Abstract
Functional imaging has become an important tool in oncology because it not only provides information about the size and localization of the tumour, but also about the pathophysiological features of the tumoural cells. One of the characteristic features of some tumour types is that their fast growth leads to deficient intratumoral vascularization, which results in low oxygen availability. To overcome this lack of oxygen, tumoural cells activate the neoangiogenic program by upregulating the transcription factor HIF-1α. Herein we report a non-invasive in vitro detection method of hypoxia using designed fluorescent peptide probes based on the oxygen-dependent degradation domain of HIF-1α. The fluorescent probe retains the oxygen-sensing capability of HIF-1α, so that it is stabilized under hypoxia and readily degraded by the proteasome under normoxia, thus providing direct information of the cellular oxygen availability.
Subject terms: Cancer, Chemical biology
Introduction
Some tumoural types tend to grow in a rapid and disorganized manner leading to poor intratumoral vascularization and low oxygen availability. In order to overcome this deficiency, tumour cells, just like their normal counterparts, promote the formation of neovessels by activating pathways that regulate this process of neoangiogenesis1. Moreover, hypoxia is a poor-prognosis microenvironmental hallmark of many solid tumours, and also influences the outcome of disseminated tumour cells contributing to their maintenance and treatment resistance2. In this context, the transcription factor HIF-1α stands out as the master regulator of neoangiogenesis, considering the number of genes whose expression controls when hypoxia is elicited, as well as its active role in the overall process3. As such, HIF-1α has been used as a prognostic marker of tumoural aggressiveness and its activity a readout of tumoural proliferation and ability to metastasize4.
The development of novel and more efficient techniques of visualization of tumoural processes has become a top priority in molecular imaging, particularly in functional imaging to monitor biological processes intimately related to cancer, such as metastasis and hypoxia5. In the last years, optical imaging methods based on fluorescence or bioluminescence have been gaining popularity in this field, thanks to the availability of a broad range of proteins and dyes6,7. In this context, a number of groups have recently developed several NIR (near-infrared) fluorescent proteins that enable real-time imaging free of autofluorescence interference, thus making possible to take a deep view into the tissues. Our group has previously described the design and in vitro and in vivo characterization of a genetically encoded biosensor. This sensor, which combines a fluorescent far-red protein (mCherry) and the firefly luciferase (FLuc) and is activated by the neoangiogenesis-related transcription factor HIF-1α, allowed us to differentiate tumoural masses with metastatic potential with high accuracy in a mouse model of metastasis8. At the same time, by fusing a fluorescent to a bioluminescent protein we obtained a bioluminescence resonance energy transfer (BRET) phenomenon, turning this fusion protein into a new class of hypoxia-sensing genetically encoded biosensor8,9. Recently, another genetically encoded biosensor consisting of the fluorescent protein GFP fused to the oxygen-dependent degradation (ODD) domain of the Droshophila homolog of HIF-1 Sima was reported10.
Although our biosensor demonstrated potential in hypoxia sensing, it was not directly applicable in a clinical setting because of the limitations inherently associated to biochemical sensors based on large protein constructs. Preeminent among those limitations are the need for transfection and over-expression, relatively low photostability, and large size that can lead to interference, poor biodistribution, or immune response11,12. In addition to avoiding those problems, peptides offer a number of advantages, including higher stability and lower immunogenicity, ease of synthesis, and the simplicity for molecular engineering, as well as better biodistribution13.
In this study, we take advantage of what we learned from our previous genetically encodable protein sensors, and describe a compact sensor consisting on a fluorescently-labelled peptide, corresponding to a small fraction of the ODD domain of HIF-1α, that mimics the behaviour of HIF-1α under hypoxia conditions, making possible its application for the monitorization of hypoxic activity with potential clinical applicability.
Results and Discussion
Biosensor design and synthesis
Hypoxia transcriptional program activation depends on hypoxia-induced stabilization of HIF-1α14. The molecular mechanism underlying this stabilization was a subject of great discussion due to the dispute between different models, such as the occurrence of hypothetical O2-binding hemoproteins or oxidases interacting with HIF-115. As we now know, HIF-1α is rapidly degraded in normoxic cells upon hydroxylation of two proline residues (Pro402 and Pro564) located in its oxygen-dependent degradation domain (ODD domain)16–18. Upon hydroxylation, these residues are recognized by von Hippel-Lindau E3 ubiquitin ligase (pVHL), leading to poly-ubiquitination and subsequent degradation of the protein19–21.
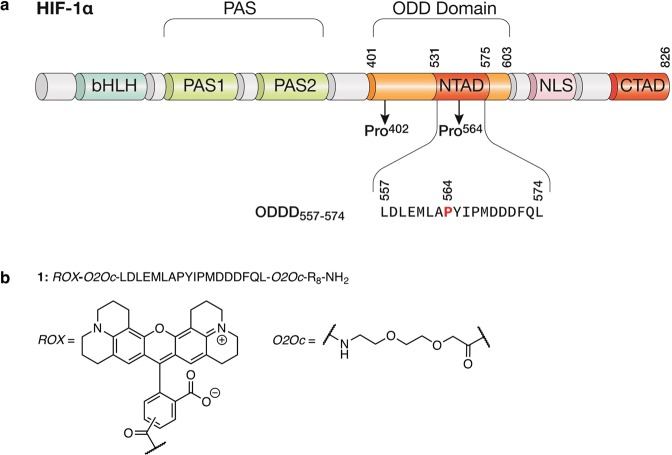
With the aim of developing new hypoxia tracers, we envisioned a fluorescent peptide that would mimic the effect of hypoxia on the half-life of HIF-1α. Since the total length of the HIF-1α ODD domain16, 203 residues as depicted in Fig. 1a, makes unpractical its chemical synthesis and the incorporation of the sensing unit, we decided to use a short 16-mer peptide derived from the HIF-1α ODD domain, Leu557 to Leu574. This peptide has been previously reported to be profusely hydroxylated in the Pro564 residue during normoxia, leading to pVHL-mediated degradation22, and thus retains the oxygen-sensing properties of HIF-1α23. As a general feature, the new sensor has three small modules with different functions: an octa-arginine peptide that mediates cell internalization24,25, a central domain from the HIF-1α degradome able to sense low oxygen levels (1: 557LDLEMLAPYIPMDDDFQL574)26 and the 5,(6)-ROX fluorochrome, a long-wavelength rhodamine characterized by a similar emission profile to mCherry27,28, good stability29,30 and high quantum yield (0.92)30, that acts as fluorescent reporter of the integrity of the peptide for in vitro and in vivo imaging (Figs 1 and S1). These three modules are connected by short PEG linkers (O2Oc)31 to avoid interference in the recognition of the HIF central domain by prolyl hydroxylase.
Figure 1.
Biosensor design rationale. (a) Domain structures of HIF-1α. The ODD domain regulates the stability of HIF-1α via recognition by the E3 ubiquitin ligase pVHL. (b) Chemical structure of sensor 1.
In contrast with other fluorescent sensors of proteins, such as those based on solvatochromic dyes that increase their emission intensity in hydrophobic environments, i.e. protein pockets32, or those based in energy transfer processes33, in this case the sensing mechanism relies on the higher lifetime of the ODD domain sequence under hypoxic conditions than under normoxia. Since the degradation of HIF-1α is triggered by hydroxylation of the Pro564 in the ODD domain, we expected that the proteolytic stability of a fluorescent peptide containing the appropriate region of ODD domain would be under the same control as HIF-1α, so that the accumulated fluorescence would reflect the changes in oxygen availability in the different tissues.
Peptide 1 was synthesized following standard Fmoc solid phase peptide synthesis (SPPS) protocols, and as a final step labelled at their deprotected N-terminus with 5(6)-ROX. Once purified by reversed-phase HPLC and lyophilized, peptide 1 was dissolved in PBS and its excitation and emission spectra were recorded showing the characteristic fluorescence profile of ROX fluorophore conjugates (Fig. S4, excitation 585 nm, emission 608 nm)30.
Internalization and stabilization of sensors by hypoxia-mimetic agents
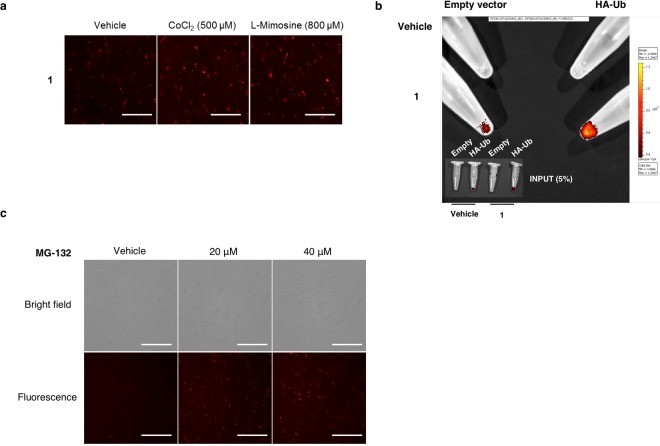
Human breast cancer cells MDA-MB 231 were treated with 1. Cells were incubated with vehicle or the hypoxia-mimetic agents: CoCl2, an inhibitor of the interaction between HIF-1α and the ubiquitin ligase von Hippel-Lindau protein (pVHL)34, and the prolyl hydroxylase inhibitor l-Mimosine35. As can be observed in Fig. 2a, the fluorescent peptide 1 is stabilized in the presence of both molecules that mimic hypoxia, thus demonstrating our hypothesis for the design of the probe based on an inhibition of the sensor degradation in a HIF-dependent hypoxic response. Although, the octa-arginine cell-penetrating peptide has been previously described as a potent inhibitor of proteasome activation36, this new fluorescent hypoxic sensor displays an improved stabilization when the level of signal in vehicle-treated cell is compared with CoCl2 and l-Mimosine-treated cells.
Figure 2.
(a) Hypoxia mimetics inhibit sensor degradation. Cells were incubated with hypoxia sensor 1 in the presence of CoCl2 (500 μM) and L-Mimosine (800 μM). Scale = 50 μm. (b) Ubiquitination of hypoxia sensor 1 is demonstrated by immunoprecipitation of HA-tagged ubiquitinated substrate. (c) Proteasome degrades sensor 1. Proteasome inhibitor MG132 inhibits in a dose response manner the degradation of hypoxia probe 1 in MDA-MB 231 cells. Scale = 100 μm.
Hypoxia targets sensors specifically via ubiquitin-proteasome degradation
One of our main concerns was the way this peptidic probe would be processed within cells. Ideally, the probe would be eliminated in the same manner as HIF-1α, i.e., degraded by the proteasome upon hydroxylation of the ODD domain. To further investigate into this issue, we transfected cells with a plasmid encoding anti-hemagglutinin (HA)-tagged ubiquitin, in order to perform an immunoprecipitation, and treated these cells with probe 1. In cells transfected with HA-tagged ubiquitin the enrichment of hypoxia sensors by (HA) immunoprecipitation shows that the degradation of the sensor is ubiquitin proteasome-regulated (Fig. 2b). Moreover, if cells incubated with hypoxic sensor 1 are treated with a proteasome inhibitor (MG-132), a dose-dependent effect on proteasome degradation of fluorescent signal is demonstrated (Fig. 2c).
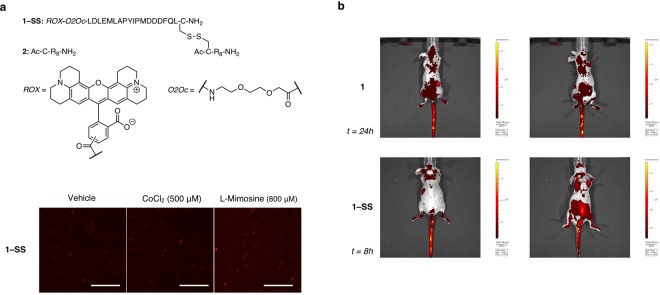
On the other hand, depending on their cargo, octa-arginines are also known to induce nuclear uptake37,38, thus probably extending the half-life of the probe within the cell and delaying its clearance. Given that disulfide bridges are sensitive to the reductive environment of the cytoplasm, we decided to synthesize a new probe, 1–SS, in which the octa-arginine peptide and the ODDD-based sensing unit are connected through a disulfide bond, with the purpose of reducing its half-life (Fig. 3a upper panel). The cleavage of the disulfide bond in the cytosol once the compound is internalized39 should give the corresponding octa-arginine and ODD domain fragments, thus facilitating the degradation of the sensing unit.
Figure 3.
(a) (upper pannel) Chemical structures of peptide 2 and probe 1-SS. (lower panel) Hypoxia mimetics also inhibit sensor 1-SS degradation. Cells were incubated with hypoxia sensor 1-SS in the presence of CoCl2 (500 μM) and L-Mimosine (800 μM). Scale = 50 μm. (b) Comparation between the clearance time of sensor 1 (up) and 1-SS (down) in vivo. Images in prone (left) and supine position (right) were taken every two hours during the first eight hours upon sensor administration and one final time at 24 hours post-injection. Images shown correspond to the last time point where specific signal from the sensor was detected.
The strategy followed for the synthesis of the N-terminus ROX-conjugated peptide 1–SS is described in detail in the methods section and supporting information (Fig. S1). In short, the precursor peptide 1-Cys, that contains the previously described ODDD sequence with an additional cysteine residue, as well as peptide 2, featuring the octa-arginine sequence with an appended cysteine residue, were synthesized following standard solid phase protocols, and HPLC-purified. Peptide 2 was then activated with 5,5-dithio-bis-(2-nitrobenzoic acid) (Ellman’s reagent) and incubated with 1-Cys to promote a disulfide exchange reaction, that provided the desired conjugate 1-SS. Probe 1-SS showed the same excitation and emission profile than peptide 1 (Fig. S5).
As was expected, probe 1–SS showed increased rate of degradation, both in vitro and in vivo. Treated MDA-MB-231 cells with hypoxia sensors in the presence of CoCl2 and l-Mimosine revealed a faster reduction of the fluorescence intensity (Fig. 3a lower panel). Along with faster clearance rates we also observed a reduction in background fluorescence and an enhanced sensitivity with increasing concentrations of CoCl2 when compared to probe 1 (Fig. S6). Indeed, this reduction was also observed when probe 1 and 1–SS were administered in vivo to mice. As can be observed in Fig. 3b, 1–SS showed a lower signal at 8 h post administration than 1 at 24 h. The fast renal elimination of fluorophore results in strong bladder fluorescence when 1–SS was administered. Additional time points can be found in Fig. S7.
In summary, we have shown that the stabilization effect of hypoxia on the half-life of HIF-1α can be exploited for the design of fluorescent sensors of hypoxia. We have therefore designed and tested a small synthetic peptide biosensor, based on the ODD domain of HIF-1α and modified with an octa-arginine cell-penetrating peptide and a fluorescent dye, whose proteolytic stability is controlled under hypoxia and the fluorescence signal depends on the oxygen availability. We have further confirmed that the probe is degraded by the proteasome upon hydroxylation of the ODD domain. Moreover, the reengineering of the initial peptide probe, by means of adding a disulfide bridge between the octa-arginine vector and the ODDD sensing unit, accelerates its degradation both in vitro and in vivo. Although other approaches have been developed around a similar concept using genetically encoded biosensors10,40 or nitroaromatic compounds41,42, our probe combines excellent cell internalization with faster clearance rates which improves its sensitivity and applicability in monitoring hypoxic environments. Therefore, these tracers could serve not only to pinpoint tumoural masses, but also to inform us about their metastatic potential and aggressiveness and, at the same time, gain a deeper view on the tumour microenvironment that modern imaging techniques do not provide.
Materials and Methods
Chemicals and instrumentation
All peptide synthesis reagents and amino acid derivatives were purchased from GL Biochem (Shanghai) and NovaBiochem. Amino acids were purchased as standard Fmoc protected amino acids: Fmoc–Leu–OH, Fmoc–Asp(OtBu)–OH, Fmoc–Glu(OtBu)–OH, Fmoc–Met–OH, Fmoc–Ala–OH, Fmoc–Pro–OH, Fmoc–Tyr(tBu)–OH, Fmoc–Ile–OH, Fmoc–Phe–OH, Fmoc–Gln(Trt)–OH, Fmoc–Arg(Pbf)–OH, Fmoc–Ser(tBu)–OH, Fmoc–Thr(tBu)–OH, and Fmoc–Cys(Trt)–OH. Fmoc–O2Oc–OH was purchased from Iris Biotech GMBH (Cat. #: FAA1435), and 5(6)-ROX, SE was purchased from Invitrogen (Cat. #: C1309). C-terminal amide peptides were synthesized on a 0.025 mmol scale using a 0.23 mmol/g loading Fmoc-PAL-PEG-PS resin from Applied Biosystems. All other chemicals were purchased from Sigma-Aldrich. All solvents were dry and synthesis grade, except DMF for peptide synthesis. Peptides were synthesized using a PS3 automatic peptide synthesizer from Protein Tecnologies.
High-Performance Liquid Chromatography (HPLC) was carried out with an Agilent 1100 series Liquid Chromatograph Mass Spectrometer system. Analytical HPLC was run using a Zorbax Eclipse XDB-C8 (5 µm) 4.6 × 150 mm analytical column (Agilent). Purification of the peptides was performed on a Jupiter Proteo 90 A (4 µm) 10 × 250 mm reversed-phase column (Phenomenex). The usual gradient used for analytical and semi-preparative HPLC was 15 → 95% CH3CN, 0.1% TFA/H2O, 0.1% TFA over 30 min. Electrospray Ionization Mass Spectrometry (ESI MS) was performed with an Agilent 1100 Series LC/MSD VL G1956A model in positive scan mode by direct injection of the purified peptide solution. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) was performed with a Bruker ultraflex III TOF/TOF, using sinapinic acid as matrix.
ROX-modified peptides appear as overlapping pair of peaks in the HPLC traces due to the use of the commercial a mixture of 5-(and 6)-Carboxy-X-Rhodamine isomers in the synthesis.
Peptide synthesis
The amino acids were coupled in 4-fold excess using 2-[(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) as coupling agent. Each amino acid was activated for 30 s in DMF before being added onto the resin. Peptide couplings were conducted for 30 to 45 min. The temporal Fmoc protecting group was removed by treating the resin with 20% 4-methylpiperidine in DMF solution for 10 min.
5(6)-ROX, Succinimidyl Ester (SE) coupling
The N-terminus-deprotected peptides 1 and 1-Cys, attached to the resins (0.025 mmol), 0.195 M N,N-diisopropylethylamine (DIEA) in DMF (1 mL, 0.195 mmol) and 5(6)-ROX, SE (12.5 mg, 0.02 mmol) were mixed and the resin suspension was shaken for 4 h. After filtration, the resin was washed with DMF (3 × 1 mL × 3 min) and CH2Cl2 (3 × 1 mL × 3 min) and dried under argon.
Peptide 2 N-terminus-acetylation
After the final Fmoc deprotection step using standard conditions (20% 4-methylpiperidine/DMF), peptide 2 (0.025 mmol) was acetylated by treatment with 20% Ac2O in DMF (2.5 mL) and 0.195 M DIEA in DMF (1.75 mL) for 45 min. After filtration, the resin was washed with DMF (3 × 5 mL × 3 min) and CH2Cl2 (3 × 5 mL × 3 min) and dried under a current of argon.
Cleavage and deprotection of semipermanent protecting groups
The resin-bound peptides 1, 1-Cys, and 2 (0.025 mmol), were placed in a 50 mL tube to which 10 mL of the cleavage cocktail (2% anisole, 3% ethanedithiol (EDT), 5% thioanisole and 90% TFA) were added. These mixtures were shaken for 3.5 h. The resins were then filtered, and the TFA filtrate was concentrated to a volume of approximately 1 mL. The residue was added to ice-cold diethyl ether (20 mL). After 10 min, the precipitate was centrifuged and washed again with 10 mL of ice-cold ether and centrifuged. The solid residue was dried under argon and redissolved in acetonitrile/water 1:1 (1 mL) and purified by semi-preparative reversed-phase HPLC. The collected fractions were lyophilized and stored at −20 °C.
Peptide 2 activation with Ellman’s reagent
Peptide 2 (10 mg, 0.007 mmol) was dissolved in 10 mM Tris·HCl, pH 7.5, 100 mM NaCl (0.65 mL). 5,5-dithio-bis-(2-nitrobenzoic acid) (5.6 mg, 0.014 mmol) was dissolved in a separate vial in 0.35 mL of 10 mM Tris·HCl, pH 7.5, 100 mM NaCl/MeCN (3:2). Both solutions were mixed, and the resulting mixture was shaken for 2 h. Then, the crude was purified by semi-preparative reversed-phase HPLC, and the collected fractions were lyophilized and stored at −20 °C.
Coupling of peptide 1-Cys with Ellman’s-conjugated peptide 2
Ellman’s-conjugated peptide 2 (5.8 mg, 0.0036 mmol) was dissolved in 10 mM Tris·HCl, pH 7.5, 100 mM NaCl (0.6 mL), and 0.4 mL of a peptide 1-Cys (5 mg, 0.0017 mmol) solution in 10 mM Tris·HCl, pH 7.5, 100 mM NaCl/MeCN (1:1), were added. The resulting mixture was shaken for 2 h, then the crude was purified by semi-preparative reversed-phase HPLC, and the collected fractions were lyophilized and stored at −20 °C.
Fluorescence spectroscopy
Fluorescence spectra were recorded with a Jobin−Yvon Fluoromax−3 (FluorEssence™), coupled to a Wavelength Electronics LFI−3751 temperature controller. All measurements were made with the following settings: increment: 1.0 nm; integration time: 0.50 s; excitation slit width: 2.0 nm; emission slit width: 2.0 nm; excitation wavelength 585 nm. The emission spectra were recorded from 595 to 700 nm. Peptide stock solutions were quantified spectroscopically taking a molar extinction coefficient of 78000 M−1 cm−1 at 579 nm43.
Cell culture and treatments
Human breast cancer cells MDA-MB 231 were maintained at 37 °C and 5% CO2 in RPMI-1640 (Sigma–Aldrich), supplemented with 10% fetal bovine serum (Fisher Scientific). Cells were seeded at a density of 2·105 cells per well in 6-multiwell plates 24 h prior treatment. For hypoxia induction, they were treated with either CoCl2 (Sigma–Aldrich) at a final concentration of 500 μM, or L-Mimosine (Sigma–Aldrich) at a final concentration of 800 μM. Treatment with each peptidic sensor (500 ng/mL) was carried out in the presence of the aforementioned inhibitors. After three hours of incubation, supernatants containing the probes and either CoCl2 or L-Mimosine were aspirated, cells were washed once with PBS 1X and fresh medium, containing again either CoCl2 or L-Mimosine, was added to the cells for an extended incubation of three hours. Treatment with proteasome inhibitor MG-132 (Sigma-Aldrich) was carried out for 6 hours at final concentrations of either 20 μM or 40 μM, upon pre-treatment of the cells with the probes, as described previously. Fluorescent probes internalization was quantified using ImageJ (NIH). Quantified fluorescence was expressed as values of corrected total cell fluorescence (CTCF), which was calculated using the formula CTCF = integrated density – (area of selected cell × mean fluorescence of background readings)44.
Immunoprecipitation
MDA-MB 231 cells were seeded at a density of 2·105 cells per well in 6-multiwell plates twenty-four hours prior transfection. Each well was transfected with 2 μg of either pcDNA3-Ub-HA or empty vector pcDNA3 (Invitrogen) using Superfect (Qiagen) as transfection reagent. Twenty-four hours upon transfection culture medium was removed, cells were washed carefully with phosphate buffered saline (PBS; Sigma-Aldrich) and fresh culture medium containing the peptidic sensor 1 was added onto the cells. PBS was used as mock treatment. Five hours upon treatment with the sensor, cells were lysed in native immunoprecipitation buffer, 20 mM Tris·HCl pH 8, 137 mM NaCl,10% glycerol,1% Nonidet P-40 (NP-40), 2 mM EDTA, containing the protease inhibitors sodium orthovanadate (1 mM), PMSF (2 mM) and aprotinin (1 μg/mL) (Sigma-Aldrich). Each immunoprecipitation contained 600 μg of whole protein extract and were incubated overnight with 5 μg of anti-hemagglutinin HA.11 (Clone 16B12) antibody (Abcam) at 4 °C. Equal amounts of rabbit immunoglobulins (Sigma-Aldrich) were used as control. HA-tagged ubiquitins were immunoprecipitated using Protein G-coupled agarose (Santa Cruz), subsequently washed with immunoprecipitation buffer and eluted under native conditions. Fluorescence images were taken using an IVIS Spectrum system (Perkin-Elmer).
Animal studies
Two female BALB/c nude mice (Janvier) of twelve weeks of age were injected in the tail-vein with 10 mg/Kg of either 1 or 1–SS prior to commence the experiment. Fluorescence pictures from injected mice and a non-treated mouse used as a negative control were taken every two hours using the IVIS system during the first 8 hours, and then one final time at 48 hours post-injection. Specific fluorescence images were acquired using the 570 nm and 620 nm filter for excitation and emission, respectively, while for background fluorescence the 465 and 620 nm were used. Substraction of background fluorescence was performed with the IVIS system software following the guidelines set out by the manufacturer. Mice were housed in specific pathogen-free (SPF) conditions, following FELASA (Federation of European Laboratory Animal Science Associations) and institutional guidelines. All animal procedures were approved and performed according to the guidelines set out by the Institutional Ethics Committee for Animal Experimentation of the Universidade de Santiago de Compostela (Protocol No 15005AE/07/FUN01/FIS02/JACP1).
Supplementary information
Acknowledgements
We thank the members of MOL Laboratory for helpful discussions and critiques of this work. This work has received financial support from the Spanish MICINN (SAF2009-08629; J.A.C.) and ISCIII, MINECO (PI15/01129), AEI/FEDER, UE (CTQ2017-89166-R; E.P.), the Consellería de Cultura, Educación e Ordenación Universitaria (GPC2014/030, INCITE08PXIB208091PR; J.A.C.), the Xunta de Galicia (Centro Singular de Investigación de Galicia accreditations 2016‐2019, ED431G/05 and ED431G/09) and the European Regional Development Fund (ERDF). C.P. thanks the Spanish MICINN for her PhD fellowship. E.P. thanks the UDC-Inditex InTalent Programme for her research contract and funding.
Author Contributions
P.I., E.P. and J.A.C. designed the experiments. C.P., P.I., L.B.-C. and E.P. carried out the experimental work. P.I., E.P. and J.A.C. analyzed data, E.P. and J.A.C. wrote the manuscript. All authors revised and edited the manuscript.
Data Availability
All data are available from the corresponding authors on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elena Pazos, Email: elena.pazos@udc.es.
Jose A. Costoya, Email: josea.costoya@usc.es
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43618-4.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Fluegen G, et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat. Cell Biol. 2017;19:120–132. doi: 10.1038/ncb3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong BW, Marsch E, Treps L, Baes M, Carmeliet P. Endothelial cell metabolism in health and disease: impact of hypoxia. EMBO J. 2017;36:2187–2203. doi: 10.15252/embj.201696150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaGory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat. Cell Biol. 2016;18:356–365. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chitneni SK, Palmer GM, Zalutsky MR, Dewhirst MW. Molecular imaging of hypoxia. J. Nucl. Med. 2011;52:165–168. doi: 10.2967/jnumed.110.075663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 7.Ettinger A, Wittmann T. Fluorescence live cell imaging. Methods Cell Biol. 2014;123:77–94. doi: 10.1016/B978-0-12-420138-5.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iglesias P, Costoya JA. A novel BRET-based genetically encoded biosensor for functional imaging of hypoxia. Biosens. Bioelectron. 2009;24:3126–3130. doi: 10.1016/j.bios.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Iglesias P, Fraga M, Costoya JA. Defining hypoxic microenvironments by non-invasive functional optical imaging. Eur. J. Cancer. 2013;49:264–271. doi: 10.1016/j.ejca.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Misra T, et al. A genetically encoded biosensor for visualising hypoxia responses in vivo. Biol. Open. 2017;6:296–304. doi: 10.1242/bio.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 12.Jensen EC. Use of fluorescent probes: their effect on cell biology and limitations. Anat. Rec. (Hoboken) 2012;295:2031–2036. doi: 10.1002/ar.22602. [DOI] [PubMed] [Google Scholar]
- 13.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov. Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 15.Semenza GL. Perspectives on oxygen sensing. Cell. 1999;98:281–284. doi: 10.1016/S0092-8674(00)81957-1. [DOI] [PubMed] [Google Scholar]
- 16.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamura T, et al. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutter CH, Laughner E, Semenza GL. Hypoxia-inducible factor 1 alpha protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc. Natl. Acad. Sci. USA. 2000;97:4748–4753. doi: 10.1073/pnas.080072497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 20.Ohh M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 21.Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hon WC, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 23.Harada H, Hiraoka M, Kizaka-Kondoh S. Antitumor effect of TAT-oxygen-dependent degradation-caspase-3 fusion protein specifically stabilized and activated in hypoxic tumor cells. Cancer Res. 2002;62:2013–2018. [PubMed] [Google Scholar]
- 24.Mitchell DJ, Steinman L, Kim DT, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Peptide Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 25.Goun EA, Pillow TH, Jones LR, Rothbard JB, Wender PA. Molecular Transporters: Synthesis of Oligoguanidinium Transporters and Their Application to Drug Delivery and Real-Time Imaging. ChemBioChem. 2006;7:1497–1515. doi: 10.1002/cbic.200600171. [DOI] [PubMed] [Google Scholar]
- 26.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 27.Lougheed T, Borisenko V, Hand CE, Woolley GA. Fluorescent gramicidin derivatives for single-molecule fluorescence and ion channel measurements. Bioconjug. Chem. 2001;12:594–602. doi: 10.1021/bc010006t. [DOI] [PubMed] [Google Scholar]
- 28.Carrigan CN, Imperiali B. The engineering of membrane-permeable peptides. Anal. Biochem. 2005;341:290–298. doi: 10.1016/j.ab.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Marras, S. A. E. Fluorescent energy transfer nucleic acid probes: designs and protocols in Methods in Molecular Biology (ed. Didenko, V. V.) Vol. 335, 3–16 (Springer, 2006). [DOI] [PubMed]
- 30.Uddin MJ, Marnett LJ. Synthesis of 5- and 6-Carboxy-X-rhodamines. Org. Lett. 2008;10:4799–4801. doi: 10.1021/ol801904k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorska K, Huang K-T, Chaloin O, Winssinger N. DNA-templated homo- and heterodimerization of peptide nucleic acid encoded oligosaccharides that mimick the carbohydrate epitope of HIV. Angew. Chem. Int. Ed. 2009;48:7695–7700. doi: 10.1002/anie.200903328. [DOI] [PubMed] [Google Scholar]
- 32.Pazos E, et al. Rational design of a cyclin A fluorescent peptide sensor. Org. Biomol. Chem. 2011;9:7629–7632. doi: 10.1039/c1ob06009k. [DOI] [PubMed] [Google Scholar]
- 33.Pazos E, et al. Cyclin A probes by means of intermolecular sensitization of terbium-chelating peptides. J. Am. Chem. Soc. 2008;130:9652–9653. doi: 10.1021/ja803520q. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Y, Hilliard G, Ferguson T, Millhorn DE. Cobalt inhibits the interaction between hypoxia-inducible factor-alpha and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-alpha. J. Biol. Chem. 2003;278:15911–15916. doi: 10.1074/jbc.M300463200. [DOI] [PubMed] [Google Scholar]
- 35.Müller HD, et al. Effects of prolyl hydroxylase inhibitor L-mimosine on dental pulp in the presence of advanced glycation end products. J. Endod. 2015;41:1852–1861. doi: 10.1016/j.joen.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Kloss A, et al. The cell-penetrating peptide octa-arginine is a potent inhibitor of proteasome activities. Eur. J. Pharm. Biopharm. 2009;72:219–225. doi: 10.1016/j.ejpb.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Khalil IA, et al. Mechanism of improved gene transfer by the N-terminal stearylation of octaarginine: enhanced cellular association by hydrophobic core formation. Gene Ther. 2004;11:636–644. doi: 10.1038/sj.gt.3302128. [DOI] [PubMed] [Google Scholar]
- 38.Vázquez O, et al. Efficient DNA binding and nuclear uptake by distamycin derivatives conjugated to octa-arginine sequences. Chembiochem. 2008;9:2822–2829. doi: 10.1002/cbic.200800345. [DOI] [PubMed] [Google Scholar]
- 39.Saito G, Swanson JA, Lee K-D. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv. Drug Deliv. Rev. 2003;55:199–215. doi: 10.1016/S0169-409X(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 40.Fujita Y, et al. In vivo imaging of brain ischemia using an oxygen-dependent degradative fusion protein probe. PLoS One. 2012;7:e48051. doi: 10.1371/journal.pone.0048051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, et al. Nitroreductase detection and hypoxic tumor cell imaging by a designed sensitive and selective fluorescent probe, 7-[(5-nitrofuran-2-yl)methoxy]-3H-phenoxazin-3-one. Anal. Chem. 2013;85:3926–3932. doi: 10.1021/ac400750r. [DOI] [PubMed] [Google Scholar]
- 42.Luo S, Zou R, Wu J, Landry MP. A probe for the detection of hypoxic cancer cells. ACS Sens. 2017;2:1139–1145. doi: 10.1021/acssensors.7b00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The molecular probes handbook–A guide to fluorescent probes and labelling technologies (ed. Johnson, I. & Spence, M. T. Z.) 76–81 (Live Technologies Corporation, Eleventh Edition 2010).
- 44.McCloy RA, et al. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding authors on reasonable request.