Abstract
Objectives: To describe a rapid workflow based on the direct detection of Escherichia coli (Ec) and Klebsiella pneumoniae (Kp) producing CTX-M extended-spectrum β-lactamase (ESBL) and/or carbapenemases (eg, KPC, VIM) from blood cultures (BCs) and the infectious disease (ID) consulting for timely appropriate antimicrobial therapy.
Methods: This observational, retrospective study included adult patients with a first episode of Ec or Kp bloodstream infection (BSI) in a large Italian university hospital, where an inpatient ID consultation team (IDCT) has been operational. Results from the BCs tested for detecting blaCTX-M, blaKPC, blaNDM, blaOXA-48-like, and blaVIM genes by the eazyplex® SuperBug CRE assay in Ec and Kp organisms had been notified for antimicrobial therapy consulting.
Results: In 321 BSI episodes studied, we found that 151 (47.0%) of Ec or Kp organisms harbored blaCTX-M and/or blaKPC and/or blaVIM (meantime from BC collection: 18.5 h). Empirical antimicrobial treatment was appropriate in 21.8% (33/151) of BSIs, namely 5.9% (3/51) of BSIs caused by KPC/VIM producers and 30.0% (30/100) of BSIs caused by CTX-M producers. After notification of results, the IDCT modified antimicrobial therapy (mean time from BC collection: 20 h) such that the proportion of appropriate treatments increased to 84.8% (128/151) of BSIs, namely 70.6% (36/51) of BSIs caused by KPC/VIM producers and 92.0% (92/100) of BSIs caused by CTX-M producers.
Conclusion: Our study shows that a rapid diagnostic-driven clinical strategy allowed for early prescription of potentially effective antimicrobial therapy in BSIs caused by CTX-M ESBL- and/or KPC/VIM carbapenemase-producing Ec and Kp organisms.
Keywords: Escherichia coli, Klebsiella pneumoniae, bloodstream infection, drug resistance, targeted therapy, infectious disease consultation
Background
Escherichia coli (Ec) and Klebsiella pneumoniae (Kp) are the leading causes of bloodstream infections (BSIs) involving gram-negative bacteria, and β-lactams are among the most used drugs for treatment of such infections. Globally, increasing attention is being focused on the growing involvement of extended-spectrum-β-lactamase (ESBL)-producing isolates of Ec and Kp in serious BSIs.1,2 This trend is due largely to the emergence of CTX-M ESBLs, a rapidly expanding group of enzymes that are being encountered with increasing frequency in both bacterial species.3,4 Carbapenems represent one therapeutic option for infections caused by multidrug-resistant (MDR) ESBL-producing organisms; however, resistance to this class of antibiotics has increased during the last decade.5 Carbapenem-resistant Enterobacteriaceae, particularly Kp producing blaKPC, are now responsible for a new dramatic scenario and are emerging as a relevant cause of health care-associated BSIs in several countries, including Italy.6–8
Detecting rapidly and accurately BSI pathogens is a critical factor for earlier implementation of effective treatment of these life-threatening infections. Although blood culture (BC) remains the gold standard for diagnosing BSIs, the time-consuming process of isolate identification/characterization with conventional culture-based methods has been improved by the development of faster technologies, allowing identification of the infecting pathogen directly from BC bottles.9 In addition to matrix-assisted laser desorption ionization–time of flight mass spectrometry (eg, MALDI BioTyperTM system [Bruker Daltonics, Bremen, Germany]), various commercial BC-based nucleic acid assays can shorten the identification time of pathogens and/or resistance genes,10 and have added value for antibiotic stewardship decisions by decreasing the time to initiating appropriate antibiotic therapy.11–15
To provide rapid pathogens’ characterization for early appropriate treatment of patients with Ec- or Kp-BSI hospitalized in a CTX-M and KPC-endemic setting, we implemented the laboratory’s BC workflow that uses the eazyplex® SuperBug CRE assay (Amplex Diagnostics GmbH, Biolife italiana SRL, Milan, Italy) when Ec or Kp organisms were identified by direct MALDI BioTyperTM analysis from positive BCs.10 Clinical experience to date shows that this panel identifies the CTX-M-1–group and CTX-M-9–group ESBLs and the VIM (−1 to −37), NDM (−1 to −7), KPC (−2 to −15), and OXA-48-like (−48, −162, −204, and −244) carbapenemases with high accuracy when applied directly to both cultured bacterial and urine samples.16–18 Our workflow was integrated with real-time result notification and inpatient infectious disease consultation team (IDCT) intervention. In the present one-year retrospective study, we assessed whether this laboratory/clinical workflow may have affected the timely prescription of appropriate antimicrobial therapy for BSIs caused by CTX-M ESBL- and/or carbapenemase-producing Ec or Kp organisms.
Methods
This study was conducted from 15 January 2015 through 15 January 2016 at a 1,200-bed university hospital (Fondazione Policlinico Universitario A. Gemelli IRCCS, Università Cattolica del Sacro Cuore, Rome, Italy), under approval by our institutional Ethics Committee (no. 0040288/16). A central microbiology laboratory, which is open from 7:00 a.m. to 7:00 p.m., Monday through Friday and from 7:00 a.m. to 4:00 p.m., Saturday serves all the hospital wards. The hospital has an IDCT comprising four ID specialists, who operate on a request basis (via the hospital’s computerized information system) by the physicians operating in medical and surgical wards (except for hematology unit and ICU, which have dedicated ID specialists). The IDCT takes charge of patients at the bedside within 24 hrs of the request.19 Eligible patients were adults (≥18 years old) with a first, clinically significant episode of BC-documented BSI for Ec or Kp. Patients were excluded if they died or were discharged before BC results were available, or if they had incomplete data.
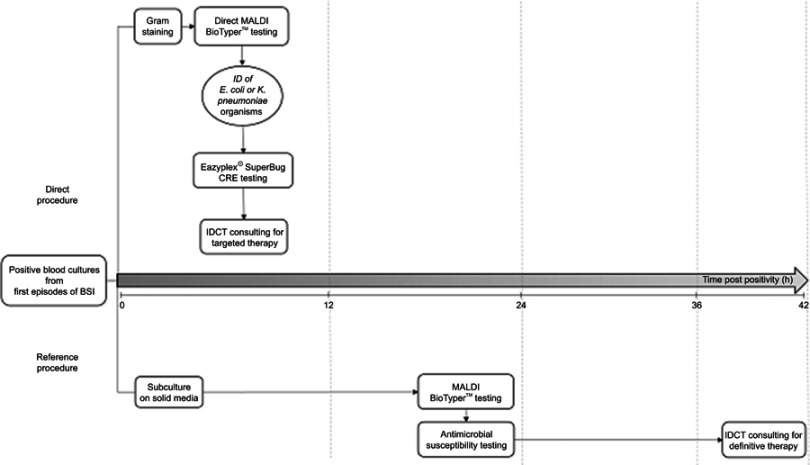
Figure 1 depicts the diagnostic workflow used in this study. When patients’ BC signaled positive in either BACT/ALERT® VIRTUOTM (bioMérieux, Marcy l’Étoile, France) or BACTECTM FX (Becton Dickinson, Sparks, MD, USA) automated systems, aliquots from BC bottle broths were subjected to the direct MALDI BioTyperTM analysis10 and, when Ec or Kp organisms were identified, the eazyplex® SuperBug CRE assay was performed. MALDI BioTyperTM identifications of Ec or Kp organisms were always concordant with those of reference (culture-based) identification methods (Figure 1). As described elsewhere,17 the eazyplex® SuperBug CRE assay relies on a loop-mediated isothermal amplification (that covers the aforementioned carbapenemase variants) and a subsequent real-time fluorescence based visualization of the amplification products. For each patient, results of the laboratory procedure were immediately available to the IDCT. The microbiologist also informed the in-charge ID specialist about that identified organisms could harbor bla genes other than those detected in the eazyplex® SuperBug CRE assay (eg, TEM/SHV ESBLs, plasmidic AmpC β-lactamases), which confer non-susceptibility to extended-spectrum cephalosporins (ESC) or carbapenems (carba). Antimicrobial minimum inhibitory concentrations (MICs) of the Ec or Kp isolates identified by the MALDI BioTyperTM analysis were determined by antimicrobial susceptibility testing (AST) broth microdilution methods and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (version 8.0, http://www.eucast.org/clinical_breakpoints/). We performed phenotypic detection of resistance mechanisms according to the EUCAST guidelines (version 2.0, http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/). To confirm antimicrobial resistance profiles, all the Ec and Kp isolates underwent PCR sequencing of the blaTEM, blaSHV, blaCTX-M, blaKPC, blaNDM, blaOXA-48-like, blaVIM, and blaAmpC genes.2,20–22
Figure 1.
Diagnostic workflow using MALDI BioTyperTM analysis/eazyplex® SuperBug CRE assay (direct procedure) or culture-based methods (reference procedure) on positive blood cultures from E. coli or K. pneumoniae bloodstream infections (BSIs). Results from both the diagnostic procedures were notified to an IDCT for streamlining the antimicrobial treatments of BSIs.
Abbreviations: ID, identification; IDCT, infectious disease consultation team.
Diagnostic and clinical definitions
Time to detection of growth (TTD) was the time from when the BC bottle entered into the BC system to when it signaled positive. The time to result (TTR) was the time elapsed between the BC system entry and completion of the aforementioned direct-detection procedure. We assessed the appropriateness of antibiotic treatments at the time of BC collection (empirical therapy) and after notification of the procedure (targeted therapy) or AST (definitive therapy) results. We defined the antimicrobial treatment as appropriate when the patient received the first (empirical) and/or subsequent (diagnostic-driven) antibiotic with known susceptibility by microbiology report.
Results
We studied 321 patients with Ec (n=214) or Kp (n=107) BSIs. The mean TTD for the patient-unique positive BCs (n=321) was 10.3 h (range: 2–90 h). The mean TTR for the 151 (47.0%) Ec or Kp organisms harboring blaCTX-M and/or blaKPC and/or blaVIM (as detected by the MALDI BioTyperTM analysis and eazyplex® SuperBug CRE assay) was 18.5 h (range: 3.3–96 h). The mean AST was 51 h (range: 36–141 h) for all the 321 Ec and Kp isolates, including 106 (33.0%) isolates that were non-susceptible to ESC but susceptible to carba (ESCR-carbaS phenotype) (Table 1). Accordingly, ESBL production was detected in 36.0% (77/214) of the Ec isolates and in 21.5% (23/107) of the Kp isolates, and all the 100 isolates carried CTX-M ESBL genes. The blaCMY-2 gene (not detectable by the eazyplex® SuperBug CRE assay) was detected in the remaining 1.8% (6/321) of Ec or Kp isolates. The proportions of the Ec or Kp isolates harboring carbapenemase genes (ESCR-carbaR phenotype) were 0.9% (2/214) and 45.8% (49/107), respectively, with blaKPC-3 being the most commonly detected gene (Table 1).
Table 1.
Antimicrobial susceptibilities for 321 Escherichia coli and Klebsiella pneumoniae BSI organisms (151 positive and 170 negative by the eazyplex® SuperBug CRE assay detection) according to the presence of resistance genes
| Type of β-lactamase in the indicated species (no. of isolates)a | No. (%) of isolates susceptible to the following antimicrobial agents | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | FEP | ETP | MEM | TZP | AK | GM | CIP | SXT | TGC | CST | |
| E. coli (214) | ||||||||||||
| blaCTX-M (77) | 0 (0.0) | 5 (6.5) | 0 (0.0) | 77 (100) | 77 (100) | 53 (68.8) | 51 (66.2) | 47 (61) | 6 (7.8) | 38 (49.3) | – | – |
| blaCMY (5)b | 0 (0.0) | 0 (0.0) | 5 (100) | 5 (100) | 5 (100) | 1 (20.0) | 4 (80.0) | 3 (60.0) | 1 (20.0) | 1 (20.0) | – | – |
| blaKPC/blaVIM (2) | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 2 |
| None (130) | 130 (100) | 130 (100) | 130 (100) | 130 (100) | 130 (100) | 114 (87.7) | 129 (99.2) | 113 (86.9) | 95 (73.1) | 83 (63.8) | – | – |
| K. pneumoniae (107) | ||||||||||||
| blaCTX-M (23) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 23 (100) | 23 (100) | 6 (26.1) | 17 (73.9) | 6 (26.1) | 2 (8.7) | 3 (13.0) | – | – |
| blaCMY (1)b | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | – | – |
| blaKPC/blaVIM (49) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 12 (24.5) | 41 (83.7) | 2 (4.0) | 9 (18.4) | 39 (78.6) | 29 (59.2) |
| None (34) | 34 (100) | 34 (100) | 34 (100) | 34 (100) | 34 (100) | 30 (88.2) | 34 (100) | 34 (100) | 30 (88.2) | 31 (91.2) | – | – |
Notes: Colistin and tigecycline susceptibility results were available only for carbapenem-resistant isolates. We did not calculate percentages in cases with less than five isolates. aWe identified the extended-spectrum β-lactamase (ESBL), plasmidic AmpC β-lactamase, and carbapenemase genes by standard PCR sequencing analysis. Specifically, E. coli isolates harbored blaCTX-M-15 (n=55), blaCTX-M-14 (n=9), blaCTX-M-27 (n=13), blaCMY-2 (n=5), blaKPC-3 (n=1), and blaVIM-1 (n=1); K. pneumoniae isolates harbored blaCTX-M-15 (n=23), blaCMY-2 (n=1), blaKPC-2 (n=2), blaCTX-M-15+ blaKPC-3 (n=4), blaKPC-3 (n=41), and blaKPC-3+ blaVIM-1 (n=2). None refers to the isolates not harboring such genes. bSix isolates harboring blaCMY genes had a negative result with the eazyplex® SuperBug CRE assay.
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ETP, ertapenem; MEM, meropenem; TZP, piperacillin-tazobactam; AK, amikacin; GM, gentamicin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; CST, colistin.
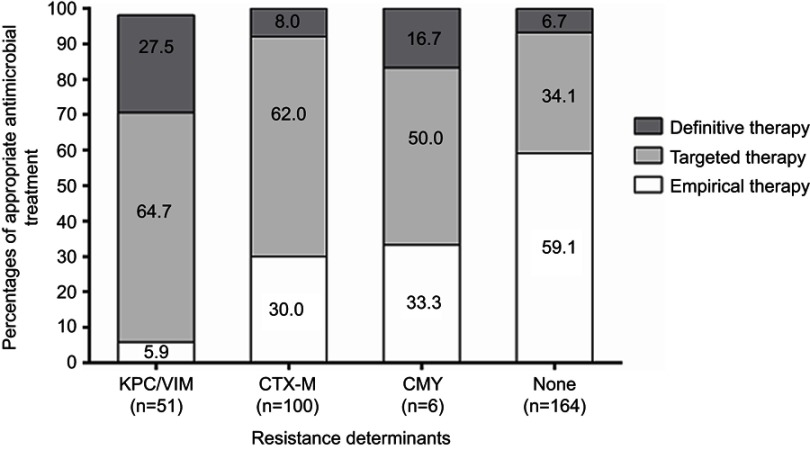
Within few hours after BCs were drawn, physicians started empirical antimicrobial therapy in all the 321 patients. The antibiotics most frequently used were piperacillin-tazobactam (24.3%) and fluoroquinolones (23.7%) followed by carba (16.2%) and ESC (8.7%); when combination therapy was used, piperacillin-tazobactam plus amikacin (9.3%) was the most frequent antibiotic regimen. In most cases (85.0%), the physician in charge of the patient made the choice of the empirical treatment, and did not use a predefined protocol. As shown in Table 2, empirical antimicrobial therapy was appropriate in 41.1% (132/321) of BSIs, with 5.9% (3/51) for KPC/VIM BSIs and 30.0% (30/100) for CTX-M BSIs (Figure 2).
Table 2.
Appropriate empirical or diagnostic-driven antimicrobial treatments stratified by the type of organisms causing bloodstream infections in 321 patients
| No. (%) of appropriate empirical treatments | No. (%) of modifications leading to appropriate treatments, according to the results by | ||
|---|---|---|---|
| MALDI BioTyperTM analysis plus eazyplex® SuperBug CRE assay | Antimicrobial susceptibility testing | ||
| Organisms causing BSI | |||
| KPC/VIM-producing E. coli (n=2) | 0/2 (0.0) | 2/2 (100) | 0/2 (0.0) |
| CTX-M-producing E. coli (n=77) | 22/77 (28.6) | 48/77 (62.3) | 7/77 (9.1) |
| CMY-producing E. coli (n=5) | 1/5 (20.0) | 3/5 (60.0) | 1/5 (20.0) |
| ESCS-carbaS E. coli (n=130) | 77/130 (59.2) | 44/130 (33.8) | 9/130 (6.9) |
| KPC/VIM-producing K. pneumoniae (n=49) | 3/49 (6.1) | 31/49 (63.3) | 14/49 (28.6)a |
| CTX-M-producing K. pneumoniae (n=23) | 8/23 (34.8) | 14/23 (60.9) | 1/23 (4.3) |
| CMY-producing K. pneumoniae (n=1) | 1/1 (100) | 0/1 (0.0) | 0/1 (0.0) |
| ESCS-carbaS K. pneumoniae (n=34) | 20/34 (58.8) | 12/34 (35.3) | 2/34 (5.9) |
| Total organisms (n=321) | 132/321 (41.1) | 154/321 (48.0) | 34/321 (10.6)a |
Notes: aThe treatment was not appropriate in 1 (0.3%) of 321 BSIs because the infecting KPC/VIM-producing K. pneumoniae organism had an AST-determined pandrug resistance phenotype.
Abbreviations: CRE, carbapenemase-resistant Enterobacteriaceae; BSI, bloodstream infection; ESCS-carbaS, extended-spectrum cephalosporin susceptible-carbapenem susceptible.
Figure 2.
Appropriateness of antimicrobial treatments in subgroups defined by the presence of the indicated resistance determinants in 321 patients with Ec or Kp bloodstream infections. Empirical or targeted antimicrobial therapy was the administration of antibiotics before and after the eazyplex® SuperBug CRE assay results were available, respectively. Definitive antimicrobial therapy was the administration of antibiotics after the antimicrobial susceptibility testing results were available. Overall, antimicrobial therapy was appropriate for any antibiotic with known susceptibility by microbiology report.
After notification of results from both the MALDI BioTyperTM analysis and eazyplex® SuperBug CRE assay, the IDCT specialists recommended a modification of the empirical antimicrobial therapy (Table 2). These actions increased the proportion of appropriate antimicrobial therapy to 89.1% (286/321), with 92.0% (92/100) for CTX-M BSIs and 70.6% (36/51) for KPC/VIM BSIs (Figure 2). The remaining 35 patients, 15 and 8 of whom were infected by carbapenemase- or CTX-M-producing Ec or Kp organisms, respectively, received antimicrobial treatments that were different from the previous empirical treatments, but the selected drug(s) were still inactive (as later documented by AST on the relative isolates). The drugs were meropenem plus colistin in 15 patients (all infected with colistin-resistant KPC-producing Kp isolates), piperacillin-tazobactam in 15 patients (8 with non-CTX M ESBL-producing isolates, 6 with ESCS-CarbaS isolates, and 1 with CMY-producing isolates), and fluoroquinolones in 5 patients.
Following release of AST results, antimicrobial therapies were further modified in the 35 aforementioned patients (Table 2), leading to a proportion of appropriate treatments up to 99.7% (320/321 patients) (Figure 2). One patient never received effective therapy, as the infecting Kp isolate was resistant to all the antibiotics tested (Table 1).
Discussion
To the best of our knowledge, this study is the first large hospital-based report that has evaluated the effects of the rapid detection of Ec and Kp isolates producing blaCTX-M or blaKPC or blaVIM on the timing of appropriate antimicrobial therapy. Several recent studies have performed similar evaluations,11,12,23,24 but all have involved a small number of patients with CTX-M- and carbaR-BSI, and none of these studies used the eazyplex® SuperBug CRE assay as a rapid resistance detection method. Here, integrating this assay with MALDI BioTyperTM analysis, our diagnostic algorithm allowed the successful identification of all Ec and Kp producing blaCTX-M, and/or blaKPC and/or blaVIM, and, more importantly, the antimicrobial streamlining in the course of BSI. Therefore, this study has some strengths. First, the results by the proposed laboratory workflow are obtainable in a mean time of 18 h, which may help clinicians to provide more therapy that is effective during the early period of clinical illness and may facilitate rapid decisions regarding isolating patients to prevent transmission. Second, the procedure underlying this workflow is simple and easy to perform and requires minimal training. Third, rapidly detecting a specific type of carbapenemase is particularly important because of the introduction of new antibiotics potentially active against certain carbapenemases into the clinical practice. In this context, our workflow is undoubtedly a positive contribution to infection control/effective treatment of patients.
Prior to the implementation of the rapid BC strategy in our hospital, we noted significant delays in initiating effective antimicrobial therapy.2,25 Conversely, based on the strategy shown here, the IDCT specialists switched 64.7% of patients with KPC/VIM BSI and 62.0% of patients with CTX-M BSI from inappropriate empirical antimicrobial regimen to effective coverage within a mean time of 20 h. In the remaining 23 patients on inappropriate empirical therapy, 15 of whom were infected by KPC producers and eight by CTX-M producers, the original regimen was discontinued, but each patient was treated with a drug against which his/her isolate had displayed resistance in AST testing. All 15 patients with KPC BSI that maintained an ineffective coverage received a colistin-based regimen. A problem of considerable importance is the increasing rate of colistin-resistance among KPC-producing Kp isolates because of its shortage of therapeutic options in patients with BSIs.26 There is an endemic widespread of such isolates in Italian healthcare facilities,27,28 and it was approximately 40% in our hospital.
The ineffective drug was piperacillin-tazobactam in all but one patients infected by CTX-M producers. Carbapenems seem to be a suitable option for treating severe infections due to those organisms; however, their increased use exerts selection pressure for carbapenem resistance. Therefore, experts have suggested carbapenem-sparing antibiotic regimens, particularly in settings such as ours where carbapenem-resistant isolates are common. Although there is emerging evidence that piperacillin-tazobactam may be a good choice for treating ESBL BSIs, its role is still controversial, particularly for BSIs from non-urinary sources.29–33 Additionally, the susceptibility of ESBL producers to piperacillin-tazobactam is less predictable than their susceptibility to carbapenems.2,4,20 Among our isolates, rates of resistance ranged from 31.2% of CTX-M-producing Ec isolates to 73.9% of CTX-M-producing Kp isolates. Therefore, physicians need to base their decisions regarding the empirical use of this antibiotic, until susceptibility results are available, on a sound knowledge of the local susceptibility patterns and the patient’s clinical condition.
This study has some well-acknowledged limitations. First, as we conducted our analysis in a single center, the results are not necessarily applicable to other hospitals, particularly those with very low rates of Enterobacteriaceae infections caused by CTX-M and/or carbapenemase producers. Second, the eazyplex® SuperBug CRE assay’s main weakness is its inability to identify ESBLs other than CTX-M, plasmidic AmpC, and some OXA-48-like enzymes, which are found among currently spreading MDR members of Enterobacteriaceae.4,28 Therefore, without the ability to detect these resistance determinants, the assay would miss a significant proportion of resistant isolates if implemented in settings where isolates harboring those genes occur.
Conclusion
Our combined use of the MALDI BioTyperTM analysis and eazyplex® SuperBug CRE assay can be a reliable, timesaving alternative to the conventional laboratory diagnosis of BSIs caused by Ec or Kp organisms with MDR determinants (ie, CTX-M ESBLs and carbapenemases). Importantly, this rapid diagnosis can enable physicians to begin appropriate antimicrobial therapy precociously. However, the applicability of our diagnostic workflow does depend on the conditions dictated by aforementioned limitations, as well as further larger studies are necessary to validate and confirm the present findings.
Acknowledgments
The authors would like to thank Maria Federica Ventriglia for her technical assistance. We also thanks all the ID consultants not included in the panel of Authors. The Italian Ministry for the University and Scientific Research in part supported the study by providing grants to T. S. and M. S. (Fondi Ateneo Linea D1 2014–2015).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008;13(47):e19044. [PubMed] [Google Scholar]
- 2.Tumbarello M, Sanguinetti M, Montuori E, et al. Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-beta-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob Agents Chemother. 2007;51(6):1987–1994. doi: 10.1128/AAC.01509-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossolini GM, D‘Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactams. Clin Microbiol Infect. 2008;14(Suppl 1):33–41. doi: 10.1111/j.1469-0691.2007.01863.x [DOI] [PubMed] [Google Scholar]
- 4.Castanheira M, Mendes RE, Jones RN, Sader HS. Changes in the frequencies of β-lactamase genes among Enterobacteriaceae isolates in U.S. hospitals, 2012 to 2014: activity of ceftazidime-avibactam tested against β-lactamase-producing isolates. Antimicrob Agents Chemother. 2016;60(8):4770–4777. doi: 10.1128/AAC.00540-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi: 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satlin MJ, Chen L, Patel G, et al. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother. 2017;61:e02349–16. doi: 10.1128/AAC.02349-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumbarello M, Viale P, Viscoli C, et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing Kp: importance of combination therapy. Clin Infect Dis. 2012;55(7):943–950. doi: 10.1093/cid/cis588 [DOI] [PubMed] [Google Scholar]
- 8.Pagano L, Caira M, Trecarichi EM, et al. Carbapenemase-producing Klebsiella pneumoniae and hematologic malignancies. Emerg Infect Dis. 2014;20(7):1235–1236. doi: 10.3201/eid2007.130094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Scola B, Raoult D, Bereswill S. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009;4:e8041. doi: 10.1371/journal.pone.0008041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiori B, D’Inzeo T, Giaquinto A, et al. Optimized use of the MALDI BioTyper system and the Filmarray BCID panel for direct identification of microbial pathogens from positive blood cultures. J Clin Microbiol. 2016;54(3):576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bork JT, Leekha S, Heil EL, Zhao L, Badamas R, Johnson JK. Rapid testing using the verigene gram-negative blood culture nucleic acid test in combination with antimicrobial stewardship intervention against gram-negative bacteremia. Antimicrob Agents Chemother. 2015;59(3):1588–1595. doi: 10.1128/AAC.04259-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sothoron C, Ferreira J, Guzman N, et al. A stewardship approach to optimize antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for gram-negative bacteria. J Clin Microbiol. 2015;53(11):3627–3629. doi: 10.1128/JCM.02161-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buehler SS, Madison B, Snyder SR, et al. Effectiveness of practices to increase timeliness of providing targeted therapy for inpatients with bloodstream infections: a laboratory medicine best practices systematic review and meta-analysis. Clin Microbiol Rev. 2016;29(1):59–103. doi: 10.1128/CMR.00053-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bookstaver PB, Nimmich EB, Smith TJ 3rd, et al. Cumulative effect of an antimicrobial stewardship and rapid diagnostic testing bundle on early streamlining of antimicrobial therapy in gram-negative bloodstream infections. Antimicrob Agents Chemother. 2017;61:e00189–17. doi: 10.1128/AAC.00189-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacVane SH, Nolte FS, Burnham C-AD. Benefits of adding a rapid PCR-based blood culture identification panel to an established antimicrobial stewardship program. J Clin Microbiol. 2016;54:2455–2463. doi: 10.1128/JCM.00996-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Findlay J, Hopkins KL, Meunier D, Woodford N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother. 2015;70(5):1338–1342. doi: 10.1093/jac/dku445 [DOI] [PubMed] [Google Scholar]
- 17.García-Fernández S, Morosini MI, Marco F, et al. Evaluation of the eazyplex® SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J Antimicrob Chemother. 2015;70(4):1047–1050. doi: 10.1093/jac/dku445 [DOI] [PubMed] [Google Scholar]
- 18.Hinić V, Ziegler J, Straub C, Goldenberger D, Frei R. Extended-spectrum β-lactamase (ESBL) detection directly from urine samples with the rapid isothermal amplification-based eazyplex® SuperBug CRE assay: proof of concept. J Microbiol Methods. 2015;119:203–205. doi: 10.1016/j.mimet.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 19.Fantoni M, Murri R, Scoppettuolo G, et al. Resource-saving advice from an infectious diseases specialist team in a large university hospital: an exportable model? Future Microbiol. 2015;10(1):15–20. doi: 10.2217/fmb.14.99 [DOI] [PubMed] [Google Scholar]
- 20.Tumbarello M, Spanu T, Sanguinetti M, et al. Bloodstream infections caused by extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae: risk factors, molecular epidemiology, and clinical outcome. Antimicrob Agents Chemother. 2006;50(2):498–504. doi: 10.1128/AAC.50.2.498-504.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagani L, Dell‘Amico E, Migliavacca R, et al. Multiple CTX-M-type extended-spectrum β-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol. 2003;41(9):4264–4269. doi: 10.1128/JCM.41.9.4264-4269.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC -lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction–based blood culture identification and susceptibility testing. Clin Infect Dis. 2015;61(7):1071–1080. doi: 10.1093/cid/civ447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker T, Dumadag S, Lee CJ, et al. Clinical impact after laboratory implementation of the verigene gram-negative bacteria microarray for positive blood cultures. J Clin Microbiol. 2016;54(7):1789–1796. doi: 10.1128/JCM.00376-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tumbarello M, Sali M, Trecarichi EM, et al. Bloodstream infections caused by extended-spectrum-beta-lactamase-producing Escherichia coli: risk factors for inadequate initial antimicrobial therapy. Antimicrob Agents Chemother. 2008;52(9):3244–3252. doi: 10.1128/AAC.00063-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capone A, Giannella M, Fortini D, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013;19(1):23–30. doi: 10.1111/1469-0691.12070 [DOI] [PubMed] [Google Scholar]
- 27.Monaco M, Giani T, Raffone M, et al. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, november 2013 to april 2014. Euro Surveill. 2014;19:e20939. doi: 10.2807/1560-7917.ES2014.19.42.20939 [DOI] [PubMed] [Google Scholar]
- 28.Giani T, Antonelli A, Caltagirone M, et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill. 2017;22:e30583. doi: 10.2807/1560-7917.ES.2017.22.31.30583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutiérrez-Gutiérrez B, Pérez-Galera S, Salamanca E, et al. A multinational, preregistered cohort study of β-lactam/β-lactamase inhibitor combinations for treatment of bloodstream infections due to extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2016;60(7):4159–4169. doi: 10.1128/AAC.00365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng TM, Khong WX, Harris PN, et al. Empiric piperacillin-tazobactam versus carbapenems in the treatment of bacteraemia due to extended-spectrum beta-lactamase-producing Enterobacteriaceae. PLoS One. 2016;11:e0153696. doi: 10.1371/journal.pone.0153696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamma PD, Han JH, Rock C, et al. Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum β-lactamase bacteremia. Clin Infect Dis. 2015;60(9):1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ofer-Friedman H, Shefler C, Sharma S, et al. Carbapenems versus piperacillin-tazobactam for bloodstream infections of nonurinary source caused by extended-spectrum beta-lactamase-producing Enterobacteriaceae. Infect Control Hosp Epidemiol. 2015;36(8):981–985. doi: 10.1017/ice.2015.101 [DOI] [PubMed] [Google Scholar]
- 33.Harris PNA, Tambyah PA, Lye DC, et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with Escherichia coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA. 2018;320(10):984–994. doi: 10.1001/jama.2018.12163 [DOI] [PMC free article] [PubMed] [Google Scholar]