Abstract
The assessment of the mechanisms and patterns of larval connectivity between geographically separated populations leads to a better understanding of benthic marine population dynamics, especially in commercially valuable species. This study investigated for the first time the fine-scale temporal genetic variability of new settlers and their origins in a benthic marine organism with one of the longest pelagic larval phases, the Caribbean spiny lobster (Panulirus argus). We genotyped newly settled postlarvae in the Florida Keys and adults of spiny lobster from the Florida Keys and throughout the Caribbean Sea. We identified strong larval connectivity between Dominican Republic, Belize, Nicaragua, the Florida Keys, and West-Florida. The larval dispersal modeling suggests that Florida’s lobster population could receive recruits from within and from other areas outside its state and national maritime boundaries. The genetic analyses refine the oceanographic model indicating that the connectivity patterns described could also result from unknown parental populations sourcing adults and postlarvae in different spawning seasons to the Florida Keys. We discuss the importance of small temporal scales to identify patterns in larval export. Our findings are significant on two levels. From the larval dispersal perspective, genetic results and biophysical modeling identify patterns of gene flow enhancing persistence of local populations. From an economic and fishery perspective, P. argus is the most important commercial species in the Caribbean and our results inform how considering larval source and sink dynamics across international boundaries could improve management plans at local, national, and regional levels.
Subject terms: Molecular ecology, Population dynamics
Introduction
The increasing number of fisheries categorized as fully or overexploited1,2 demonstrates the need to more fully understand population dynamics of key fisheries to prevent or reverse stock decline and collapse. Fisheries data are used to inform how much is removed from the ecosystem, but knowing how much is recruited, the origin of those recruits, and the spatiotemporal variability in those patterns is further information required to assess population dynamics and to determine the suitable spatial scales to manage fished populations. This is especially critical for commercially important species with a broad distribution, complex life cycles, and that suffer high removal rates through chronic fishing pressure3,4.
The Caribbean spiny lobster, Panulirus argus (Latreille, 1804), occurs throughout the Caribbean basin and north to Bermuda and is the most important commercial fishery in the Caribbean Sea5,6. Amongst marine invertebrates, P. argus has one of the longest larval dispersal phase, lasting up to nine months in the plankton7,8. Spiny lobster planktonic larvae have limited horizontal swimming ability and are therefore subject to widespread dispersal by currents9, but also undertake ontogenetic vertical migration during their dispersal phase10. The final larval stage metamorphoses into a non-feeding postlarva that actively swims from oceanic waters to the coast wherein they settle in shallow protective habitats9. The complex pelagic phase in the life cycle of P. argus has made it a species difficult to study for population dynamics.
Larval dispersal knowledge has benefited from the use of direct genetic methods, which have provided new evidence of different patterns of local retention, self-recruitment, or asymmetrical gene flow in marine organisms (e.g. reef fishes11–13, pink abalone14, California red rock lobster15, and demersal fish16). Nonetheless, larval transport and recruitment are known to be affected by the variability of oceanographic currents and biological factors, such as the larval dispersal duration, spawning abundance and characteristics, and larval behaviours. Hence, complicating the full assessment of benthic population dynamics. For example, in the emperor fish, Lethrinus nebulosus, extensive gene flow was predicted in biophysical modelling of passive particles, however, common history of spawning and cohesiveness of larval transport pathways resulted in a non-random genetic structure17. In P. argus, the predicted average distance a larva settles from its release location is reduced by over 60% when larval diel vertical migration is incorporated into a biophysical transport model10. Another factor affecting the assessment of larval dispersal is the effect of climate change; in the European lobster, Homarus gammarus, the increase of water temperature is expected to increase temperature-dependent mortality of larvae, as a result of a forward shift in hatching and poor quality and abundance of food18. The effects of climate change on the larval development and therefore recruitment to fishery strongly depend on the biology and phenology of the species and its vulnerability to environmental stressors19.
Previous studies have contributed to the understanding of spiny lobster postlarvae recruitment and connectivity, suggesting that local populations depend on asymmetric larval supply from many potential source regions across the Caribbean but also identifying that self-recruitment occurs20–25. However, the direct genetic comparison between adults and recruits has yet to be investigated. In this study, we used both indirect and direct genetic methods to investigate the fine-scale temporal variation in larval connectivity and recruitment by estimating the extent of relatedness between postlarvae settled in the Florida Keys and adult spiny lobsters from geographically remote populations across the Caribbean Sea. Indirect oceanographic modeling methods were used to investigate the transport and retention patterns of spiny lobster larvae in the Caribbean region. The modelling results were compared with the genetic results.
Results
Genotyping
A total of 2030 postlarvae, from the lower and middle Florida Keys, and 799 adult spiny lobsters, from 15 locations throughout Florida and the Caribbean Sea, were genotyped for 14 microsatellite loci. Departures from Hardy-Weinberg equilibrium (HWE) were consistent in both adult and postlarvae databases for several loci, thus genetic diversity estimates for combined postlarvae and adult data were estimated and summarized (Table S1). All loci were retained for further analyses given the high relatedness resolving power estimated by the polymorphic information content (PIC), (Table S1).
Temporal genetic differentiation
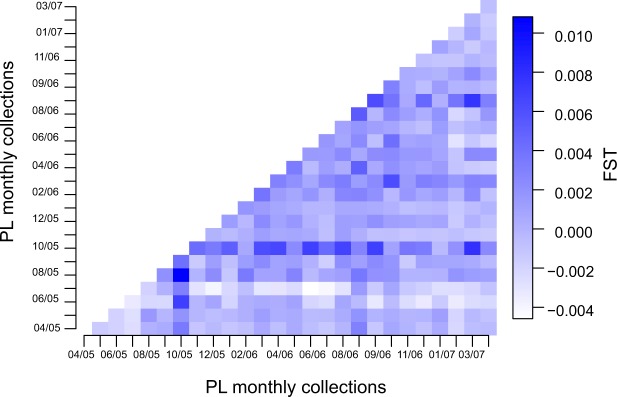
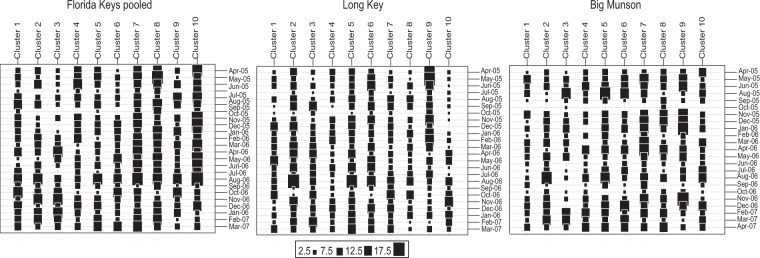
Genetic differentiation of postlarvae monthly samples was overall low, FST values ranged from −0.004 to 0.0079, but showed a complex pattern of genetic differentiation where some months were slightly less differentiated than others (Fig. 1). The DAPC cluster analysis suggested an optimum of 10 genetic clusters describing the postlarvae genotype variability, while individual postlarvae from each monthly sample were assigned in different proportions to each of these clusters throughout the time of the study (Fig. 2).
Figure 1.
Heatmap of pairwise FST values estimated from microsatellite data between all monthly Panulirus argus postalarvae collections. This illustrates the complex temporal genetic variation of settlers in the Florida Keys. The darker color indicate stronger genetic differentiation.
Figure 2.
Cluster analysis as implemented in DAPC of microsatellite loci genotypes of Panulirus argus postlarvae collected in the Florida Keys (two sites pooled), Long Key (middle Florida Keys) and Big Munson (lower Florida Keys). The proportional contribution of each genotypically described cluster in each monthly sample is indicated by the size of the black boxes.
Postlarvae source populations
The parentage assignment test for all 2030 postlarvae was resolved for 226 individual postlarvae (11%); of these, 28.3% (64 individuals, 3% of the total postlarvae), were assigned to the Florida Keys adult population (Table 1). The weighted proportion of these assignments to each potential parental population (i.e. adult samples included in this study) indicated postlarvae settlers had a strong connectivity with adult populations from the Dominican Republic (0.0514), Belize (0.0406), West Florida (0.0362), and Nicaragua (0.0313); but also to local adult populations such as Lower Keys and Middle Keys (0.0346 and 0.0327, respectively) (Fig. 3).
Table 1.
Summary of number of Panulirus argus recruits assigned to each source-population (parentage assignment) as estimated in CERVUS.
| NC | BE | GOM | W-FL | DT | lFK | mFK | E-FL | BH | BZ | NI | DR | CR | SK | VZ | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N adult | 40 | 49 | 16 | 39 | 50 | 119 | 77 | 49 | 54 | 48 | 22 | 36 | 102 | 49 | 49 | |
| Assignments | Long key | 5 | 4 | 4 | 15 | 9 | 15 | 7 | 5 | 6 | 12 | 4 | 21 | 21 | 0 | 8 |
| Big Munson | 0 | 2 | 0 | 6 | 0 | 25 | 17 | 1 | 5 | 8 | 3 | 0 | 10 | 9 | 4 | |
| % | Long key | 0.4409 | 0.3527 | 0.3527 | 1.3228 | 0.7937 | 1.3228 | 0.6173 | 0.4409 | 0.5291 | 1.0582 | 0.3527 | 1.8519 | 1.8519 | 0.0000 | 0.7055 |
| Big Munson | 0.0000 | 0.2232 | 0.0882 | 0.0882 | 0.0000 | 2.7902 | 1.8973 | 0.1116 | 0.5580 | 0.8929 | 0.3348 | 0.0000 | 1.1161 | 1.0045 | 0.446429 | |
| weighted % | Long key | 0.0110 | 0.0072 | 0.0220 | 0.0339 | 0.0159 | 0.0111 | 0.0080 | 0.0090 | 0.0098 | 0.0220 | 0.0160 | 0.0514 | 0.0182 | 0.0000 | 0.0144 |
| Big Munson | 0.0000 | 0.0046 | 0.0055 | 0.0023 | 0.0000 | 0.0234 | 0.0246 | 0.0023 | 0.0103 | 0.0186 | 0.0152 | 0.0000 | 0.0109 | 0.0205 | 0.0091 | |
| total | 0.0110 | 0.0118 | 0.0276 | 0.0362 | 0.0159 | 0.0346 | 0.0327 | 0.0113 | 0.0201 | 0.0406 | 0.0313 | 0.0514 | 0.0291 | 0.0205 | 0.0235 |
Adult population as follow: NC = North Carolina, BE = Bermuda, GOM = Gulf of Mexico, W-FL = Western Florida, lFK = Lower Florida Keys, mFK = Middle Florida Key, E-FL = Eastern Florida, BH = Bahamas, BZ = Belize, NI = Nicaragua, DR = Dominican Republic, CR = Saint Croix, SK-Saint Kitts, VZ = Venezuela. Adult sample size at each location: N adult, number of postlarvae assigned to each adult population: Assignments, percentage of postlarvae assigned to each location: %, and percentage of postlarvae assigned to each location weighted by adult sample size: weighted %.
Figure 3.
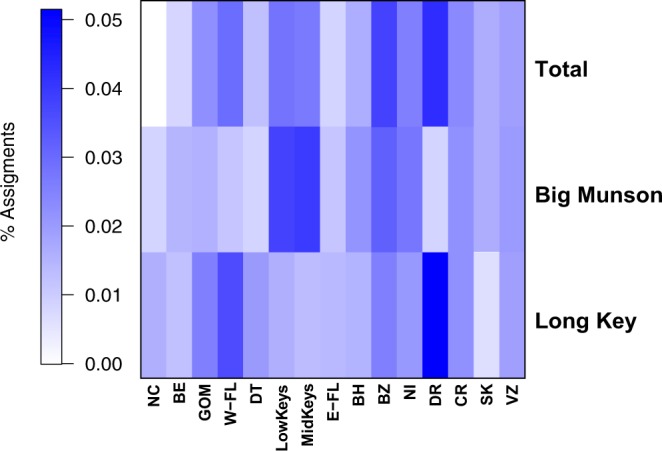
Heat-map of the weighted percentage of assignments of Panulirus argus postlarva to adult population as implemented in the parentage analysis in CERVUS. Acronyms of adult populations are: NC = North Carolina, BE = Bermuda, GOM = Gulf of Mexico, W-FL = Western Florida, Low-FK = Lower Florida Keys, Mid-FK = Middle Florida Keys, E-FL = Eastern Florida, BH = Bahamas, BZ = Belize, NI = Nicaragua, DR = Dominica Republic, CR = Saint Croix, SK = Saint Kitts, VZ = Venezuela. The percentage of postlarvae assigned to parental populations was weighted by adult sample size and total number of postlarvae collected.
Larval dispersal model
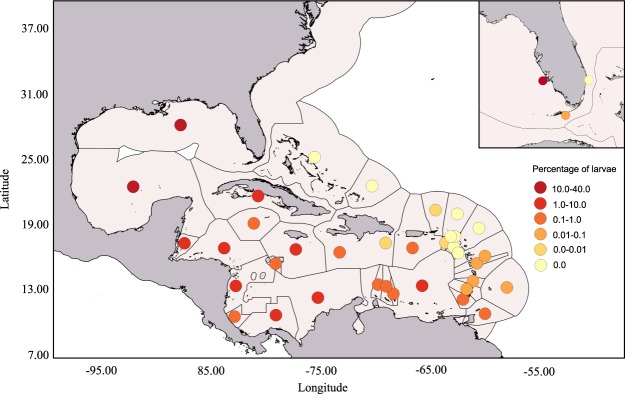
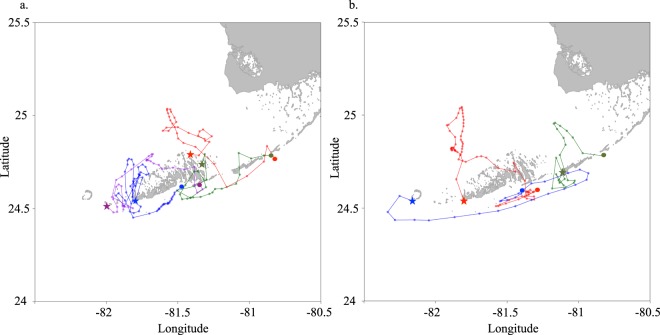
To investigate the large-scale connectivity, a large-scale Caribbean wide model was used to predict release (spawning) locations for the postlarvae which settled at the two collection sites (Lower Florida Keys: 24.617°N, 81.387°W; Middle Florida Keys: 24.803°N, 80.84°W). The sources of larvae were particularly important in the west coast of Florida, northwest Cuba, the Caribbean coast of Mexico, Belize, and Honduras (Fig. 4). Within the Florida region, the local model predicted the postlarvae that settled in the Lower and Middle Florida Keys mainly originated from habitats near the Keys (see Fig. S1 in Supporting Information). Trajectories of these larvae that originated and settled in the Keys showed two main paths: northward and southward the Keys (e.g. Fig. 5). The releasing locations of the larvae were consistently located west of their settlement locations.
Figure 4.
Distribution of Panulirus argus larvae after 196 days of simulated backward transport in the Caribbean region using the low resolution model (HYCOM, horizontal resolution 8 km). Virtual larvae were released from two locations in the Florida Keys: Long Key (middle Florida Keys) and Big Munson (lower Florida Keys). See Fig. 6 for the location of the release polygons (black arrows). The percentage of larvae was averaged in each Exclusive Economic Zone of the Caribbean region and are shown as heat colored circles.
Figure 5.
Examples of larval trajectories of Panulirus argus during the simulated backward transport from settlement locations using the high resolution model (ROMS, horizontal resolution ~2.8 km). Virtual larvae were released in September 2007 (a) and October 2007. (b) The predicted origin location of each larva is marked with a star (i.e. release) and the circle indicates its settlement location.
Discussion
The present study is the first to investigate fine-scale genetic relatedness between adults and new settlers of spiny lobster using a parentage analysis as a proxy to assess the reproductive connectivity of postlarvae in the Florida Keys with adult populations across the wider Caribbean.
The parental assignment based on the extent of alleles shared between Florida Keys postlarvae and adult spiny lobsters, indicated a high degree of connectivity to Dominican Republic, Belize, Nicaragua, West Florida and the Florida Keys. The backward larval dispersal modeling conducted at both Caribbean wide and Florida spatial scales predicted comparable results to the genetic analyses performed. Larval dispersal between the Florida Keys and Belize, Western Florida, Gulf of Mexico, and within the Florida Keys itself was indeed predicted by the models. These results are overall consistent with the major larval exporter countries previously suggested by Kough et al.23 (i.e. Venezuela, Nicaragua, Belize and Dominican Republic). Although including all the potential sources of postlarvae in our analysis could better resolve the larval origins and connectivity of the Florida Keys recruits, it remains difficult to achieve given the broad distribution of the species.
Previous modeling studies have suggested that larval recruitment in the northern regions of the Caribbean are highly dependent on distant sources supplied by strong northward currents, while recruitment dynamics in the southern regions of the Caribbean are more influenced by retentive oceanographic structures, dominated by persistent gyres10,20. In this regard, high connectivity between Florida, southwest Cuba, and the Mexican Caribbean might be expected considering the proximity of these sites to the Yucatan Current where advection is likely to occur despite of the occurrence of persisting recirculation observed in southwest Cuba and the Campeche Bank (see Briones-Fourzán et al.20). Similarly, Zeng et al.’s26 dispersal model for bonefish larvae (53 days of pelagic phase duration) showed high connectivity between Belize, Mexico, Cuba, and the Florida Keys, but also showed that the Lower and Middle Key have the highest retention rate as seen in our local model. In a recent study, Garavelli et al.27 showed that the highest values of local retention and self-recruitment in the Caribbean wide region were observed in Mexico and Florida. These high retention rates were driven by the availability of favorable habitat and the populations in each region were found self-persistent, meaning that they did not rely on external larval supply to maintain their population27. Therefore, despite potential significant influx from other geographically close sources, the estimated percentage of postlarvae assigned to the Florida Keys could remain the same.
Along the Florida Keys the coastal counter-current and the Tortugas Gyre, when present, could play an important role in the spiny lobster postlarvae transport, survival, and recruitment28,29. The strong genetic relatedness between postlarvae and adults from the Florida Keys supports the hypothesis that the southwest Florida shelf area plays an important role in the local retention and self-recruitment of larvae originated in the Florida Keys. The local retention and self-recruitment are also assisted by elevated nutrient levels from the upwelling in the interior and fronts of the eddies and gyres that surround the Florida Keys29,30. This upwelling enhances larval food supply and potentially may increase larval survival28. The Florida regional model simulation confirmed the retention role of the southwest Florida shelf (Fig. 5), with postlarvae coming from both the northern and southern parts of the Keys and predicted drift patterns similar to drifter trajectories in the same region shown in Yeung et al.29. This result is also consistent with the recent observations describing the southward movement of reproductively active females in the Keys31 that would spawn within the counter current area. Lee and Williams32 identified several “recruitment conveyors” forming a network of retention pools (gyres) on both sides of the Florida Keys (Fig. S2). The longest retention pool associated with a location that includes adult spiny lobsters has a 6–8 months retention time and is located north of the Dry Tortugas. But Lee and Williams32 also showed the likeliness of offshore spawned larvae to be transported to the coastal zone and channels between the Keys by the bottom Ekman transport29. Lobster larvae can then migrate to the retention pool on the south Florida shelf. In addition, larvae could be released on the Southwest Florida Shelf north of the Florida Keys.
More exhaustive genetic analysis would be needed to allow the differentiation between parent-offspring relationships from sibling relationships from multiple year classes. Therefore, the most parsimonious explanation for our results is that a same parent population was the source of adults and postlarvae sampled in the locations in the Florida Keys across different spawning seasons. Indeed, the low levels of relatedness to adult populations downstream from the Florida Keys in East Florida (0.0113), North Carolina (0.0110), and Bermuda (0.0118) support this hypothesis, most likely due to the lack of significant reverse flow near the Gulf Stream, which is incompatible with the concept of a parental relationship.
Self-recruitment has also been suggested in a congeneric species, P. interruptus, in the Mexican Pacific coast where a high proportion of kinship among adult lobsters was found15. However, for P. argus in Florida, the strong spawner vs. recruit correlation suggestive of self-recruitment33, was reassessed and patterns observed seem to represent broader adult abundance from Caribbean wide origin34. Thus, the contribution of local Florida spawning stock to local post-larval recruitment remains unresolved by using adult lobster abundance and postlarvae recruitment patterns alone.
The use of direct genetic methods at fine temporal scales allowed the identification of complex genetic variability in recruits (Table 1, Fig. S3) yielded by asymmetrical gene flow, where some locations (e.g. Florida Keys, Belize and US Virgin Islands) consistently provide disproportionately more larvae to the Florida Keys than other populations during the time of the study (Fig. S4). In the rock lobster, P. cygnus, the combination of temporal genetic variation and dissimilar recruitment patterns at two sites resulted in genetically different cohorts, but was also different from following time replicates35. Similarly, our results from the DAPC clustering analyses showed temporal variation of the inferred clusters and that clusters 7–10 were more represented than cluster 1–6 throughout the two years of the study (Fig. 2). This temporal genetic variability is normally neglected using indirect genetic analyses, as previously estimated between P. argus adult populations36,37 and in this study (Fig. S3), but is shown here to contribute to population dynamics and structuring. Our results provide evidence supporting the hypothesis that spiny lobster populations are not as vagrant and assorted as previously thought, as indicated by the low but significant levels of genetic differentiation between some populations in Central America38, and the significant differences found between geographically close basins but not between most distant basins25. Likewise, across the range of the American lobster, Homarus americanus, a hierarchical genetic structure revealed significant differences between north (Gulf of St. Lawrence) and south (Gulf of Maine) regions and among populations within these regions39.
In marine invertebrates with pelagic larval phase, FST values are typically very low (<0.05), which challenges the inference of genetic structure40. The low genetic differentiation as estimated by FST may have resulted from significant gene flow or larval connectivity over evolutionary time scales that homogenize allele frequencies but, does not necessarily mean that populations are well mixed on ecological time scales. This supports the hypothesis that stepping-stone import of foreign genotypes will result in panmixia over an evolutionary time scale41. However, to correctly describe the ecological processes at work to inform management decision making, it is crucial to detect the fine-scale temporal variability of settlement and identify the consistent and key sources of recruits into local populations which may vary through time.
Population modeling studies on P. argus in Honduras suggested that a reserve network at country scale will result in a significant increase of both persistence and yield of the species42. This is consistent with the hypothesis that Caribbean spiny lobster populations are influenced by both open and closed recruitment dynamics. Thus, management actions at national level to conserve spawning stock biomass may deliver improvements at a country scale given the correct oceanographic conditions even for species with long pelagic larval phases. Including the study of genetic variability at these shorter temporal and finer spatial scales is key to elucidating patterns in larval export and determining the importance of self-recruitment which may otherwise be masked in larger scale studies. Understanding these patterns can help assess the potential contribution from different levels of management intervention, from local to international, for marine species with a broad distribution and complex dispersal patterns.
Methods
Sample collection
From April 2005 to March 2007, postlarvae from Florida were collected on the seventh day of each lunar month (n = 50 targeted per lunar month), using five artificial collectors placed parallel to the shore near inter-island channels and anchored near shore in shallow waters (<2 m) from two long-term monitoring sites near Big Munson, in the Lower Florida Keys (24.617°N, 81.387°W), and the south side of Long Key, in the Middle Keys (24.803°N, 80.84°W) (Fig. 6). Whole individual postlarvae were preserved into vials containing 90% ethanol and were identified by sample location, date, and individual identification number.
Figure 6.
Map of the Caribbean basin showing sampling locations of adult lobsters Panulirus argus. NC = North Carolina, BE = Bermuda, GOM = Gulf of Mexico, W-FL = Western Florida, DT = Dry Tortugas, lFK = Lower Florida Keys, mFK = Middle Florida Keys, E-FL = Eastern Florida, BH = Bahamas, BZ = Belize, NI = Nicaragua, DR = Dominican Republic, CR = Saint Croix, SK-Saint Kitts, VZ = Venezuela.
We collected tissues from adult spiny lobsters from 15 sites throughout the U.S. and Caribbean regions including North Carolina, the northernmost limit of species distribution, the Gulf of Mexico Flower Gardens Banks, the Dry Tortugas, Florida Lower Keys, the Upper Keys, Fort Pierce (East Florida), Panama City (West Florida), Bermuda, the Bahamas, Belize, Nicaragua, St. Kitts, St. Croix (U.S.Virgin Islands), the Dominican Republic, and Venezuela (Fig. 6). Approximately fifty specimens were collected from each site. Adult samples were collected from harvested individuals. Tissue was obtained from the last segment of a middle walking leg and was excised with sterile scissors. Depending on field conditions, the entire segment or dissected muscle was preserved in 90% ethanol.
Microsatellite genotyping
Total genomic DNA was extracted and purified from tissue samples using PUREGENE® DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, Minnesota) following manufacturer guidelines. Individual lobsters (postlarvae and adults) were genotyped for 14 bi-parentally inherited microsatellite DNA markers developed for this species43,44. PCR conditions were as follow: 94 °C for 2 min, 32 cycles of 94 °C for 40 s, 58 °C for 40 s, 72 °C for 45 s, followed by 72 °C for 7 min. PCR products were visualized on an ABI 3130 genetic analyser and genotyped by using GeneMapper® software v3.7 (Applied Biosystems, Inc.). Genotypes across all 14 microsatellite loci were tested for the presence of allelic dropout and null alleles using the program MICRO-CHECKER45. Genetic diversity within each location and monthly collections were assessed using standard measures (number of alleles and allelic richness) estimated in FSTAT 2.9.346. Observed heterozygosity (Ho), expected heterozygosity (He), departures from Hardy-Weinberg equilibrium (HWE) and the power of resolution of the microsatellite loci (PIC) were examined in CERVUS 3.0.747.
Genetic differentiation among recruits (indirect method)
Temporal genetic differentiation among monthly postlarvae samples was examined based on differences in allele frequency to estimate convectional pairwise FST using ARLEQUIN 3.548. Pairwise genetic differentiation was also estimated between postlarvae and adult population samples using convectional pairwise FST using ARLEQUIN 3.548. To determine genetic differentiation between postlarvae samples, we used a discriminant analysis of principal components (DAPC)49, a multivariate method implemented in the adegenet package50 for R51. The postlarvae collection date was used as a prior. The DAPC approach proposes a distribution of postlarvae into predefined groups in relation to the discriminant function of principal components. The optimum number of clusters was defined by k-means algorithm that uses the Bayesian information criterion. This approach is preferred for species exhibiting potentially high gene flow, as DAPC maximizes genetic separation among groups and minimizes variation within groups49.
Inferring larval connectivity (direct method)
A single-parent parentage analysis was conducted to investigate the extent of connectivity between postlarvae settled in the Florida Keys and adult lobsters from other locations, both local within Florida and from other countries internationally. This direct method compared multi-loci genotypes of adults with multi-loci postlarvae genotypes to assign individual postlarvae to a candidate parent. This analysis is a proxy to infer the potential sourcing population, rather than an actual parentage analyses to assign postlarvae to parent populations (recruit-spawner). Given that lobsters are long-lived animals, there is potential for generation overlap that might make it difficult to distinguish between parent-offspring and siblings from a different year class. The parentage analysis was conducted as implemented in CERVUS 3.0.747. The probability for the most likely parent for each postlarva was defined by taking the natural log (log base e) of the overall likelihood ratio (LOD scores). The critical LOD value was determined with a 95% confidence level running parentage simulations for 10,000 offspring while considering 98 candidate parents (half of the adult sample size collected in the Florida Keys) and a proportion of candidate mothers estimated at 0.016, based on the female spawning stock assessment for the Florida Keys in 200433. To increase the strictness of the parentage analyses, we only considered genotype comparisons of more than 10 microsatellite loci and a maximum two pair loci mismatched. The critical LOD score estimated (LOD = 2.5) was the cut-off for assigning the single parent, which is a conservative approach that accounts for some unsampled putative parents in the population52. The proportion of postlarvae assigned to different source populations were weighted by the total number of postlarvae and the corresponding adult sample size.
Larval dispersal model
To estimate the spawning locations of P. argus larvae settling in the Florida Keys, we modeled the larval dispersal of the species using the offline Lagrangian tool Ichthyop v3.253. Ichthyop v3.2 allows the coupling between a hydrodynamic model and an individual-based model and is based on an Euler advection scheme. In the model, each particle represents a virtual larva and is characterized by its longitude, latitude, and depth in three dimensions. Two simulations were performed: the first one to investigate the transport patterns at large spatial scale in the Caribbean region and the second to investigate the retention patterns around Florida by using a higher spatial resolution ocean model. The large scale simulation used the 8-km horizontal resolution HYCOM consortium global model54. The HYCOM model fields were extracted daily for the intra America Seas region and converted to Ichthyop’s input format. At the Florida scale, we used the Regional Oceanic Modeling System (ROMS) high-resolution (~2.8 km) simulation of the south Florida shelf, which includes tides and is described in Criales et al.55. Two polygons, encompassing the postlarvae collection sites (Lower Florida Keys: 24.617°N, 81.387°W; Middle Florida Keys: 24.803°N, 80.84°W), i.e. settlement locations, were defined. We adapted the size of the polygons depending on the resolution of each model used. In the large-scale model, each polygon was 64 km2 and 25 km2 in the Florida scale model. Ten thousand particles were released from each polygon at 5 m depth once a month from July to December 2007. The dispersal duration was set to 196 days with 152 days of pre-competency period8. An ontogenetic vertical migration behaviour was included in the model following Callwood56. The P. argus virtual larvae were tracked backward in time from their settlement locations to estimate their origin and the percentage of larvae was averaged over the number of polygon per nation in the Caribbean region.
Supplementary information
Acknowledgements
This work was funded by NOAA MARFIN Grant no. NA05NMF4331076 awarded to the Florida Fish and Wildlife Conservation Commission, ISG Summit Foundation grant 504595. We thank Biol. Samantha Schmitt for the laboratory work and data management. This is contribution no. 1111 of the Smithsonian Marine Station at Fort Pierce.
Author Contributions
I.S.-G. designed the study, performed the genetic analyses and drafted the manuscript, L.G. and L.M.C. directed the biophysical modelling and contributed to manuscript drafting, M.T. generate the genotype data and contributed to data analyses, S.J.B. contributed to manuscript structure and proof-read it, T.M. helped in interpreting results and contributed to manuscript revision, J.H. got funds to support sample collection and the molecular analyses. All authors reviewed various versions of the manuscripts.
Data Availability
Microsatellite genotypes generated and analysed during the current study will be available in the DRYAD repository once the manuscript is accepted. 10.5061/dryad.27812c7.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43435-9.
References
- 1.Pauly D, Watson R, Alder J. Global trends in world fisheries: impacts on marine ecosystems and food security. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005;360:5–12. doi: 10.1098/rstb.2004.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello C, Gaines SD, Lynham J. Can catch shares prevent fisheries collapse? Science. 2008;321:1678–81. doi: 10.1126/science.1159478. [DOI] [PubMed] [Google Scholar]
- 3.Truelove Nathan K., Box Stephen J., Aiken Karl A., Blythe-Mallett Azra, Boman Erik M., Booker Catherine J., Byfield Tamsen T., Cox Courtney E., Davis Martha H., Delgado Gabriel A., Glazer Bob A., Griffiths Sarah M., Kitson-Walters Kimani, Kough Andy S., Pérez Enríquez Ricardo, Preziosi Richard F., Roy Marcia E., Segura-García Iris, Webber Mona K., Stoner Allan W. Isolation by oceanic distance and spatial genetic structure in an overharvested international fishery. Diversity and Distributions. 2017;23(11):1292–1300. doi: 10.1111/ddi.12626. [DOI] [Google Scholar]
- 4.Pinsky ML, Palumbi SR. Meta-analysis reveals lower genetic diversity in overfished populations. Mol. Ecol. 2014;23:29–39. doi: 10.1111/mec.12509. [DOI] [PubMed] [Google Scholar]
- 5.FAO. First meeting of the OSPESCA/WECAFC/CFMC Working group on Caribbean spiny lobser (2015).
- 6.Hunt, J. H. Status of the fishery for Panulirus argus in Florida. In Spiny Lobster management (eds Phillips, B. F., Cobb, J. S. & Kittaka, K.) 189–199 (Fishing New Books, 2000).
- 7.Lewis JB. The phyllosoma larvae of the spiny lobster, Panulirus argus. Bull.Mar.Sci. 1951;1:89–103. [Google Scholar]
- 8.Goldstein JS, Matsuda H, Takenouchi T, Butler MJ. The complete development of larval Caribbean siny lobster Panulirus argus (Latreille, 1804) in culture. J. Crustac. Biol. 2008;28:306–327. doi: 10.1163/20021975-99990376. [DOI] [Google Scholar]
- 9.Phillips, B. F., Booth, J. D., Cobb, J. S., Jeffs, A. G. & McWillliam, P. Larval and postlarval ecology. In Lobsters: Biology, Management, Aquaculture and Fisheries (ed. Phillips, B. F.) 231–262 (Blackwell Scientific Press, 2006).
- 10.Butler, Paris CB, Goldstein JS, Matsuda H, Cowen RK. Behavior constrains the dispersal of long-lived spiny lobster. Mar. Ecol. Prog. Ser. 2011;422:223–237. doi: 10.3354/meps08878. [DOI] [Google Scholar]
- 11.Jones GP, Planes S, Thorrold SR. Coral reef fish larvae settle close to home. Curr. Biol. 2005;15:1314–1318. doi: 10.1016/j.cub.2005.06.061. [DOI] [PubMed] [Google Scholar]
- 12.Christie MR, Johnson DW, Stallings CD, Hixon MA. Self-recruitment and sweepstakes reproduction amid extensive gene flow in a coral-reef fish. Mol. Ecol. 2010;19:1042–1057. doi: 10.1111/j.1365-294X.2010.04524.x. [DOI] [PubMed] [Google Scholar]
- 13.Saenz-Agudelo P, Jones GP, Thorrold SR, Planes S. Patterns and persistence of larval retention and connectivity in a marine fish metapopulation. Mol. Ecol. 2012;21:4695–4705. doi: 10.1111/j.1365-294X.2012.05726.x. [DOI] [PubMed] [Google Scholar]
- 14.Díaz-Viloria N, Próo SAG-D, Cruz P, Perez-Enriquez R. Assessment of Self-Recruitment in a Pink Abalone (Haliotis corrugata) Aggregation by Parentage Analyses. J. Shellfish Res. 2013;32:105–113. doi: 10.2983/035.032.0116. [DOI] [Google Scholar]
- 15.Iacchei M, et al. Combined analyses of kinship and FST suggest potential drivers of chaotic genetic patchiness in high gene-flow populations. Mol. Ecol. 2013;22:3476–3494. doi: 10.1111/mec.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ottmann D, et al. Long-term aggregation of larval fish siblings during dispersal along an open coast. Proc. Natl. Acad. Sci. 2016;113:14067–14072. doi: 10.1073/pnas.1613440113. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Berry O, England P, Marriott RJ, Burridge CP, Newman SJ. Understanding age-specific dispersal in fishes through hydrodynamic modelling, genetic simulations and microsatellite DNA analysis. Mol. Ecol. 2012;21:2145–2159. doi: 10.1111/j.1365-294X.2012.05520.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmalenbach I, Franke HD. Potential impact of climate warming on the recruitment of an economically and ecologically important species, the European lobster (Homarus gammarus) at Helgoland, North Sea. Mar. Biol. 2010;157:1127–1135. doi: 10.1007/s00227-010-1394-8. [DOI] [Google Scholar]
- 19.Pecl GT, et al. Rapid assessment of fisheries species sensitivity to climate change. Clim. Change. 2014;127:505–520. doi: 10.1007/s10584-014-1284-z. [DOI] [Google Scholar]
- 20.Briones-Fourzán P, Candela J, Lozano-Àlvarez E. Postlarval settlement of the spiny lobster Panulirus argus along the Caribbean coast of Mexico: Patterns, influence of physical factors, and possible sources of origin. Limnol. Oceanogr. 2008;53:970–985. doi: 10.4319/lo.2008.53.3.0970. [DOI] [Google Scholar]
- 21.Butler MJ, Herrnkind WF. A test of recruitment limitation and the potential for artificial enhancement of spiny lobster (Panulirus argus) populations in Florida. Can. J. Fish. Aquat. Sci. 1997;54:452–463. doi: 10.1139/f96-281. [DOI] [Google Scholar]
- 22.Chollett I, et al. A case for redefining the boundaries of the Mesoamerican Reef Ecoregion. Coral Reefs. 2017;36:1039–1046. doi: 10.1007/s00338-017-1595-4. [DOI] [Google Scholar]
- 23.Kough, A. S., Paris, C. B. & Butler, M. J. Larval Connectivity and the International Management of Fisheries. PLoS One8 (2013). [DOI] [PMC free article] [PubMed]
- 24.Truelove NK, et al. Genetic analysis reveals temporal population structure in Caribbean spiny lobster (Panulirus argus) within marine protected areas in Mexico. Fish. Res. 2015;172:44–49. doi: 10.1016/j.fishres.2015.05.029. [DOI] [Google Scholar]
- 25.Truelove NK, et al. Biophysical connectivity explains population genetic structure in a highly dispersive marine species. Coral Reefs. 2017;36:233–244. doi: 10.1007/s00338-016-1516-y. [DOI] [Google Scholar]
- 26.Zeng Xiangming, Adams Aaron, Roffer Mitchell, He Ruoying. Potential connectivity among spatially distinct management zones for Bonefish (Albula vulpes) via larval dispersal. Environmental Biology of Fishes. 2018;102(2):233–252. doi: 10.1007/s10641-018-0826-z. [DOI] [Google Scholar]
- 27.Garavelli L, White JW, Chollett I, Chérubin LM. Population models reveal unexpected patterns of local persistence despite widespread larval dispersal in a highly exploited species. Conserv. Lett. 2018;11:e12567. doi: 10.1111/conl.12567. [DOI] [Google Scholar]
- 28.Lee TN, Clarke ME, Williams E, Szmant AF, Berger T. Evolution of the Tortugas Gyre and its influence on recruitment in the Florida Keys. Bull. Mar. Sci. 1994;54:621–646. [Google Scholar]
- 29.Yeung C, Jones DL, Criales MM, Jackson TL, Richards WJ. Influence of coastal eddies and counter-currents on the influx of spiny lobster, Panulirus argus, postlarvae into Florida Bay. Mar. Freshw. Res. 2001;52:1217–1232. doi: 10.1071/MF01110. [DOI] [Google Scholar]
- 30.Richardson DE, et al. Sailfish (Istiophorus platypterus) spawning and larval environment in a Florida Current frontal eddy. Prog. Oceanogr. 2009;82:252–264. doi: 10.1016/j.pocean.2009.07.003. [DOI] [Google Scholar]
- 31.Bertelsen RD. Characterizing daily movements, nomadic movements, and reproductive migrations of Panulirus argus around the Western Sambo Ecological Reserve (Florida, USA) using acoustic telemetry. Fish. Res. 2013;144:91–102. doi: 10.1016/j.fishres.2012.12.008. [DOI] [Google Scholar]
- 32.Lee TN, Williams E. Mean distribution and seasonal variability of coastal currents and temperature in the Florida Keys with implications for larval recruitment. Bull. Mar. Sci. 1999;64:35–56. [Google Scholar]
- 33.Ehrhardt NM, Fitchett MD. Dependence of recruitment on parent stock of the spiny lobster, Panulirus argus, in Florida. Fish. Oceanogr. 2010;19:434–447. doi: 10.1111/j.1365-2419.2010.00555.x. [DOI] [Google Scholar]
- 34.Butler, M. J. et al. Patterns of spiny lobster (Panulirus argus) postlarval recruitment in the Caribbean: A CRTR Project. In Proceedings of the Gulf and Caribbean Fisheries Institute 360–369 (2010).
- 35.Johnson MW, Wernham J. Temporal variation of recruits as a basis of ephemeral genetic heterogeneity in the western rock lobster Panulirus cygnus. Mar. Biol. 1999;135:133–139. doi: 10.1007/s002270050610. [DOI] [Google Scholar]
- 36.Sarver SK, Silberman JD, Walsh PJ. Mitochondrial DNA sequence evidence supporting the recognition of two subspecies or species of the Florida spiny Panulirus argus. J. Crustac. Biol. 1998;18:177–186. doi: 10.2307/1549532. [DOI] [Google Scholar]
- 37.Silberman JD, Sarver SK, Walsh PJ. Mitochondrial DNA variation and population structure in the spiny lobster Panulirus argus. Mar. Biol. 1994;120:601–608. doi: 10.1007/BF00350081. [DOI] [Google Scholar]
- 38.Truelove NK, et al. Genetic evidence from the spiny lobster fishery supports international cooperation among Central American marine protected areas. Conserv. Genet. 2015;16:347–358. doi: 10.1007/s10592-014-0662-4. [DOI] [Google Scholar]
- 39.Benestan L, et al. RAD genotyping reveals fine-scale genetic structuring and provides powerful population assignment in a widely distributed marine species, the American lobster (Homarus americanus) Mol. Ecol. 2015;24:3299–3315. doi: 10.1111/mec.13245. [DOI] [PubMed] [Google Scholar]
- 40.Hedgecock D. Determining parantage and relatedness from genetic markers sheds light on patterns of marine larval dispersion. Mol. Ecol. Notes. 2010;19:845–847. doi: 10.1111/j.1365-294X.2010.04525.x. [DOI] [PubMed] [Google Scholar]
- 41.Kimura M, Weisss GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chollett I, et al. A Genuine Win-Win: Resolving the “Conserve or Catch” Conflict in Marine Reserve Network Design. Conserv. Lett. 2017;10:555–563. doi: 10.1111/conl.12318. [DOI] [Google Scholar]
- 43.Diniz FM, Maclean N, Paterson IG, Bentzen P. Polymorphic tetranucleotide microsatellite markers in the Caribbean spiny lobster, Panulirus argus. Mol. Ecol. Notes. 2004;4:327–329. doi: 10.1111/j.1471-8286.2004.00683.x. [DOI] [Google Scholar]
- 44.Tringali MD, Seyoum S, Schmitt SL. Ten di- and trinucleotide microsatellite loci in the Caribbean spiny lobster, Panulirus argus, for studies of regional population connectivity. Mol. Ecol. Resour. 2008;8:650–652. doi: 10.1111/j.1471-8286.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- 45.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 46.Goudet, J. FSTAT 2.9.3.2. (2002).
- 47.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 48.Excoffier L, Lischer H. An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinform. Online. 2015;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 49.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jombart T. Adegenet: a R package for multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 51.R Core Team. R: A Language and Environment for Statistical Computing (2016).
- 52.Schunter C, Garza JC, Macpherson E, Pascual M. SNP development from RNA-seq data in a nonmodel fish: How many individuals are needed for accurate allele frequency prediction? Mol. Ecol. Resour. 2014;14:157–165. doi: 10.1111/1755-0998.12155. [DOI] [PubMed] [Google Scholar]
- 53.LETT C, et al. A Lagrangian tool for modelling ichthyoplankton dynamics. Environ. Model. Softw. 2008;23:1210–1214. doi: 10.1016/j.envsoft.2008.02.005. [DOI] [Google Scholar]
- 54.Chassignet EP, et al. US GODAE: Global Ocean Prediction with the HYbrid Coordinate Ocean Model (HYCOM) Oceanography. 2009;22:64–75. doi: 10.5670/oceanog.2009.39. [DOI] [Google Scholar]
- 55.Criales MM, Cherubin LM, Browder JA. Modeling larval transport and settlement of pink shrimp in South Florida: dynamics of behavior and tides. Mar. Coast. Fish. 2015;7:148–176. doi: 10.1080/19425120.2014.1001541. [DOI] [Google Scholar]
- 56.Callwood, K. Use of larval connectivity modeling to determine settlement habitats of Panulirus argus in the Bahamas as a pre-cursor to marine protected area network planning. Open Access Theses (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microsatellite genotypes generated and analysed during the current study will be available in the DRYAD repository once the manuscript is accepted. 10.5061/dryad.27812c7.