on gene replication at molecular level.
Figure 3. Interference of GO with DNA replication and its mutagenic effects.
(a) GO treatment inhibited the replication of PKCζ gene. 5 ng of plasmid DNA was used as templates and the reaction mixture was 25 μL. (lanes 1–5: the control and GO treatments at 1, 10, 50, and 100 ng/mL). Full-length gels are presented in Supplementary Figure 3. (b) Automated DNA sequence analysis of gene mutation.
Table 1. Mutation effect of GO on PKCζ gene that assessed by the clone experiment.
| Group | Experiment No. | Clone number | Total gene length (bp) | Mutation number (bp) | Mut. Ratea (‰) | Mean mut. rate (‰) |
|---|---|---|---|---|---|---|
| Control | 1 | 32 | 19200 | 7 | 0.36 | 0.38 |
| 2 | 32 | 19200 | 7 | 0.36 | ||
| 3 | 32 | 19200 | 8 | 0.42 | ||
| GO | 1 | 32 | 19200 | 10 | 0.52 | 0.52b |
| 2 | 32 | 19200 | 10 | 0.52 | ||
| 3 | 33 | 19800 | 10 | 0.51 |
aMut. rate (mutation rate) was calculated by dividing the mutation number observed by the total gene length.
bCompared to the control, GO-treatment group at 10 ng/mL showed statistically significant difference in mutation rate (p < 0.05).
GO interfered with gene expression and induced DNA-damage response, cell apoptosis, and cell cycle changes at cellular level
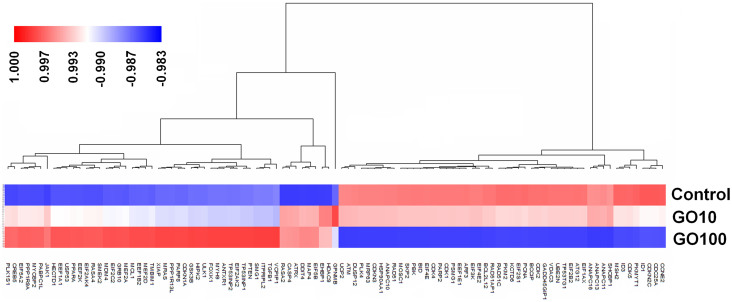
Microarray experiment was used to examine the impact of GO treatment on gene expression at cellular level. Among 30,000 genes tested (the Affymetrix GeneChip Human Genome U133 Plus 2.0 array), GO treatments at 10 μg/mL and 100 μg/mL induced marked changes of 186 and 3693 gene expression patterns, respectively. 101 genes showing 2-fold or greater expression changes in both two GO treatment groups were then grouped by the complete-kink hierarchical clustering method using Genesight 6.4 (Fig. 4)30. These genes mainly mediated DNA-damage control, cell apoptosis, cell cycle, and metabolism (Table 2).
Figure 4. Hierarchical clustering analysis of genes with significantly differential expression between the control and GO treatment groups at doses of 10 μg/mL (GO10) and 100 μg/mL (GO100).
Table 2. Genes with differential expressions grouped into potentially relevant functional classes of biologic activitya.
| Signal pathway | Deferentially expressed genes with 2-fold or greater change |
|---|---|
| DNA damage | DDIT4, MYH9, TP53INP1, TP53INP2, HECTD1, SMG1, VCPIP1, MDM4, HIPK2, ATM, UBE2N, RAD51, RAD51AP1, RAD51C, PBK, MSH2, MRP63, KCTD6, PSMG1, DUSP12, TP53TG1 |
| Cell cycle | USP53, MAP4, GPNMB, PLK1S1, TGFβ1, ATRX, HDAC9, SMEK2, FOXK1, MYCBP2, CDKN1A, BCCIP, CCNE2, ID1, ID3, CDK1, CDK2, CDK4, CDK5, CDKN2C, CDKN3, CDC25A, MOSC1, SKP2, PCNA, PKMYT1, PLK4, ANAPC10, ANAPC11, ANAPC13, ANAPC16, GADD45GIPI, ARF3 |
| Cell apoptosis | CASP4, TMBIM1, ITPRIPL2, XIAP, GSK3B, RASA2, RASA4, CREB5, PARP8, MCL1, ULK1, PTEN, MRAS, JAK1, ANTXR1, BCL2L12, HSP90AA1, BID, PARP2, ATG12 |
| Translational control | PPP1R13L, PPP1R9A, PPPRARA, EEF2K, EEF1B2, PABPC1L, EEF1A1, EIF2C2, EIF5A2, EIF5B, EIF2AK2, EIF2AK4, EIF1AX, EEF1E1, EIF2B2, EIF4E, EIF4E2, EIF3K, EIF2S1 |
| Cellular metabolism | GRB10, EHBP1, MEF2A, MEF2D, PKM2, UCP2, VDAC3, SHCBP1 |
aRegular and boldface italic font respectively indicate significantly higher and lower expressed genes in both GO treatment groups (10 μg/mL and 100 μg/mL), compared to the control group.
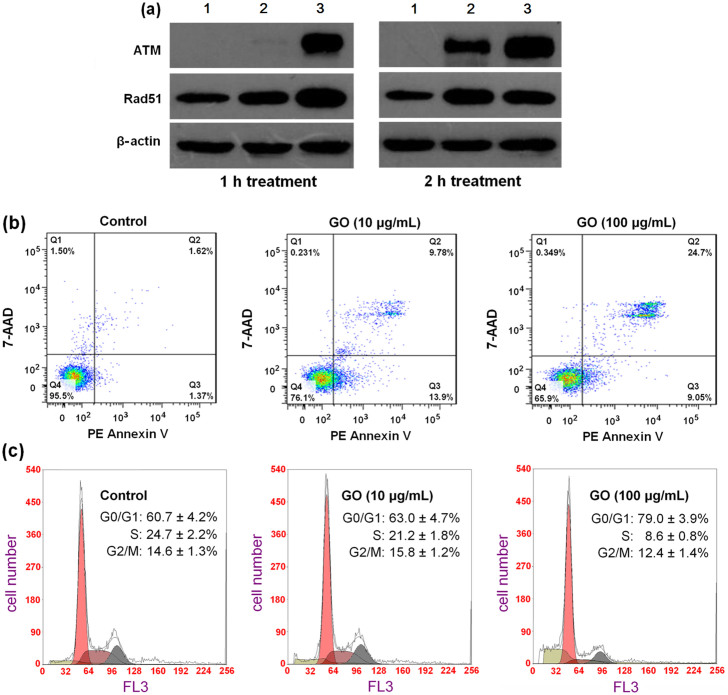
To preliminarily confirm above gene expression results, we used western blot analysis and flow cytometry assay to assess DNA-damage, cell apoptosis and cell cycle changes at cellular level. ATM and Rad51, as DNA repair proteins, remain inactive under the normal conditions and become active upon DNA damage caused by irradiation, oxidative stress, etc. Therefore, the transient increases of ATM and Rad51 expressions are often observed at the beginning of DNA damage22,23. The results of western blot analysis (Fig. 5a) showed that GO treatments at 10 and 100 μg/mL induced aberrant increases of ATM and Rad51 expressions in cells during a short term (1 and 2 h), confirming the occurrence of DNA damage. Fig. 5b shows the results of the apoptosis evaluation of cells treated with GO for 24 h. Cell apoptosis (Fig. 5b, Q1 + Q2) was clearly observed even at a low GO concentration of 10 μg/mL. With the increase of GO concentration, the fraction of late apoptotic cells (Fig. 5b, Q2) significantly increased, further confirming that GO treatment induced cell apoptosis. Fig. 5c shows the cell cycle distributions determined by the flow cytometry. At GO concentration of 100 μg/mL, the proportion of G0/G1 phase cells increased to 79.0 ± 3.9%, significantly higher (p < 0.01) than that of the negative control (60.7 ± 4.2%), which indicated that cells were arrested at the G0/G1 phase. At the same time, the proportion of cells at S-phase decreased from 24.7% to 8.6 ± 0.8%, suggesting that GO significantly inhibited the DNA synthesis. The above results suggested that GO treatment at 100 μg/mL evidently inhibited the transition from G0/G1 to S phase in cell cycle.
Figure 5. The induction effects of GO on DNA-damage, cell apoptosis and cell cycle arrest.
(a) Western blot analysis of ATM and Rad51 protein expressions in MDA-MB-231 cells treated with GO for 1 and 2 h. (lanes 1–3: the control and GO treatments at 10 and 100 μg/mL). Full-length blots are presented in Supplementary Figure 4. (b) GO treatment induced cell apoptosis. Cells stained with PE annexinV/7-AAD and analyzed by flow cytometry. Q1: dead cells; Q2: late apoptotic cells; Q3: early apoptotic cells; Q4: live cells. (c) GO treatment changed cell cycle distribution based on flow cytometry analysis. All experiments were repeated at least three times. * indicates P < 0.05 for comparison with the control at GO concentration of 0 μg/mL.
GO induced micronucleated polychromatic erythrocytes (MNPCEs) in mice
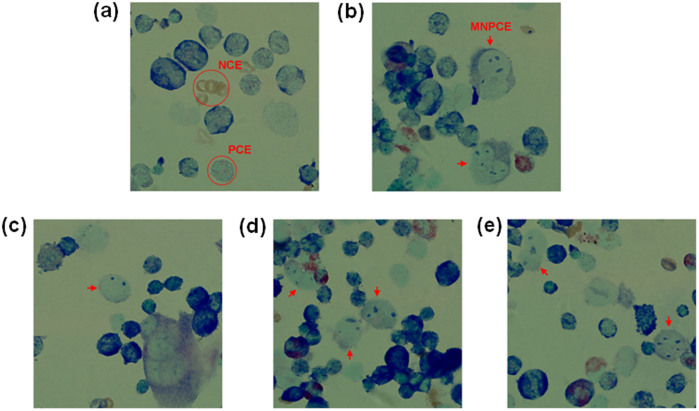
Next, we tested the hypothesis that GO could induce mutagenesis at animal level by using micronucleus test. Micronucleus test, developed by Schemid in 197531, is a classical assay for mutagenesis. It measures formation of micronucleated polychromatic erythrocytes (MNPCEs) and directly assesses the frequency of structural or numerical chromosome aberrations. Cyclophosphamide, a potent mutagen, induced formation of MNPCE at 50 mg/kg (Fig. 6b). The micronuclei were round or oval in shape and had a diameter about 1/20 to 1/5 that of PCEs, consistent with previous report32. Intravenous injection of GO at 1, 2, 4 mg/kg for 5 consecutive days, corresponding respectively to the total doses of 5, 10 and 20 mg/kg, clearly induced the formation of MNPCE in mice (Fig. 6c–e). As shown in Table 3, the frequencies of MNPCEs in GO treated mice exhibited dose-dependent property, and increased from 2.4 ± 0.74 to 7.0 ± 1.31 per 1000 erythrocytes with GO dose increasing from 5 to 20 mg/kg. There were significant differences between GO treatment groups and the negative control group (1.1 ± 0.64 MNPCEs per 1000 erythrocytes), further confirming the mutagenic effect of GO. Moreover, the calculated ratio of PCE/NCE in bone marrow preparations showed no statistically significant differences when compared to the negative control group, indicating that GO at 20 mg/kg didn't directly kill erythrocytes (Table 3). Together, the results suggested that GO may not be a dangerous cytotoxin, but is a potential inducer of mutagenesis.
Figure 6. Representative pictures of mouse bone marrow cells in microbucleus test.
(20× magnification, Giemsa staining). (a) The negative control with physiological saline treatment. (b): the positive control with cyclophosphamide treatment. (c–e) GO treatments at 5, 10 and 20 mg/kg, respectively. PCE: polychromatic erythrocyte; NCE: normochromatic erythrocyte; MNPCE: micronucleus (MN) in polychromatic erythrocyte (red arrow).
Table 3. Polychromatic erythrocytes with micronuclei (MNPCE) observed in bone marrow cells of female (F) and male (M) mice treated with GO and respective controls.
| Number of MNPCE per animala | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Groups | Dose (mg/kg) | F1 | F2 | F3 | F4 | M1 | M2 | M3 | M4 | MNPCE (Mean ± SD) | PCE/NCEb (Mean ± SD) |
| Negative | 0 | 1 | 2 | 1 | 1 | 2 | 1 | 0 | 1 | 1.1 ± 0.64 | 1.88 ± 0.25 |
| GO1 | 5 | 3 | 3 | 1 | 3 | 2 | 3 | 2 | 2 | 2.4 ± 0.74* | 1.78 ± 0.27 |
| GO2 | 10 | 3 | 4 | 5 | 3 | 3 | 2 | 3 | 5 | 3.5 ± 1.07** | 1.67 ± 0.24 |
| GO4 | 20 | 6 | 8 | 5 | 9 | 7 | 6 | 7 | 8 | 7.0 ± 1.31*** | 1.69 ± 0.34 |
| Positive | 50 | 12 | 10 | 13 | 9 | 10 | 8 | 9 | 11 | 10.3 ± 1.67*** | 1.72 ± 0.18 |
aTwo thousand cells were analyzed per animal, for a total of 16,000 cells per group.
bPCE/NCE: the ratio of polychromatic erythrocytes to normochromatic erythrocytes, and determined on total of 1000 erythrocytes counted.
Signifcantly different from negative control (*P < 0.05, **P < 0.01, and ***P < 0.001).
Discussion
Several reports have shown that treatments with carbon nanomaterials, such as nanodiamonds and multiwalled carbon nanotubes, can elevate the expressions of p53, MOGG-1, and Rad51, reflecting the chromosomal DNA damage22,23. However, it is currently not clear whether this DNA damage induced by carbon nanomaterials can induce mutagenesis. Due to the unique nanosheet structure of GO and its potential biomedical applications7,8,9,10,11,12,13,20,26,33, we paid attentions to its mutagenic effects in this study, and the results confirmed that small sized GO induced mutagenesis both in vitro and in vivo.
At the molecular level, GO interacted with genomic DNA and interfered with DNA replication fidelity. We believed the nanosheet structure of GO made it easily interact with genomic DNA. The highly planar GO nanosheet is very similar to the planar aromatic ring structures of many DNA intercalators, such as berberine, ethidium bromide and proflavine. It was thus deduced that GO could also insert between base pairs of double strand DNA and interfere with the flow of genetic information like those DNA intercalators34,35,36. The previous investigations have confirmed that GO nanosheets can intercalate efficiently into DNA molecules24. We therefore believed the interaction between GO and DNA was one of the main reasons for the mutagenic effect of GO. In addition, GO and other carbon nanomaterials can induce the cellular generation of reactive oxygen species (ROS), which can also result the potential of DNA damage18,22,23. Perhaps, there were other indirect mechanisms, because GO could also react with other cell components such as proteins and polysaccharides due to its high surface activity.
At the cellular level, GO altered the expression of a large repertoire of genes at the concentration of 10 μg/mL. Among these genes, ataxia telangiectasia mutated (ATM) protein kinase and recombination protein A (Rad51) (Fig. 4) are critical components of a DNA-damage response network configured to maintain genomic integrity. ATM- and/or Rad51 differentially expressed cells are characterized by genomic instability including chromosome breaks, chromosome gaps, translocations, and aneuploidy37,38,39. Caspase 4-apoptosis-related cysteine peptidase (CASP4) (Fig. 4) plays a central role in the execution-phase of cell apoptosis. Up-regulation of CASP4 expression often induces cell apoptosis40. CDK2 and CDK4 (Fig. 4), members of cyclin-dependent kinases (serine/threonine kinases), play important roles in regulating the cell cycle transition from G1 to S phase, and their down-expressions often increase G0/G1 cell cycle arrest41. The next cell experiments confirmed GO treatment could induce DNA-damage and cell apoptosis even at a low dose of 10 μg/mL. However, the significant cell cycle changes were only observed at GO dose of 100 μg/mL, which was potentially due to the adequate DNA-repair effect of ATM and RAD51 proteins.
A number of studies have investigated the biocompatibility and biosafety of GO, showing that GO induced cytotoxicity, oxidative stress, and apoptosis16,17,18,19,20 at cellular level, and also caused the lung granuloma formation in mice after a single tail vein injection at a high dose (20 mg/kg) due to its accumulation in the lung17. However, our results firstly confirmed that GO induced the formation of MNPCE at the animal level and its potence at the dose of 20 mg/kg was equivalent to that of cyclophosphamid (50 mg/kg), which is a classic mutagene. The mutagenic potential of GO is perhaps a more dangerous threat than the adverse effects listed above, because it is also possible to induce germline mutation that threatens the health of next generation. In a conclusion, the dosage of GO needs to be specially considered during its in vivo applications, such as the uses as drug carriers and the photothermal therapeutic agents. According to the previous reports20,21, surface chemical modification is likely to improve the biocompatibility of GO, but the breaking of chemical bonds can also cause GO release in vivo. Thus, extra consideration on the stability of GO with surface modification is also required for its biomedical applications.
Methods
Preparation and characterization of GO
GO was prepared from purified natural graphite according to a modified Hummers method25, following our previous protocol7. The content of carboxylic acid groups of GO was determined by the acid-base titration method that we previously reported, and the value was about 8.1%. The morphology of synthesized GO was characterized by transmission electron microscopy (TEM, FEI, TECNAI-20). The detail preparation and characterization methods were demonstrated in Supplementary information. GO was then dispersed in deionized water at the concentration of 1 mg/mL, sonicated at 30 W for 3 min by a probe type sonifier (Automatic Ultrasonic Processor UH-500A, China) using a pulse function (pulse on 2.0 s, pulse off 2.0 s), and the sonication was repeated three times. Next, GO dispersion was centrifuged at 5000 rpm for 5 min to obtain GO aqueous dispersion. The morphology of small sized GO in above dispersion was characterized by atomic force microscope (AFM, Veeco, Nanoscope IV, Digital Instruments), and the size and size distribution at GO concentration of 0.25 mg/mL were monitored a Malvern Zetasizer Nano ZS instrument.
Cytotoxicity assay
The cytotoxicity of GO was assessed by Cell Counting Kit-8 (CCK8) assay (Cell Counting Kit, Dojindo Laboratories, Kumamoto, Japan). Briefly, MDA-MB-231 human breast cancer cells (ATCC, Rockville, MD) were seeded into 96-well plates at an initial density of 5 × 103 cells/well in 100 μL of culture medium and cultured for 24 h. The original media were then removed and replaced with culture media containing GO with concentrations from 10 μg/mL to 500 μg/mL, and the cells were continually incubated for 48 h. The absorbance of each well was then measured using a multiwell spectrophotometer reader (Bio-tek EPOCH, ELX 800) at 450 nm. The cytotoxicity was expressed as a percentage of the control.
The interaction between DNA and GO nanosheets
Genomic DNA was extracted from MDA-MB-231 cells based on the protocol from the manufacture (TaKaRa DV811A, China). At 37°C, 0.1 μg genomic DNA was incubated with GO at different concentrations (0, 200, 300, 400, 500, and 600 μg/mL) for 2 h, or at GO concentration of 600 μg/mL for different times (0, 10, 40, 80 min, and 2 h). Each reaction was quenched by adding 2 μL of a loading buffer solution (0.05% bromophenol blue, 1% SDS, and 50% glycerol, pH 8.0) and then subjected to electrophoresis on a 0.8% agarose gel containing 50 μg of ethidium bromide (EB) in 40 mL of TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM EDTA, pH 8.3) at 100 V for approximately 1 h. Agarose gel electrophoresis was carried out with JUNYI-SPAT electrophoresis apparatus (Beijing Junyi-Dongfang, China). The agarose gels were imaged using a GelDoc-ItTM Imaging System (UVP Inc., America).
The effects of GO nanosheets in DNA replication
Polymerase chain reaction (PCR) was used to examine the effects of GO in DNA replication using human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) gene as a template. Briefly, genomic DNA above extracted (0.05 μg) was incubated with different doses of GO (0, 200, 400, 600, 800, and 1000 ng/mL) at 37°C for 2 h, and then amplified in a 25 μL reaction solution containing High Fidelity PCR buffer (10×) with 15 mM MgCl2, 200 μM of each dNTP, 1.25 units of High Fidelity PCR Enzyme Mix, and 0.4 μM of each primer (forward: 5′-GCCACTAGGCGCTCACTGTTCT-3′, reverse: 5′-TGGGGTCGGGTCAACGCTAGG-3′). PCR reaction was carried out in one cycle at 94°C for 3 min and subsequently 30 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 1 min using a Thermocycler Mastercycler Gradient (Eppendorf, Germany). 5 μl of each PCR product was electrophoresed on a 1% agarose gel, which was then imaged using the GelDoc-ItTM Imaging System (UVP Inc., America).
Mutation assay
The effect of GO on the fidelity of gene replication was examined by PCR of PKCζ followed by sequencing. Plasmids expressing PKCζ were amplified by PCR at various GO treatments. PCR reactions were performed in a total volume of 25 μL containing 5 ng PKCζ plasmid, 1 μL of 20 μM primer for PKCζ (forward: CTGAGGAGCACGCCAGGTT, reverse: ACGGGCTCGCTGGTGAACT, 620 bp), 12.5 μL of 5 U/μL TAKARA EXTaq polymerase (Millipore, USA), and GO with concentrations of 0, 1, 10, 50, and 100 ng/mL, respectively. After purification and extraction from 1% agarose gel using a Genomic DNA Extraction kit (TaKaRa, China), PCR products were cloned into pMD19T vectors following the manufacturer's protocol (TaKaRa, China), and then transformed into Escherichia coli (E. coli) DH5α. Next, about 30 clones per group were selected randomly via white-blue screening and sequenced using bi-directional sequencing method (Beijing Genomics Institute, China). All experiments were repeated three times.
Microarray experiment
Microarray experiment was next used to examine the impact of GO treatment on gene expression. MDA-MB-231 cells were divided into three groups and incubated with GO at concentrations of 0, 10, and 100 μg/mL, respectively. Each sample group contained 5 replicates. After incubation for 48 h, the total cellular RNA of each sample was isolated using QIAGEN RNeasy Mini Kit (Catalog no. 74104) in accordance with the manufacturers' instructions. Integrity of RNA was evaluated using a spectrophotometer (NanoDrop 2000; Thermo Scientific, USA) and analyzed on a bioanalyzer (2100; Agilent, Santa Clara). Subsequently, all cRNAs were hybridized to gene expression profiles (GeneChip Human Genome U133 Plus 2.0 Arrays) according the standard Affymetrix protocol. Integrative comparative and statistical analysis was performed to determine the differentially expressed genes between GO-treatment groups (10 and 100 μg/mL) and control group. RNA expression data CEL files were generated using Affymetrix GeneChip Command Console (AGCC) Software and normalized using Partek Genesignt 6.4 version software. Normalized data were imported into dChip (http://biosun1.harvard.edu/complab/dchip/) for differential expression analysis. Two-group comparative analysis was performed using selection variables of at least 2-fold change and t-test at P < 0.05. Samples were permuted 100 times through dChip to assess the false discovery rate. Genes with differential expressions in both two GO treatment groups were then selected and clustered by the hierarchical method30.
Western blot analysis
MDA-MB-231 cells were incubated with GO at concentrations of 0, 10, and 100 μg/mL. After 1 and 2 h respectively, the cells were harvested and lysed in RIPA buffer (0.15 M NaCl, 1% NP-40, 0.05% deoxycholic acid, 0.1% SDS, 50 mM Tris-HCl (pH 7.4)) in the presence of protease inhibitors (Roche, according to manufacturer's instructions). Western blot was used to analyze the DNA repair proteins ATM and Rad51 by probing with an anti-rad51, anti-ATM monoclonal antibody (Cell Signaling Inc.).
Apoptosis analysis
MDA-MB-231 cells were incubated with GO at concentrations of 0, 10, and 100 μg/mL for 24 h. The cells were then washed twice with cold PBS and resuspended cells in 1× Binding Buffer. Double staining with PE-Annexin V and 7-AAD was carried out using the PE Annexin V Apoptosis Detection Kit (BD Pharmingen™, Franklin Lakes, NJ), according to the manufacturer's recommendations, and the plates were analyzed within an hour after staining by flow cytometry (Beckman Coulter, USA). Cells were discriminated into viable cells, dead cells, early apoptotic cells, and apoptotic cells by using CellQuest software (BD Biosciences); then the percentages of early apoptotic cells from each experiment were compared.
Cell cycle analysis
Flow cytometry assay was used to assess the changes in cell cycle after GO treatment. MDA-MB-231 cells were spread onto 60-mm tissue culture dishes and incubated to reach 50% confluence. After treatment with different doses of GO for 48 h, cells were digested and washed with PBS (pH 7.4), and then fixed in 70% cold ethanol for 1 h at −20°C. After centrifugation, the cells were washed twice with ice-cold PBS and treated with RNase (0.1 mg/mL), and then stained by propidium iodide (40 μg/mL). Flow cytometric analysis was performed using an EPICS XL flow cytometer (Beckman Coulter, USA), and the cell cycle was analyzed with Cell Quest software.
Micronucleus assay
Next, micronucleus test was used to examined the mutagenic potential of GO. Experiments were carried out using Kunming mice with the weight of 25–30 g that purchased from Wei Tong Li Hua experimental animal (Beijing, China). All experimental procedure was approved by the local animal care committee and was carried out in according to the guidelines of the Ethical Committee for Animal Experiments of Tianjin Medical University. The mice were divided into 5 experimental groups of eight animals each (4 females and 4 males) and kept in a climate-controlled environment (25 ± 4°C, 55 ± 5% humidity). Three group mice were administered intravenously with GO solution at 1, 2 and 4 mg/kg once a day for five consecutive days, corresponding respectively to the total doses of 5, 10 and 20 mg/kg. The other two group mice were respectively received physiological saline and cyclophosphamide (50 mg/kg) as the negative and positive controls. After humanitarian executions, femur bones of mice were excised and the bone marrow flushed into test tubes, and then the micronucleus assay was carried out following standard protocols, as recommended by Schmid31 and Krishna42. For analysis of the micronucleated cells, 2000 polychromatic erythrocytes per animal were scored to determine the mutagenic property of GO. To detect possible cytotoxic effects, the poychromatic erythrocyte/normochromatic erythrocyte ratio in 200 erythrocytes per mouse was calculated.
Statistical analysis
Each experiment was repeated three times in duplicate. The results were presented as mean ± SD. Statistical differences were evaluated using the t-test and considered significance at P < 0.05.
Supplementary Material
Supplementary information
Acknowledgments
We thank Dr. Yongsheng Chen (Institute of Polymer Chemistry, College of Chemistry, Nankai University, Tianjin, China) for kindly providing GO. This project was supported by grant from the 973 program (2011CB933100), National Science Fund for Distinguished Young Scholars (81125019), National Natural Science Foundation of China (81371671), and the special program of China Postdoctoral Science Foundation (201104308).
Footnotes
The authors declare no competing financial interests.
Author Contributions N.Z. and Y.W. proposed and supervised the project. Y.L. and Y.W. prepared and characterized small sized GO. Y.L. performed cell proliferation experiment, western bloting and the interaction between DNA and GO nanosheets, Y.L. and J.W. performed mutation assay, cell cycle analysis, the effects of GO nanosheets on microarray experiment, and micronucleus assay under supervision of N.Z. and Y.W. R.Y. and B.W. helped with the preparation and characteriztion of small sized GO, and R.Y. also hepled cell cycle analysis and micronucleus assay. X.Y. and J.Y. performed Raman, thermogravimetric and X-ray diffraction analyses. N.Z. and Y.W. analyzed all data and wrote the manuscript. All the authors participated in discussions of the research.
References
- Novoselov K. S. et al. Electric field effect in atomically thin carbon films. Science 306, 666–669 (2004). [DOI] [PubMed] [Google Scholar]
- Geim A. K. & Novoselov K. S. The rise of graphene. Nat. Mater. 6, 183–191 (2007). [DOI] [PubMed] [Google Scholar]
- Geim A. K. Graphene: status and prospects. Science 324, 1530–1534 (2009). [DOI] [PubMed] [Google Scholar]
- Huang Y., Liang J. J. & Chen Y. S. An overview of the applications of graphene-based materials in supercapacitors. Small 8, 1805–1834 (2012). [DOI] [PubMed] [Google Scholar]
- Balandin A. A. Superior thermal conductivity of single-layer graphene. Nano Lett. 9, 902–907 (2008). [DOI] [PubMed] [Google Scholar]
- Lee C., Wei X. D., Kysar J. W. & Hone J. Measurement of the elastic properties and intrinsic strength of monolayer graphene. Science 321, 385–388 (2008). [DOI] [PubMed] [Google Scholar]
- Yang X. Y. et al. High-efficiency loading and controlled release of doxorubicin hydrochloride on graphene oxide. J. Phys. Chem. C 112, 17554–17558 (2008). [Google Scholar]
- Liu Z., Robinson J. T., Sun X. & Dai H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 130, 10876–10877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. et al. Graphene in mice: ultrahigh in vivo tumor uptake and efficient photothermal therapy. Nano Lett. 10, 3318–3323 (2010). [DOI] [PubMed] [Google Scholar]
- Wu J. et al. Graphene oxide used as a carrier for adriamycin can reverse drug resistance in breast cancer cells. Nanotechnology 23, 355101 (2012). [DOI] [PubMed] [Google Scholar]
- Yang K. et al. Graphene in mice: ultrahigh in vivo tumor uptake and efficient photothermal therapy. Nano Lett. 10, 3318–3323 (2010). [DOI] [PubMed] [Google Scholar]
- Robinson J. T. et al. Ultrasmall reduced graphene oxide with high near-infrared absorbance for photothermal therapy. J. Am. Chem. Soc. 133, 6825–6831 (2011). [DOI] [PubMed] [Google Scholar]
- Yang K. et al. Multimodal imaging guided photothermal therapy using functionalized graphene nanosheets anchored with magnetic nanoparticles. Adv. Mater. 24, 1868–1872 (2012). [DOI] [PubMed] [Google Scholar]
- Tung V. C., Allen M. J., Yang Y. & Kaner R. B. High-throughput solution processing of large-scale graphene. Nat. Nanotechnol. 4, 25–29 (2009). [DOI] [PubMed] [Google Scholar]
- Lu W. B. et al. High-yield, large-scale production of few-layer graphene flakes within seconds: using chlorosulfonic acid and H2O2 as exfoliating agents. J. Mater. Chem. 22, 8775–8777 (2012). [Google Scholar]
- Lv M. et al. Effect of graphene oxide on undifferentiated and retinoic acid-differentiated SH-SY5Y cells line. Nanoscale 4, 3861–3866 (2012). [DOI] [PubMed] [Google Scholar]
- Wang K. et al. Biocompatibility of graphene oxide. Nanoscale Res. Lett. 6, 8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. L. et al. In vitro toxicity evaluation of graphene oxide on A549 cells. Toxicol. Lett. 200, 201–210 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Distribution and biocompatibility studies of grapheme oxide in mice after intravenous administration. Carbon 49, 986–995 (2011). [Google Scholar]
- Liu Z., Robinson J. T., Sun X. M. & Dai H. J. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 130, 10876–10877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. et al. In vivo pharmacokinetics, long-term biodistribution and toxicology of PEGylated graphene in mice. ACS Nano 5, 516–522 (2011). [DOI] [PubMed] [Google Scholar]
- Xing Y. et al. DNA damage in embryonic stem cells caused by nanodiamonds. ACS Nano 5, 2376–2384 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu L., Chang D. W., Dai L. & Hong Y. DNA damage induced by multiwalled carbon nanotubes in mouse embryonic stem cells. Nano Lett. 7, 3592–3597 (2007). [DOI] [PubMed] [Google Scholar]
- Ren H. et al. DNA cleavage system of nanosized graphene oxide sheets and copper ions. ACS Nano 4, 7169–7174 (2010). [DOI] [PubMed] [Google Scholar]
- Becerril H. A. et al. Evaluation of solution-processed reduced graphene oxide films as transparent conductors. ACS Nano 2, 463–470 (2008). [DOI] [PubMed] [Google Scholar]
- Yang X. Y. et al. High-efficiency loading and controlled release of doxorubicin hydrochloride on graphene oxide. J. Phys. Chem. C 112, 17554–17558 (2008). [Google Scholar]
- Shan C. et al. Water-soluble graphene covalently functionalized by biocompatible poly-L-lysine. Langmuir 25, 12030–12033 (2009). [DOI] [PubMed] [Google Scholar]
- Marcano D. C. et al. Improved synthesis of graphene oxide. ACS Nano 4, 4806–4814 (2010). [DOI] [PubMed] [Google Scholar]
- Shang J. et al. The Origin of fluorescence from graphene oxide. Sci. Rep. 2, 792 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreo J. A., Valencia A. & Dopazo J. A hierarchical unsuperuised growing neural network for clustering gene expression patterns. Bioinformatics 17, 126–136 (2001). [DOI] [PubMed] [Google Scholar]
- Schmid W. The Micronucleus Test. Mutat. Res. 31, 9–15 (1975). [DOI] [PubMed] [Google Scholar]
- Gollapudi B. B. & McFadden L. G. Sample size for the estimation of polychromatic to normochromatic erythrocyte ratio in the bone marrow micronucleus test. Mutat. Res. 347, 97–99 (1995). [DOI] [PubMed] [Google Scholar]
- Sun X. et al. Nano-graphene oxide for cellular imaging and drug delivery. Nano Res. 1, 203–212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S. Structural considerations in the interaction of DNA and acridines. J. Mol. Biol. 3, 18–30 (1961). [DOI] [PubMed] [Google Scholar]
- Lerman L. S. Acridine mutagens and DNA structure. J. Cell. Physiol. 64, S1–S18 (1964). [PubMed] [Google Scholar]
- Graves D. E. & Velea L. M. Intercalative binding of small molecules to nucleic acid. Curr. Org. Chem. 4, 915–929 (2000). [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3, 155–168 (2003). [DOI] [PubMed] [Google Scholar]
- Lavin M. F. et al. ATM signaling and genomic stability in response to DNA damage. Mutat. Res. 569, 123–132 (2005). [DOI] [PubMed] [Google Scholar]
- Tarsounas M., Davies A. A. & West S. C. RAD51 localization and activation following DNA damage. Phil. Trans. R. Soc. Lond. B. 359, 87–93 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph E. K. & Levine J. D. Caspase signaling in neuropathic and inflammatory pain in the rat. Eur. J. Neurosci. 20, 2896–2902 (2004). [DOI] [PubMed] [Google Scholar]
- Berthet C. & Kaldis P. Cdk2 and Cdk4 cooperatively control the expression of Cdc2. Cell Division 1, 10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna G. & Hayashi M. In vivo rodent micronucleus assay: protocol, conduct and data interpretation. Mutat. Res. 455, 155–166 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information