Abstract
The metagenomic approach was applied to characterize variations of microbial structure and functions in raw (RW) and treated water (TW) in a drinking water treatment plant (DWTP) at Pearl River Delta, China. Microbial structure was significantly influenced by the treatment processes, shifting from Gammaproteobacteria and Betaproteobacteria in RW to Alphaproteobacteria in TW. Further functional analysis indicated the basic metabolic functions of microorganisms in TW did not vary considerably. However, protective functions, i.e. glutathione synthesis genes in ‘oxidative stress’ and ‘detoxification’ subsystems, significantly increased, revealing the surviving bacteria may have higher chlorine resistance. Similar results were also found in glutathione metabolism pathway, which identified the major reaction for glutathione synthesis and supported more genes for glutathione metabolism existed in TW. This metagenomic study largely enhanced our knowledge about the influences of treatment processes, especially chlorination, on bacterial community structure and protective functions (e.g. glutathione metabolism) in ecosystems of DWTPs.
Modern drinking water (DW) treatment usually is a multistep process, including flocculation, sedimentation, filtration and disinfection, to reduce the carrying particles and microorganisms in raw water (RW)1. Among these processes, disinfection is a key step in DW treatment plants (DWTPs) to eliminate pathogenic microorganisms by applying various disinfectants, such as chlorine, monochloramine, and ozone2. Although microorganisms can be effectively removed after treatment, some of them may survive and proliferate in DW distribution system (DWDS), and subsequently induce several serious problems3,4, including biofilm growth, nitrification, microbially mediated corrosion, and pathogens persistence. Thus, fully investigation of microbial structure and functions in RW and treated water (TW) of DWTPs is necessary and will facilitate the enhancement of treatment efficiency, the development of anti-pathogen strategies, and the optimization of DWDS.
Recently, high-throughput sequencing (HTS) techniques have shown great advantages on analyzing the microbial community for its unprecedented sequencing depth5. Compared with traditional shotgun sequencing method, HTS techniques, such as 454 pyrosequencing and Illumina sequencing, are proven to be more time-saving and cost-effective6, and have been applied for investigating microbial structure and/or functions in various complex environments, such as fresh water7, sea water8, soil9, and human guts10. Recently, several studies have applied HTS technique to evaluate microbial community in DWTPs and DWDS11,12,13. These studies successfully assessed the microbial community before and after treatment and provided useful information for optimizing the DW treatment processes. However, they did not study the variation of microbial functions, which might be crucial for understanding treatment processes more comprehensively. Up to now, only one metagenomic work evaluated the microbial structure and functions after DW treatment14. However, their study primarily focused on the disinfection effects by two independent methods, rather than the comparison of the RW and TW metagenomes and evaluation the influences of treatments in DWTPs.
Thus, the aim of the present study was to address the following questions: 1) What would be the impact of treatment on the DW microbial structure? 2) What kind of protective functions will play roles in protecting the microorganisms against the chlorination? 3) Will the antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs) be significantly removed in DW treatment processes? To answer the above questions, we collected the microorganisms in RW and TW of a DWTP at Pearl River Delta (PRD), China, separately extracted genomic DNA and conducted Illumina sequencing. About 13 Gb DNA reads were generated and then used to investigate the microbial community, functional profiles, as well as occurrence of ARGs and MGEs in RW and TW of PRD_DWTP.
Results
Metagenomes summary and repeatability
After the metagenomic data were extracted, the reads quality was evaluated by FastQC pipelines. Although the reads quality in TW was not as good as that in RW, the overall quality for TW metagenomes was still acceptable, judged from the quality scores across all bases in TW reads (Figure S1). For RW, 98 ± 0.55% reads could pass the quality check (QC) pipelines of MG-RAST (Table 1). However, only 55 ± 28% reads in TW remained after QC. This was consistent with another study14, in which the reads of two TW metagenomes had lower QC pass ratios (35 and 18%). However, the possible reasons behind this were still unclear. Among reads passing QC, the contents of rRNA in DW were between 0.036 and 0.14%. This varying range was comparable with metagenomes in marine ecosystems15 and activated sludge16, indicating that waterborne metagenomes might contain similar rRNA contents. The unknown reads, which failed to be identified as rRNA genes or annotated as protein with known functions, occupied quite large portion in both of RW (76 ± 3.3%) and TW (84 ± 1.8%) samples. This may suggest that a certain amount of novel reads in PRD_DW samples were captured.
Table 1. Metagenome summary of RW and TW.
| RW (%) | TW (%) | ||||
|---|---|---|---|---|---|
| Metagenomes | RW11 | RW12-1 | RW12-2 | TW11 | TW12 |
| Total Reads | 11,907,016 | 41,924,342 | 36,078,336 | 11,918,058 | 34,555,046 |
| Past QC | 11,799,760 (99) | 41,109,708 (98) | 35,446,306 (98) | 8,973,420 (75) | 12,326,212 (36) |
| Ribosomal RNAa | 15,992 (0.14) | 51,390 (0.13) | 44,052 (0.12) | 3,251 (0.036) | 7,673 (0.062) |
| Annotated Proteina | 3,268,476 (28) | 8,980,700 (22) | 7,847,216 (22) | 1,545,699 (17) | 1,810,357 (15) |
| Unknowna | 8,515,292 (72) | 32,077,618 (78) | 27,555,038 (78) | 7,424,470 (83) | 10,508,182 (85) |
a taking the reads which past QC pipelines as 100%.
To test the repeatability of DNA extraction and Illumina sequencing, the sample of RW12 was divided into two parts as technical duplicates (RW12_1 and RW12_2). DNA in these two duplicates were separately extracted and then sequenced. The results indicated the repeatability was quite well, since high correlation coefficients could be obtained between two duplicates, even at species level (Figure S2).
Taxonomic analysis
Although eukaryotic, archaeal and viral reads (including predicted proteins and rRNA genes) could be detected in RW and TW samples, most of the reads were related to the Bacteria domain (86–96% of total annotated reads, Figure S3). Comparing with RW (23 ± 2 phyla), significantly few (P = 0.019) bacterial phyla were assigned in TW (12 ± 2 phyla), indicating that the diversity of TW community in PRD_DWTP significantly decreased (Figure S4). This could be confirmed by the significantly decreased alpha diversity (P < 0.001), based on the calculation by MG-RAST, in the TW (312 ± 17 species) compared with the RW (1,017 ± 60 species). Similar results could also be obtained by Chao and Shannon indexes in RW and TW metagenomes of the DWTP (Table S1).
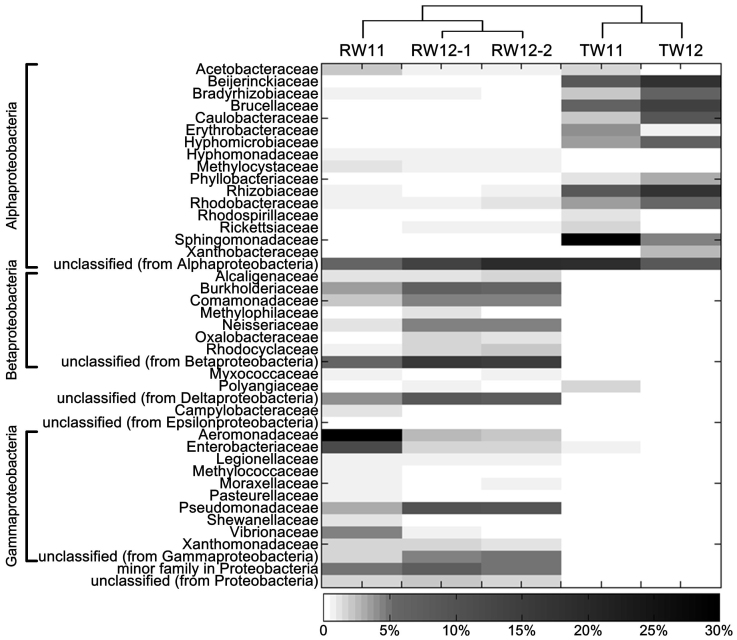
The bacterial structure in Proteobacteria was further analyzed (Figure 1). In RW, Gamma, Beta, and Alphaproteobacteria were the top 3 classes in Proteobacteria (Figure 1). For TW, the abundances of Gamma- (P = 0.043) and Betaproteobacteria (P = 0.022) sharply decreased. While, Alphaproteobacteria increased significantly (P = 0.002) after treatment, as several families in this class became more abundant, including Sphingomonadaceae, Beijerinckiaceae and Rhizobiaceae. This revealed that these families remained after treatment and thus became more abundant in TW of PRD_DWTP, especially for Sphingomonadaceae (from 0.30% to 19% in average, Figure 1).
Figure 1. Percentage (also called relative distribution) of different families in the phylum of Proteobacteria based on the rRNA reads annotated using SILVA SSU database for RW and TW.
The reads number which annotated as Proteobacteria was taken as 100%. The families, which accounted for more than 0.5% in either RW or TW, are shown in the figure. The clustering among samples was according to Gower distance and drawn on the PAST software (version 1.99).
Functional analysis
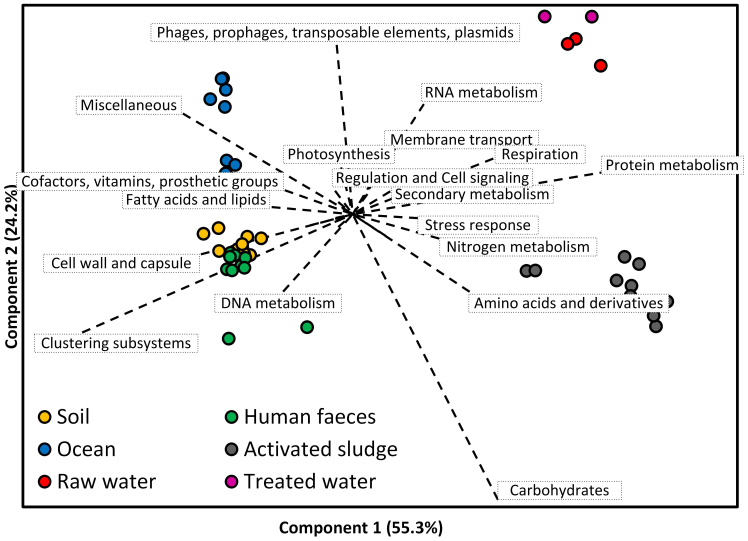
Several Level 1 subsystems of SEED had the largest quantity of annotated reads in RW and TW (Figure S5), including ‘protein metabolism’, ‘carbohydrates’, ‘amino acids and derivatives’ and ‘clustering-based subsystems’. Previous studies also obtained similar major subsystems in metagenomes of soil9, freshwater7, and activated sludge16. To identify specific functions for DW metagenomes in PRD_DWTP, PCA analysis of Level 1 subsystems in different ecosystems was conducted (Figure 2). Metagenomes from different ecosystems separately distributed, revealing functional differences in different ecosystems. Several subsystems, including ‘protein metabolism’, ‘RNA metabolism’, ‘respiration’ and ‘membrane transport’, positively correlated to the DW samples. These functions may play more roles in DW metagenomes than in other ecosystems. Moreover, DW samples clustered together, indicating the unique characteristics of drinking water ecosystems.
Figure 2. The principal component analysis of five ecosystems using the percentage of annotated reads in Level 1 SEED subsystems.
The ecosystems of soil, human faeces and ocean were analyzed by using public data on MG-RAST. The metagenomes of activated sludge17 were also analyzed on MG-RAST. The metagenomic information of these 4 ecosystems is shown in Table S2.
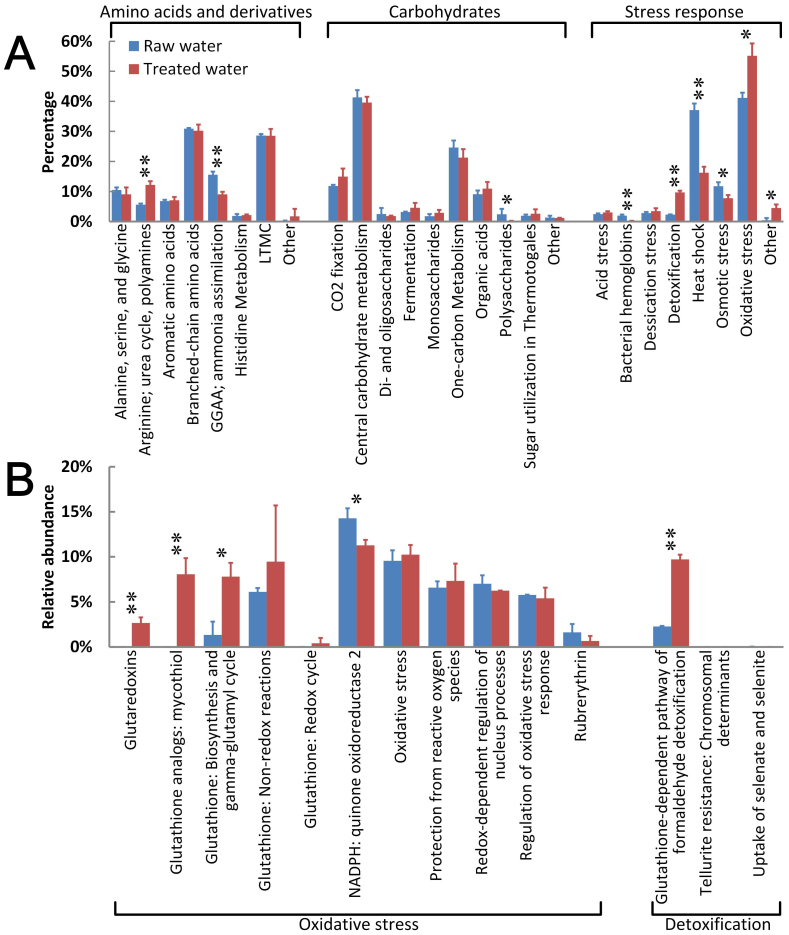
Several dominant subsystems of ‘amino acid and derivatives’ and ‘carbohydrates’, which related to the basic cellular processes that are essential to bacteria, were further analyzed. Their Level 2 subsystems showed high similarity in relative abundances (Figure 3A), suggesting that the treatment processes might not significantly affect the synthesis of amino acid and carbohydrates. However, the protective functions, e.g. ‘stress response’, largely changed after DW treatment (Figure 3B). Comparing with RW, the abundances of genes in ‘oxidative stress’ (P = 0.011) and ‘detoxification’ (P = 0.030) significantly increased in TW, as the genes related to glutathione synthesis largely increased after treatment in PRD_DWTP (Figure 3B).
Figure 3. Average percentages of Level 2 subsystems in ‘amino acids and derivatives’, ‘carbohydrates’, and ‘stress response’ (A) and relative abundances of Level 3 subsystems in ‘oxidative stress’ and ‘detoxification’ (B) in RW and TW.
For Level 2 subsystems (A), the reads number which annotated to the belonging Level 1 subsystems was taken as 100%. For ‘oxidative stress’ and ‘detoxification’ (B), the reads number which annotated to the ‘stress response’ was taken as 100%. The asterisks indicate the significant differences between RW and TW (*: P < 0.05; **: P < 0.01). Here, GGAA represents ‘Glutamine, glutamate, aspartate, asparagine’. LTMC represents ‘Lysine, threonine, methionine, and cysteine’.
Glutathione metabolism
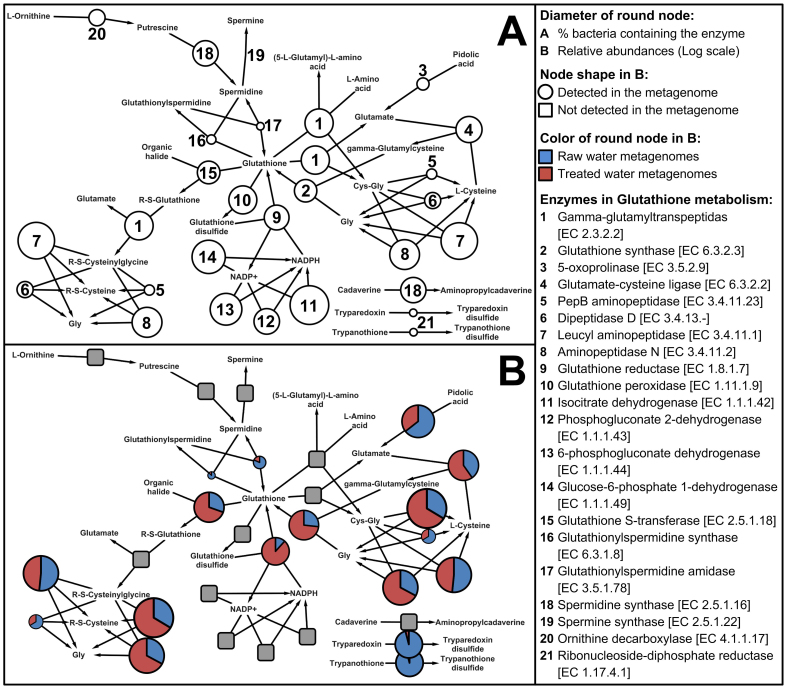
To further study the glutathione synthesis in metagenomes of RW and TW in PRD_DWTP, the pathway of glutathione metabolism was reconstructed by using 1,205 bacterial species (non-redundant) in KEGG collection18. Compared to 40 enzymes in original KEGG pathway constructed by prokaryotes and eukaryotes, the reconstructed pathway contained 21 enzymes produced by bacteria only (Figure 4A). Several enzymes, including isocitrate dehydrogenase, leucyl aminopeptidase and glucose-6-phosphate 1-dehydrogenase, could be produced by most of bacteria, suggesting they might play significant roles for glutathione metabolism in many ecosystems. In DW metagenomes of PRD_DWTP, 12 enzymes were detected (Figure 4B), revealing the bacteria in PRD_DW system only contained partial reactions for glutathione metabolism. Interestingly, several differences were observed when comparing DW samples (Figure 4B) with the established pathway for bacteria (Figure 4A). For instance, quite few bacteria contained enzymes of 5-oxoprolinase and pepB aminopeptidases. While genes for these enzymes could be abundantly found in DW metagenomes of PRD_DWTP, indicating the glutathione metabolism in DW might be different with that in other ecosystems.
Figure 4. Modified KEGG pathway for glutathione metabolism in bacteria domain only.
The pathway A was constructed by 1,205 bacterial species (non-redundant) in KEGG. The pathway B contained average relative abundances of annotated enzymes detected in RW and TW metagenomes.
Two enzymes involving in the glutathione synthesis had relative higher abundances in DW ecosystem of PRD_DWTP, i.e. glutathione synthase and glutathione reductase. However, the reaction of glutathione synthesis by glycine and gamma-glutamylcysteine via glutathione synthase should be the major pathway in DW samples, as abundant enzymes for biosynthesis of glycine (PepB aminopeptidase, leucyl aminopeptidase and aminopeptidase N) and gamma-glutamylcysteine (glutamate-cysteine ligase) could be detected (Figure 4B). Moreover, since the enzymes for the production of NADPH and glutathione disulfide were absent, the importance of glutathione reductase might significantly decrease for glutathione synthesis in DW metagenomes of PRD_DWTP. For most of detected enzymes, significant more genes could be found in TW than RW (Figure 4B). This was consistent with the above observation that genes related to glutathione synthesis were significantly increased after treatment in PRD_DWTP (Figure 3B). The quantification of glutathione synthesis genes were partially confirmed by qRT-PCR, which also showed significant higher abundances of glutathione synthase and glutamate-cysteine ligase genes in TW (Figure S6).
ARGs and MGEs analysis
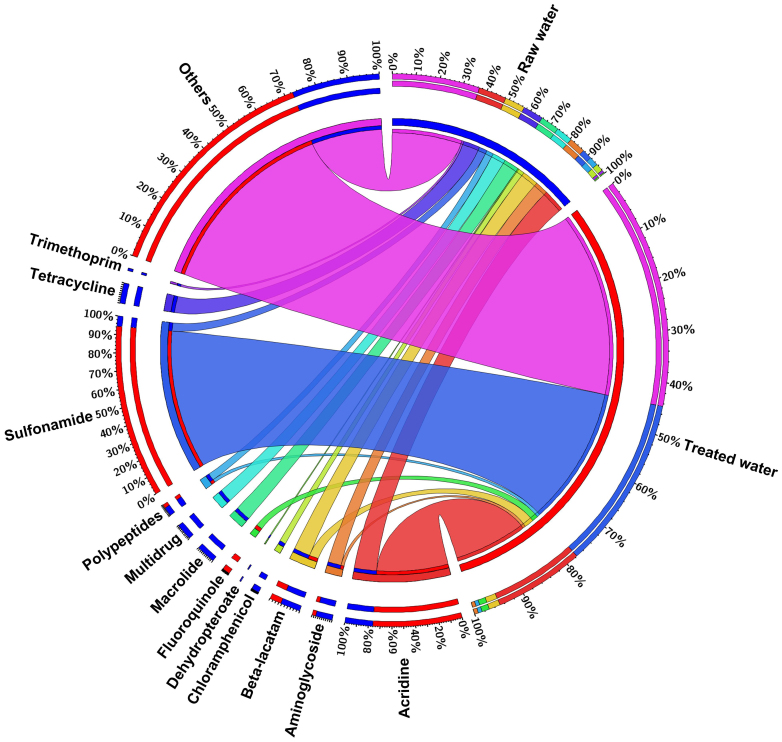
ARGs were detected in RW and TW samples of PRD_DWTP by aligning the reads against related databases. The level (annotated reads number/total reads number) of 10 and 183 ppm for detected ARGs in RW were discovered through comparison against ARDB and CARD database, respectively, and abundance of 2 ppm for ARGs were annotated via ARDB&CARD database. While, more reads in TW could be annotated according to ARDB, CARD and ARDB&CARD databases (Table 2). To be more rigorous, following analysis were based on the results annotated via ARDB&CARD database. Although the level of ARGs showed an increase trend, the diversity of ARGs decreased from 10 to 7 types, accompanying by the percentages of ARGs varied significantly after treatment in PRD_DWTP (Figure 5). In RW, the acridine resistance genes had the highest level, followed by genes resistant to beta-lactam and tetracycline. In TW, the top ARGs were genes resistant to sulfonamide and acridine. Acridine resistance genes showed higher abundance in both DW samples and owned a slight increase with 5% after disinfection. Noticeably, as the dominant ARGs, sulfonamide resistance genes increased from 3.5% to the highest level as 33% after treatment in PRD_DWTP, which may be due to its property as a key component of integrons (all of the sulfonamide resistance genes detected in this study were sul1)19. This could be further convinced by the increasing abundance of integrons in RW and TW with 10 and 58 ppm, respectively (Table 2). This showed much higher abundance than a previous study, in which sulfonamide resistance genes only occupied 5.5% in TW20.
Table 2. The level and diversity of ARGs and MGEs in RW and TW.
| RW | TW | ||||
|---|---|---|---|---|---|
| Database | Level (ppm)a | Diversityb | Level (ppm) | Diversity | |
| ARGs | ARDB | 10 | 13 | 18 | 9 |
| CARD | 183 | 22 | 241 | 16 | |
| ARDB&CARD | 2 | 10 | 8 | 7 | |
| MGEs | Integron | 10 | 60 | 58 | 18 |
| VF | 30 | 83 | 10 | 30 | |
| IS | 19 | 77 | 258 | 29 | |
| Plasmid | 1,650 | 467 | 8,650 | 304 | |
a Level: annotated reads number/total reads number; ppm: parts per million.
b Diversity: assigned categories of ARGs and MGEs by using annotated reads.
Figure 5. Average relative distribution of ARGs in RW and TW against ARDB&CARD database.
The mobility of ARGs usually depends on the MGEs, including integrons, IS, and plasmids21. Totally, the levels of 10 and 58 ppm in RW and TW of PRD_DWTP were annotated to known integronase genes and gene cassettes, respectively. Their levels showed large increase after treatment (Table 2). The plasmids and IS owned a similar trend with larger number detected in TW. However, the level of annotated VF, in TW (10 ppm) was lower than those in RW (30 ppm), revealing that the treatment might remove more VF. Although treatment processes in PRD_DWTP demonstrated different removal efficiencies for different MGEs, they all showed a significantly lower diversity after disinfection, like ARGs (Table 2).
Discussion
In the present study, the majority of cleaned reads (72–85%, Table 1) failed to be annotated as known genes. Other metagenomic studies also obtained similar percentage of unknown reads in various ecosystems, including desert/non-desert soil (77–87%)22, permafrost (~89%)23, grassland (~66%)9 and so on, suggesting low annotation ratio of metagenomic reads is quite common in metagenomic research. This phenomenon might be mainly attributed to several possible explanations, such as limitation of available rRNA/protein database for annotation, short length of obtained reads, algorithms and criterions for alignment, etc. Although minority of obtained reads could be annotated, previous studies also evaluated the general functions (e.g. SEED subsystems) as well as several specific functions (e.g. carbon and nitrogen cycle genes) of microbial communities in various ecosystems9,22,23, revealing that the metagenomes could be applied, at least to some extent, to analyze the detailed microbial structure/functions, even with such low annotation portion of metagenomic reads. Moreover, in the present study, an individual method, i.e. qRT-PCR, was also applied to verify the functional results (glutathione synthesis genes, Figure S6) obtained by metagenomic analysis (Figure 4). The good consistency of results obtained by metagenomic and qRT-PCR methods further proved the validity of metagenomes to study the microbial structure and functions in DW ecosystem.
The taxonomic analysis revealed that DW treatment in the PRD_DWTP could significantly influence the microbial structure, as indicated by the large drop of several dominant phyla in RW after treatment except Proteobacteria (Figure S4). Although previous studies showed that Proteobacteria often dominated in freshwater ecosystems, including DW systems13,24,25, our study revealed the decrease of other phyla which have not been reported before. Furthermore, the dominant classes in Proteobacteria obviously shifted from Gamma and Betaproteobacteria in RW to Alphaproteobacteria in TW (Figure 1), implying bacteria in Alphaproteobacteria may tolerate more chlorination during DW treatment. This revealed more than the results reported in previous studies, that, one only showed the dominant Betaproteobacteria class in RW decreased significantly after chlorination24 and another observed the large increase of Alphaproteobacteria after DW treatment13. Among the dominant families in Alphaproteobacteria of TW, the survival of Sphingomonadaceae family might be associated with its high resistance to chlorination26. Thus, the bacteria in Sphingomonadaceae family are often abundantly found in DW systems27,28, as we observed in PRD_DWTP (Figure 1).
Previous studies indicated that some bacteria, which might resist to chlorination at a certain degree, could survive after disinfection and proliferate in the DWDS29,30. This is consistent with the results of the present study, since the functional analysis strongly suggested the microorganisms in TW of PRD_DWTP contained higher protective genes responding to the selective pressure of chlorination, such as glutathione related genes (Figure 3 and 4). Glutathione has been proven to directly increase bacterial resistance to chlorine compounds31 and is also indirectly implicated in the regulation of other oxidation resistant systems, such as OxyR, SoxR and SOS systems32. Noticeably, starvation could stimulate the glutathione synthesis and subsequently enhance bacterial chlorine resistance32. This may explain the poor efficiency of residual disinfectants in DW to inactivate pathogens33, especially considering the oligotrophic conditions in the DWDS networks. Moreover, it was reported that glutathione could be primarily found in Gram-negative bacteria and eukaryotes34. While, the genes for glutathione biosynthesis in eukaryotes were proposed to have been transferred from bacteria via the progenitor of mitochondria during evolutionary35. This widely accepted theory strongly suggests the Alphaproteobacteria, the modern relatives of the mitochondrial progenitor34, may commonly contain glutathione biosynthesis genes. This might be one of possible explanations for the taxonomic observation that Alphaproteobacteria became dominant in the TW metagenomes of PRD_DWTP (Figure 1), rather than other bacterial classes.
After treatment in PRD_DWTP, the levels of most detected ARGs decreased significantly (Figure 5), suggesting the DW treatment could significantly remove most of ARGs in RW. This might be mainly attributed to the effective removal of the corresponding bacteria, which carried those ARGs, during treatment36. However, as several specific ARGs largely accumulated after treatment in PRD_DWTP (Figure 5), the level of total ARGs in TW significantly increased (Table 2), revealing the chlorination may increase bacterial resistances for specific antibiotics. This is consistent with the previous observation that bacteria in swine wastewater displayed higher antibiotic resistances after chlorination37. This phenomenon might be mainly caused by the co-selection of chlorine/chloride and antibiotic resistance38. For example, Pseudomonas aeruginosa could join in the course of benzalkonium chloride to promote resistance to several antibiotics39. The co-selection mechanism was reported that chlorination could induce the mutation and substitution of bacterial structure genes, e.g. gyrA, nfxB and mexR, which are important mechanisms of resistance, encoding different ARGs to ciprofloxacin and fluoroquinolone40.
Similar to ARGs, the levels of MGEs in TW of PRD_DWTP also increased (Table 2). This might be partially attributed to the bacterial stress response which can enhance the plasmid production when facing environmental stresses41 and thus may increase the plasmids copy number in the cells of remaining bacteria after chlorination20. For other MGEs, the mechanisms behind are still unclear. Since MGEs were reported to play important roles in ARGs horizontal gene transfer42, the increase of ARGs and MGEs in surviving bacteria might enlarge the risks for ARGs horizontal transfer among bacteria in DWDS and cause serious threats to human being. Therefore, more efforts should be made in future to assess the occurrence and fate of ARGs and MGEs in DW systems and comprehensively evaluate their potential risks for human health.
HTS has been proven to be a powerful technique to characterize various ecosystems including DW systems11,12,13,14. In the present study, we successfully characterized the taxonomic and functional profiles of bacteria in PRD_DWTP before and after treatment via metagenomic analysis. However, there are still some limitations in our study. First of all, although it was suggested that reads length of 100 bp was long enough to resolve microbial community differences5, Illumina reads with 100 bp length were still relatively short for accurate identification of the DW community at deeper levels (genus or species). Second, large amount of dead cells were generated after disinfection. Although released DNA from broken cells could dissolve and might not be retained by the applied filters, the DNA from dead but intact cells might contaminate the TW samples. This is possible to bring bias to the presented results. Thirdly, it should be noticed that the rise of functional genes for oxidative stress, detoxification and glutathione metabolism after treatment were just potential trends and did not guarantee any increase of the expression of these genes in TW ecosystems, as the current analyses were based on DNA instead of RNA. Thus, metatranscriptomic studies based on RNA should be conducted in future to accurately evaluate the active taxonomy and functions in DW systems.
Methods
Water samples
RW and TW were collected from a DWTP which has a production capacity of 135,000 m3/day, located at Pearl River Delta area, China. RW was from a reservoir and was treated by flocculation, sedimentation, sand filtration and chlorination (Figure S8). Torayvino high-performance cartridge-type water purifiers (Toray Industries Inc., Japan) were equipped on the RW and TW taps in the DWTP, since they are featured for easy operation and effective collection for microorganisms. After purifiers' equipment, the RW and TW were filtrated for 3 and 72 h, respectively. And the microorganisms in the resulting 86 L of RW and 2070 L of TW were then collected by controlling the flow rate at 8 mL/s. After collection, the purifiers were immediately transported to laboratory. Upon the arrival, the hollow fiber filter in purifier was immersed into 200 mL ultrapure water and then treated by ultrasonication (8200E-1, Branson Ultrasonics Corp., US) for 30 min to detach the microbial cells. Visual microscopy indicated that the collected microorganisms could be effectively detached from filter surface by the ultrasonication (Figure S9). The cells in water were collected by filtration using mixed cellulose esters membrane (HAWP04700, Millipore Corp., US) with a pore size of 0.45 μm. Previous studies supported 0.45 μm membrane could effectively collect microorganisms in the water20. The membrane was stored at −20°C before DNA extraction. Five samples of RW and TW collected in July of 2011 and 2012 were named as RW11, RW12-1, RW12-2, TW11, and TW12. Biological duplicated were designed by applying RW and TW samples from different years. Technical duplicates were conducted as RW12-1 and RW12-2 from the same RW sample.
DNA extraction and Illumina sequencing
Genomic DNA of microorganisms in RW and TW was separately extracted by FastDNA SPIN Kit for Soil (MP Biomedicals, Illkirch, France) according to the instruction. The concentration and purity of DNA was evaluated by NanoDrop spectrophotometer (ND-1000, Thermo Fisher Scientific, US). DNA of ~10 μg for each sample was used for library construction. In detail, DNA fragmentation was carried out by Covaris S2 (Covaris, 01801-1721). The fragments were then processed by end reparation, A-tailing, adapter ligation, DNA size-selection, PCR reaction and products purification. Finally, a ~180 bp DNA fragment reads library was constructed and then sequenced by Illumina HiSeq 2000 (BGI, China). The base-calling pipeline (Version Illumina Pipeline - 0.3) was used to process the raw fluorescence images and call reads. Raw reads with >10% unknown nucleotides or with >50% low quality nucleotides (quality value < 20) were discarded10.
Acquired Illumina reads were filtered by using Meta Genome Rapid Annotation using Subsystem Technology (MG-RAST, http://metagenomics.nmpdr.org) QC pipelines43 to remove the replicated reads, since the platforms of HTS occasionally produce large numbers of reads that are nearly identical44. Only one representative read in the clusters of replicated reads, whose first 50 base pairs were identical, was preserved. The reads which contained 5 or more ambiguous base were then removed. The filtered reads were used for the following bioinformatic analysis.
Bioinformatic analysis
Illumina reads of RW and TW samples were annotated by using MG-RAST online server (version 3.3)43. The reads were distributed into different categories, including rRNA reads, protein reads (with known or unknown functions), and unknown reads, according to the results of similarity comparison with rRNA and protein databases.
For taxonomic analysis, SILVA Small Subunit (SSU) database (version 104)45 was used as annotation source for 16S rRNA reads to analyze the bacterial populations in RW and TW samples by using an E-value cutoff of 10−5, minimum identity cutoff of 60%, and minimum alignment length cutoff of 15 aa.
For functional analysis, SEED Subsystems46 and KEGG18 databases were applied to explore the microbial functions in DW samples. Similarity search between protein reads and the SEED/KEGG databases was conducted by using an E-value cutoff of 10−5, minimum identity cutoff of 60%, and minimum alignment length cutoff of 15 aa. The annotated reads were sorted into 28 Level 1 subsystems to provide overall profile of microbial functions and were then compared with other 42 metagenomes from 4 ecosystems (Table S2). The Level 2 and 3 subsystems, which belong to subsystems of ‘amino acids and derivatives’, ‘carbohydrates’, and ‘stress response’, were further analyzed to investigate specific shift of microbial functions after treatment. The KEGG was used to construct glutathione pathway in bacteria domain and to evaluate the specific reactions for glutathione metabolism in DW metagenomes.
ARGs and MGEs analysis
ARG reads on Antibiotic Resistance Genes Database (ARDB, 7828 reads, http://ardb.cbcb.umd.edu)47 and Comprehensive Antibiotic Resistance Database (CARD, 3380 reads, http://arpcard.mcmaster.ca) were downloaded. The sub-databases of ARDB and CARD were created according to the antibiotic categories42 (Figure S10). Then reads in ARDB were aligned against CARD using BLAST with an E-value cutoff of 10−6 to develop the core database of ARDB and CARD, referring as ARDB&CARD. A protein read in the ARDB and CARD was annotated as a shared resistance gene in the ARDB&CARD, according to its BLAST hit (blastp) for the alignment with amino acid read identity as 100%. Then reads in RW and TW were aligned against ARDB, CARD and ARDB&CARD using BLAST with an E-value cutoff of 10−6, respectively. A read was annotated as a resistance gene according to its BLAST hit (blastx) for the alignment with amino acid read identity above 90% for at least 25 aa42. The BLAST results were sorted into the sub-database by a script. The sorting results of RW and TW were then compared to evaluate the occurrence, level and elimination of ARGs during treatment.
MGEs, including integrons, insertion sequences (IS), VF and plasmids, which play important roles in transporting ARGs in environments, were also conducted. The metagenomes were searched for signatures of known MGEs in the reference databases, including INTEGRALL for integrons48, ISfinder for IS49, VFDB for VF50, and NCBI RefSeq for plasmids42. A read was assigned to an integron, IS, VF, plasmid if the BLAST hit (blastn) with a nucleotide read identity over 90% or at least 50 bases42. After the level of MGEs being explored, the diversity of integron, IS, VF and plasmid in RW and TW were also counted by script according to annotated accession numbers, and then compared.
Quantitative RT-PCR
The results from metagenomic data were confirmed by quantitative RT-PCR (qRT-PCR) on a MyiQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The relative concentration of two glutathione synthesis genes, i.e. gshA (glutamate-cysteine ligase, EC 6.3.2.2) and gshB (glutathione synthase, EC 6.3.2.3), were measured by qRT-PCR using selected primers (Table S3). Reactions were conducted in PCR tubes strip with a final volume of 25 μL, containing 12.5 μL 2 × iQSYBRGreen Super-Mix (Bio-Rad, Hercules, CA), 0.5 μL of each primer (10 μM) and 1 μL template DNA (~25 ng DNA). Thermal cycling were conducted using the following protocol: 94°C for 3 min, followed by 40 cycles of 94°C for 5 s, annealing at 56°C for 30 s and 72°C for 1 min. Each reaction was run in triplicate. The quantification of target genes was done with the software iCycler iQversion 5.0 (Bio-Rad, Hercules, CA). The cycle threshold (Ct) value was used to calculate and compare the relative gene concentrations in RW and TW. The RW and TW samples in 6 months of 2012 (Feb., Apr. Jun., Aug., Oct., and Dec.) were applied.
Supplementary Material
Supporting Information
Acknowledgments
The authors thank the Hong Kong GRF (HKU 7201/11E) for the financial support on this study. Yuanqing Chao, Liping Ma, Ying Yang and Feng Ju thank HKU for the postgraduate studentship. Prof. Xu-Xiang Zhang thanks HKU for the postdoctoral fellowship. The technical assistance of Ms. Vicky Fung is greatly appreciated.
Metagenomes accession numbers: DW metagenomes studied in the present study were deposited in the NCBI Sequence Read Archive database with accession numbers of SRR835363, SRR850211, SRR850212, SRR850456 and SRR850459.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.C. and L.M. conducted the experiments, analyzed the data, and wrote the manuscript. Y.Y. conducted the experiments and analyzed the data. F.J. analyzed the data. X.X.Z. conducted the experiments. W.M.W. provided important suggestions. T.Z. designed the experiments and modified this manuscript. All authors reviewed the manuscript.
References
- Betancourt W. Q. & Rose J. B. Drinking water treatment processes for removal of Cryptosporidium and Giardia. Vet. Parasitol. 126, 219–234 (2004). [DOI] [PubMed] [Google Scholar]
- Ramseier M. K., von Gunten U., Freihofer P. & Hammes F. Kinetics of membrane damage to high (HNA) and low (LNA) nucleic acid bacterial clusters in drinking water by ozone, chlorine, chlorine dioxide, monochloramine, ferrate(VI), and permanganate. Water Res. 45, 1490–1500 (2011). [DOI] [PubMed] [Google Scholar]
- Berry D., Xi C. W. & Raskin L. Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol. 17, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- Pronk M., Goldscheider N. & Zopfi J. Particle-size distribution as indicator for fecal bacteria contamination of drinking water from karst springs. Environ. Sci. Technol. 41, 8400–8405 (2007). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porazinska D. L. et al. Evaluating high-throughput sequencing as a method for metagenomic analysis of nematode diversity. Mol. Ecol. Resour. 9, 1439–1450 (2009). [DOI] [PubMed] [Google Scholar]
- Breitbart M. et al. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro CiEnegas, Mexico. Environ. Microbiol. 11, 16–34 (2009). [DOI] [PubMed] [Google Scholar]
- DeLong E. F. et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311, 496–503 (2006). [DOI] [PubMed] [Google Scholar]
- Delmont T. O. et al. Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J. 6, 1677–1687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J. J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P. Y. et al. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Appl. Environ. Microbiol. 76, 5631–5635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S., Moon E., Kim T. S., Hong S. & Park H. D. Pyrosequencing demonstrated complex microbial communities in a membrane filtration system for a drinking water treatment plant. Microbes Environ. 26, 149–155 (2011). [DOI] [PubMed] [Google Scholar]
- Pinto A. J., Xi C. W. & Raskin L. Bacterial community structure in the drinking water microbiome is governed by filtration processes. Environ. Sci. Technol. 46, 8851–8859 (2012). [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V., Revetta R. P. & Domingo J. W. S. Metagenomic analyses of drinking water receiving different disinfection treatments. Appl. Environ. Microbiol. 78, 6095–6102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A. et al. Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environ. Microbiol. 11, 111–125 (2009). [DOI] [PubMed] [Google Scholar]
- Ye L., Zhang T., Wang T. T. & Fang Z. W. Microbial structures, functions, and metabolic pathways in wastewater treatment bioreactors revealed using high-throughput sequencing. Environ. Sci. Technol. 46, 13244–13252 (2012). [DOI] [PubMed] [Google Scholar]
- Ju F., Guo F., Ye L., Xia Y. & Zhang T. Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 10.1111/1758-2229.12110 (2013). [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M. & Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucl. Acids Res. 40, D109–D114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. P. et al. Occurrence, abundance and elimination of class 1 integrons in one municipal sewage treatment plant. Ecotoxicology 20, 968–973 (2011). [DOI] [PubMed] [Google Scholar]
- Shi P. et al. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res. 47, 111–120 (2013). [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Tsafnat G., Coiera E. & Iredell J. R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784 (2009). [DOI] [PubMed] [Google Scholar]
- Fierer N. et al. Cross-biome metagenomic analysis of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 109, 21390–21395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackelprang R. et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480, 368–371 (2011). [DOI] [PubMed] [Google Scholar]
- Kormas K. A., Neofitou C., Pachiadaki M. & Koufostathi E. Changes of the bacterial assemblages throughout an urban drinking water distribution system. Environ. Monit. Assess. 165, 27–38 (2010). [DOI] [PubMed] [Google Scholar]
- Poitelon J. B. et al. Assessment of phylogenetic diversity of bacterial microflora in drinking water using serial analysis of ribosomal sequence tags. Water Res. 43, 4197–4206 (2009). [DOI] [PubMed] [Google Scholar]
- Koskinen R. et al. Characterization of Sphingomonas isolates from Finnish and Swedish drinking water distribution systems. J. Appl. Microbiol. 89, 687–696 (2000). [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Harrington G. W., Xagoraraki I. & Goel R. Factors affecting bulk to total bacteria ratio in drinking water distribution systems. Water Res. 42, 3393–3404 (2008). [DOI] [PubMed] [Google Scholar]
- Vaz-Moreira I., Nunes O. C. & Manaia C. M. Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl. Environ. Microbiol. 77, 5697–5706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton C. D., LeChevallier M. W. & Falkinham J. O. Survival of Mycobacterium avium in a model distribution system. Water Res. 38, 1457–1466 (2004). [DOI] [PubMed] [Google Scholar]
- Wang Y. Y., Claeys L., van der Ha D., Verstraete W. & Boon N. Effects of chemically and electrochemically dosed chlorine on Escherichia coli and Legionella beliardensis assessed by flow cytometry. Appl. Microbiol. Biotechnol. 87, 331–341 (2010). [DOI] [PubMed] [Google Scholar]
- Chesney J. A., Eaton J. W. & Mahoney J. R. Bacterial glutathione: A sacrificial defense against chlorine compounds. J. Bacteriol. 178, 2131–2135 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saby S., Leroy P. & Block J. C. Escherichia coli resistance to chlorine and glutathione synthesis in response to oxygenation and starvation. Appl. Environ. Microbiol. 65, 5600–5603 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payment P. Poor efficacy of residual chlorine disinfectant in drinking water to inactivate waterborne pathogens in distribution systems. Can. J. Microbiol. 45, 709–715 (1999). [PubMed] [Google Scholar]
- Copley S. D. & Dhillon J. K. Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome. Biol. 3, research0025.1–0025.16 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey R. C., Newton G. L., Arrick B., Overdankbogart T. & Aley S. B. Entamoeba histolytica - a eukaryote without glutathione metabolism. Science 224, 70–72 (1984). [DOI] [PubMed] [Google Scholar]
- Xi C. W. et al. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl. Environ. Microbiol. 75, 5714–5718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley J. J., Qiang Z. M., Adams C. D., Surampalli R. & Mormile M. R. Disinfection of swine wastewater using chlorine, ultraviolet light and ozone. Water Res. 40, 2017–2026 (2006). [DOI] [PubMed] [Google Scholar]
- Loughlin M. F., Jones M. V. & Lambert P. A. Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49, 631–639 (2002). [DOI] [PubMed] [Google Scholar]
- Mc Cay P. H., Ocampo-Sosa A. A. & Fleming G. T. A. Effect of subinhibitory concentrations of benzalkonlum chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology 156, 30–38 (2010). [DOI] [PubMed] [Google Scholar]
- Varon E., Janoir C., Kitzis M. D. & Gutmann L. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43, 302–306 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn G. & Wegrzyn A. Stress responses and replication of plasmids in bacterial cells. Microb. Cell Fact. 1, 2 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E. et al. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE 6, e17038 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V., Teal T. K. & Schmidt T. M. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3, 1314–1317 (2009). [DOI] [PubMed] [Google Scholar]
- Pruesse E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl. Acids Res. 35, 7188–7196 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucl. Acids Res. 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. & Pop M. ARDB-Antibiotic Resistance Genes Database. Nucl. Acids Res. 37, D443–D447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A. et al. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098 (2009). [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J. & Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucl. Acids Res. 34, D32–D36 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Chen L. H., Sun L. L., Yu J. & Jin Q. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucl. Acids Res. 36, D539–D542 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information