The presence of dfrA35 associated with ISCR2 in Escherichia coli from animals, as well as its presence in other E. coli strains from different sources and countries and in Acinetobacter, highlights the global spread of this gene and its potential for further dissemination. The genetic link of ISCR2-dfrA35 with other antibiotic and disinfectant resistance genes showed that multidrug-resistant E. coli may be selected and maintained by the use of either one of several antimicrobials.
KEYWORDS: Escherichia coli, animals, antibiotic resistance, gene cassettes, trimethoprim
ABSTRACT
Whole-genome sequencing of trimethoprim-resistant Escherichia coli strains MF2165 and PF9285 from healthy Swiss fattening calves revealed a so far uncharacterized dihydrofolate reductase gene, dfrA35. Functionality and association with trimethoprim resistance were demonstrated by cloning and expressing dfrA35 in E. coli. The DfrA35 protein showed the closest amino acid identity (49.4%) to DfrA20 from Pasteurella multocida and to the Dfr determinants DfrG (41.2%), DfrD (40.8%), and DfrK (40.0%) found in Gram-positive bacteria. The dfrA35 gene was integrated within a florfenicol/chloramphenicol-sulfonamide resistance ISCR2 element (floR-ISCR2-dfrA35-sul2) next to a Tn21-like transposon that contained genes with resistance to sulfonamides (sul1), streptomycin (aadA1), gentamicin/tobramycin/kanamycin (aadB), and quaternary ammonium compounds (qacEΔ1). A search of GenBank databases revealed that dfrA35 was present in 26 other E. coli strains from different origins as well as in Acinetobacter.
IMPORTANCE The presence of dfrA35 associated with ISCR2 in Escherichia coli from animals, as well as its presence in other E. coli strains from different sources and countries and in Acinetobacter, highlights the global spread of this gene and its potential for further dissemination. The genetic link of ISCR2-dfrA35 with other antibiotic and disinfectant resistance genes showed that multidrug-resistant E. coli may be selected and maintained by the use of either one of several antimicrobials.
OBSERVATION
Trimethoprim is a synthetic folic acid antagonist that was introduced as an antimicrobial drug in human and veterinary medicine in the late 1960s. It is most commonly used in combination with sulfonamides: both antibiotics sequentially inhibit bacterial synthesis of tetrahydrofolic acid, which is a cofactor essential for the synthesis of thymidine and purine, the fundamental bases of DNA and RNA (1–4). Sulfonamides are analogues of para-aminobenzoic acid (PABA) and compete with PABA to bind to the dihydropteroate synthase (DHPS), thereby inhibiting the synthesis of dihydrofolic acid. Trimethoprim binds to the dihydrofolate reductase (DHFR), thereby blocking the conversion of dihydrofolic acid into tetrahydrofolic acid (1, 2). The use of trimethoprim/sulfonamides in both veterinary and human medicine has selected for a resistant bacterial population. In Switzerland, 28% of the Escherichia coli strains isolated from humans and up to 25% of E. coli strains from cattle and pigs have been reported to be resistant to the combination of these antimicrobials (5).
Resistance to sulfonamides and trimethoprim has been associated with five main mechanisms, including (i) a permeability barrier, (ii) a naturally insensitive intrinsic DHFR, (iii) spontaneous chromosomal mutations in the intrinsic DHPS (folP) and DHFR (folA) genes involved in the folic acid pathways, (iv) increased production of the sensitive target enzyme by upregulation of gene expression or gene duplication, and (v) the acquisition of alternative DHPS (sul) and DHFR (dfr) genes with integrons, plasmids, and transposons (6, 7). To date, three different alternative sul genes (sul1, sul2, and sul3) and 40 different types of alternative dfr genes have been described in Gram-positive and Gram-negative bacteria (8–10). However, the trimethoprim resistance mechanism remained unknown for 8 of 56 trimethoprim-resistant E. coli strains isolated from rectal swabs of healthy veal calves in 2017 in Switzerland (11). We selected two genetically diverse E. coli strains (MF2156 and PF9285) from two different farms based on rep-PCR profile to further investigate the nature of the trimethoprim resistance in these strains. The whole-genome sequences of both strains were screened for DHFR homologs followed by proof of functionality.
Identification and localization of a new DHFR.
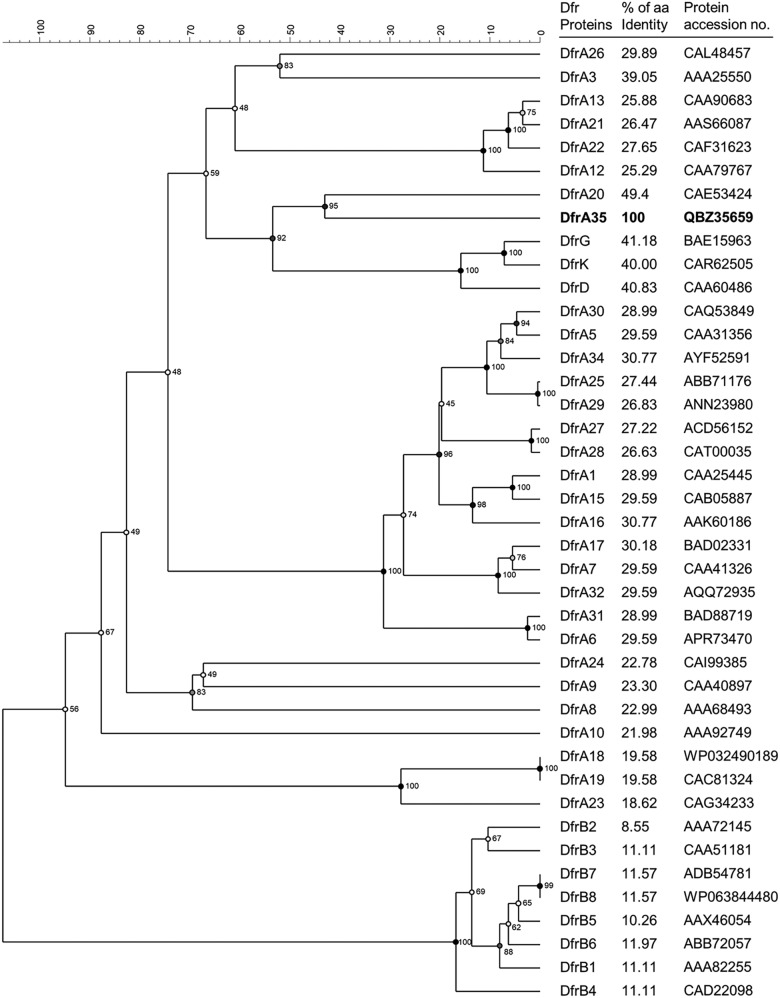
Genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Inc., Venlo, The Netherlands), and purified using the AMPure XP PCR purification system (Beckman Coulter Life Sciences, Indianapolis, IN). DNA was sequenced on an Illumina HiSeq platform (2 × 150 paired ends) (Eurofins, Constance, Germany) and on a R9.4 SpotON flow cell and library kit (Oxford Nanopore Technologies, Oxford, United Kingdom) to obtain long reads and facilitate genome assembly. Genome assembly and read mapping of the Illumina reads against the MinION scaffolds were performed as previously described (12). Analyses of the complete genome using RESFinder 3.1 (Center for Genomic Epidemiology, DTU, Denmark) (20% coverage, 30% identity) confirmed the absence of any known acquired dfr gene in both strains MF2156 and PF9285. Comparison of the chromosomal folA gene with that of the trimethoprim-susceptible E. coli strain K-12 MG1655 (GenBank accession no. NC_000913) showed no mutation in this gene in both strains, suggesting the presence of an alternative mechanism. Search for a possible new acquired dfr gene within the complete genomes of MF2156 and PF9285 using blastx (https://blast.ncbi.nlm.nih.gov/) and DfrA1 (NCBI accession no. CAA25445) as the reference revealed the presence of a 177-amino-acid DHFR homologue (513 bp) in both strains. This putative new DHFR shared the closest amino acid identity (49.4%) with DfrA20 from Pasteurella multocida (GenBank accession no. CAE53424) (13) and was next closely related to Dfr proteins DfrD, DfrG, and DfrK, which have been identified so far only in Gram-positive bacteria (Fig. 1). The new gene was named dfrA35 following the nomenclature used for Gram-negative bacteria (9).
FIG 1.
Phylogenetic tree of all known Dfr proteins, including the novel protein DfrA35. The tree was obtained by multiple alignment of amino acid sequences (without fast alignment) and the UPGMA clustering method with Jukes and Cantor correction using Bionumerics 7.6 (Applied Maths, Kortrijk, Belgium): multiple alignment with an open gap penalty (OG) of 100%, a unit gap penalty (UG) of 0%, a gap penalty of 100%, and 1,000 bootstrap values (nodes). Scala results are shown as distances. The percentages of amino acid sequence identity between DfrA35 and other Dfr proteins were determined by multiple sequence alignment with clustalW 2.1 (cost matrix Blosum) using Geneious prime 2019.1.1 (Biomatters, Ltd., Auckland, New Zealand).
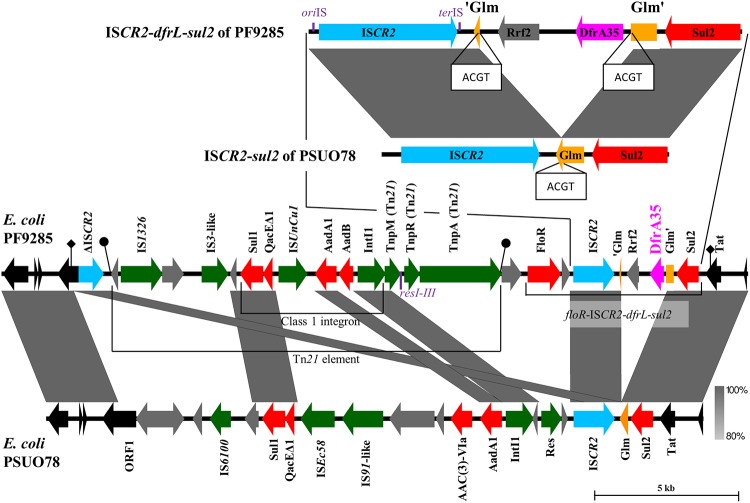
The dfrA35 gene was located on a 22,977-bp fragment integrated in the same location of the chromosome in both E. coli strains MF2156 and PF9285. Both ends of the integrated element were identified by comparative analysis of sequences of strains MF2156 and PF9285 with those from E. coli strain PSU078, which shared an identical flanking region (GenBank accession no. CP012112). The fragment was delimited by a truncated ISCR2 (ΔISCR2) interrupting a putative restriction endonuclease subunit S gene on one side and by a sul2 gene interrupting a putative Tat pathway signal sequence protein gene on the other side. The fragment contains an ISCR2 element carrying the dfrA35 gene, the florfenicol/chloramphenicol export gene floR, and the sulfonamide resistance gene sul2 (floR-ISCR2-dfrA35-sul2) and a 14,231-bp Tn21-like element (Fig. 2). Given the comparison to PSUO78, it is likely that the dfrA35 region and the Tn21-like element were mobilized into the chromosome either simultaneously or separately by homologous recombination. The dfrA35 gene was integrated with a gene of the Rrf2 transcriptional regulator family into the phosphoglucosamine mutase gene glm of a floR-ISCR2-sul2 element, which has been previously reported in Stenotrophomonas maltophilia (14, 15). Integration of the dfrA35-rrf2 fragment split the glm gene into two pieces, generating a duplication of the ACGT integration sequence (Fig. 2). The dfrA35-rrf2 has likely been trapped by the gene-capturing machinery described for ISCR elements and mobilized by rolling circle transposition, but dfrA35 does not seem to be a single cassette as many other dfr genes are (14, 16, 17). The Tn21-like element contained the characteristic features defining the Tn21 subgroup with a transposase gene, tnpA, a resolvase gene, tnpR, in the same orientation, the res sites preceding tnpR, and two 38-bp inverted repeats at both ends (18, 19). It also contained a class I integron In290 (INTEGRALL database), including the integrase gene intI1, the sulfonamide resistance gene sul1, the streptomycin/spectinomycin resistance gene aadA1 [ant(3′')-Ia], the gentamicin/tobramycin/kanamycin resistance gene aadB [ant(2'')-Ia], the quaternary ammonium compound (QAC) efflux transporter gene qacEΔ1, as well as the transposase genes of IS1326 and of a new transposase gene related to IS3. Duplication of ISCR2 sequences suggests that insertion and movement of the different pieces of the element may have also occurred by homologous recombination (Fig. 2).
FIG 2.
Schematic representation of the integration of dfrA35 into ISCR2-sul2 (ISCR2-dfrA35-sul2) and its genetic linkage with a Tn21-like transposon in E. coli PF9285. Shown are open reading frames (ORFs) and functions. The ORF of DfrA35 is indicated by a magenta arrow and represents the dehydrofolate reductase for trimethoprim resistance. ORFs of other antibiotic resistance proteins are in red: Sul1 and Sul2, dihydropteroate synthase for sulfonamide resistance; AadA1, streptomycin/spectinomycin 3″-adenylyltransferase ANT(3″)-Ia; AadB, aminoglycoside-2″-O-nucleotidyltransferase ANT(2″)-Ia; FloR, florfenicol/chloramphenicol export protein; QacEΔ1, quaternary ammonium compound efflux transporter; aminoglycoside 3-N-acetyltransferase. ORFs of hypothetical proteins are represented by gray arrows. ORFs of transposases associated with insertion sequences (IS), transposon Tn21 (Tnp), as well as integrase (IntI1) and resolvase (Res) are indicated by green arrows, except for the ORFs of ISCR2 and ΔISCR2, which are indicated in deep sky blue. The res sites I, II, and III of Tn21 as well as the oriIS and terIS sequences of ISCR2 are indicated in purple. The putative phosphoglucosamine mutase Glm as well as the 3′-end truncated part (Glm′) and the 5′-end truncated part (′Glm) are indicated in orange. ORFs representing the core genomes of strains PF9285 (GenBank accession no. CP038791) and PSUO78 (GenBank accession no. CP012112) are indicated by black arrows with Tat as a putative Tat pathway signal sequence protein and ORF1 as a putative restriction endonuclease subunit S protein. Arrows with black diamonds attached indicate the limit of the 22,977-bp region in PF9285 as determined by the end of common sequences between the E. coli PF9285 and PSUO78 chromosomes interrupted by ΔISCR2 on the left side and by the beginning of the sul2 core sequences on the right side (positions 989354 to 1012331 in CP038791). Inverted repeats of the Tn21 element are indicated by oval arrows with black circles (IV-L, GGGGGCACCTCAGAAAACGGAAAATAAAGCACGCTAAG; and IV-R, CTTAGCGTGCTTTATTTTCCGTTTTCTGAGACGACCCC). Direct repeats of ACGT duplicated by the insertion of the rrf2-dfrA35 fragment into pgm are indicated in white boxes. The figure was created using Microsoft PowerPoint and Easyfig 2.2.2 (22).
As expected by its chromosomal location and genetic context, transferability of the dfrA35 gene could not be observed experimentally by filter mating using strains MF1256 and PF9285 as donors and the rifampin-, sodium azide-resistant strain E. coli J53dR as the recipient (20). Selection was performed on Mueller-Hinton (MH) agar plates containing trimethoprim (30 µg/ml) with either sodium azide (100 µg/ml) or rifampin (50 µg/ml) for 48 h at 37°C.
Expression of dfrA35 in E. coli.
Functionality and association of dfrA35 with trimethoprim resistance were determined by cloning an 878-bp region containing the 513-bp dfrA35 gene and a 265-bp region upstream of the dfrA35 start codon. This fragment was amplified by PCR using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, MA) and primers dfrA35-F (5′-TGGTGCGCAAATATTTCGGC-3′) and dfrA35-R (5′-ACGTTAACCCGAAAAAGCGA-3′) (annealing temperature, 56°C; extension time, 30 s) and cloned into the PCR-Blunt II-TOPO vector following the manufacturer’s instructions (Zero blunt TOPO PCR cloning kit; Thermo Fisher Scientific).
The ligated vector insert DNA was transformed into chemically competent cells of E. coli One Shot TOP10 (Thermo Fisher Scientific). Transformants obtained on LB plates containing 50 µg/ml kanamycin after 24 h of incubation at 37°C were tested for the presence of the dfrA35 gene by PCR using Taq polymerase and internal primers dfrA35int-F (5′-GCATTTACCGGCCGATATGC-3′) and dfrA35int-R (5′-ACACGCAGCACCTCTTCATT-3′) (annealing temperature, 56°C; extension time, 30 s). The resulting dfrA35-containing plasmids, pT2156c19 and pT9285c17, were isolated using the PureLink Quick plasmid miniprep kit (Thermo Fisher Scientific) and analyzed by Sanger sequencing to confirm the veracity of the sequence and that dfrA35 was inserted in the opposite direction of the TOPO promoter Plac and was therefore under the control of its own promoter. The MIC of trimethoprim for the parent strains MF2156 and PF9285, recipient strain TOP10, and transformant strains TOP10/pT2156c19 and TOP10/pT9285c17 was determined on a microtiter plate using the 2-fold-dilution technique (concentration range, 1 to 512 µg/ml) following CLSI guidelines (21). The MIC for trimethoprim increased in E. coli TOP10 expressing dfrA35 to 128 µg/ml compared to the nontransformed TOP10 strain (≤0.25 µg/ml), almost reaching the level of the MIC observed in the parent strains MF2156 and PF9285 (256 µg/ml). The MIC of 13 other antibiotics was determined using EUVSEC Sensititre plates (Thermo Fisher Scientific), showing an association between the other antibiotic resistance genes found on strains MF2156 and PF9285 and their phenotype (Table 1).
TABLE 1.
Characteristics of Escherichia coli strains and MICs of antibiotics
| E. coli strain (sequence type) | Origin and characteristics | Reference | Antibiotic resistance gene(s)a | MIC (µg/ml) ofb
: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AZI | CHL | CIP | CST | FOT | GEN | MERO | NAL | SMX | TAZ | TET | TGC | TMP | ||||
| MF2156 (ST3057) | Rectal swab of calf 1, farm 1 | This study | aadA1, aadB, blaTEM-1D, dfrA35, floR, sul1, sul2, tet(A), qacEΔ1 | >64 | ≤2 | 128 | ≤0.015 | ≤1 | ≤0.25 | 8 | ≤0.03 | ≤4 | >1,024 | ≤0.5 | >64 | 0.5 | 256 |
| PF9285 (ST10) | Rectal swab of calf 1, farm 2 | This study | aadA1, aadB, aph(3′)-Ia, blaTEM-1B, catA1, dfrA35, floR, strA, strB, sul1, sul2, tet(A), qacEΔ1 | >64 | 4 | >128 | ≤0.015 | ≤1 | ≤0.25 | 8 | ≤0.03 | ≤4 | >1,024 | ≤0.5 | 32 | ≤0.25 | 256 |
| TOP10 | OneShot TOP10 cells for transformation | Thermo Fisher Scientific | None | 2 | 4 | ≤8 | ≤0.015 | ≤1 | ≤0.25 | ≤0.5 | ≤0.03 | ≤4 | ≤8 | ≤0.5 | ≤2 | ≤0.25 | ≤0.25 |
| TOP10/pT2156c19 | TOP10 with dfrA35 from MF2156 cloned into vector pCR-BluntII-Topo | This study | dfrA35 | 2 | 4 | ≤8 | ≤0.015 | ≤1 | ≤0.25 | ≤0.5 | ≤0.03 | ≤4 | ≤8 | ≤0.5 | ≤2 | ≤0.25 | 128 |
| TOP10/pT9285c17 | TOP10 with dfrA35 from PF9285 cloned into vector pCR-BluntII-Topo | This study | dfrA35 | 2 | 4 | ≤8 | ≤0.015 | ≤1 | ≤0.25 | ≤0.5 | ≤0.03 | ≤4 | ≤8 | ≤0.5 | ≤2 | ≤0.25 | 128 |
Genes and functions: aadA1, streptomycin/spectinomycin adenyltransferase; aadB, gentamicin/tobramycin/kanamycin nucleotidyltransferase; aph(3′)-Ia, kanamycin/neomycin/paromomycin/ribostamycin/lividomycin/gentamicin B phosphotransferase; blaTEM-1B and blaTEM-1D, ampicillin β-lactamase; catA1, chloramphenicol acetyltransferase; dfrA35, trimethoprim dihydrofolate reductase; floR, florfenicol/chloramphenicol exporter; strA, strB, streptomycin phosphotransferase; sul1 and sul2, sulfonamide dihydropteroate synthase; tet(A), tetracycline exporter; qacEΔ1, quaternary ammonium compound multidrug exporter.
Antibiotics: AMP, ampicillin; AZI, azithromycin; CHL, chloramphenicol; CIP, ciprofloxacin; CST, colistin; FOT, cefotaxime; GEN, gentamicin; MERO, meropenem; NAL, nalidixic acid; SMX, sulfamethoxazole; TAZ, ceftazidime; TET, tetracycline; TGC, tigecycline; TMP, trimethoprim. Note that the MICs for TMP have been highlighted in boldface.
Spread of dfrA35 in association with ISCR2 and sul2.
PCR screening of 8 additional strains from calves with no known trimethoprim resistance gene revealed dfrA35 in 5 of them. The gene was also located in these 5 strains on an ISCR2-dfrA35-sul2 fragment, as determined by Taq PCR using primers ISCR2-F (5′-CGCCTGCATTGAAGACCCTA-3′) and Sul2-F (5′-TGTCTGTTTCGCGCAAATCC-3′) (annealing temperature, 56°C; extension time, 3 min). Searching for DfrA35 in the NCBI database using blastp revealed its presence in 26 other E. coli strains as well as in an Acinetobacter sp. strain originating from different sources (cattle, dogs, a horse, and humans) and countries, which indicates that the dfrA35 gene has potential for dissemination among bacterial species even if transfer could not be demonstrated experimentally. In 18 of them, the available sequences allowed us to determine that dfrA35 was also linked to sul2, and 6 sequences revealed the presence of the ISCR2-dfrA35-sul2 element (see Table S1 in the supplemental material).
List of sequenced bacteria containing dfrA35 found in the NCBI GenBank database (accessed 30 November 2018). Download Table S1, PDF file, 0.03 MB (31.6KB, pdf) .
Copyright © 2019 Wüthrich et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
This study identified a novel functional trimethoprim resistance gene (dfrA35) in E. coli from calves. This gene appeared to be widespread in other E. coli strains as well as in Acinetobacter, where it was also mainly directly flanked by ISCR2 and/or sul2. In the two analyzed E. coli strains from cattle, the ISCR2-dfrA35-sul2 sequence was also linked to floR and was part of a larger multiple antibiotic and QAC resistance element. The dissemination of this element may further jeopardize the efficacy of antibiotics and disinfectants in both veterinary and human medicine.
Accession number(s).
The complete chromosome of E. coli strain PF9285 containing the 22,977-bp fragment carrying dfrA35 and flanking regions (positions 989354 to 1012331) has been deposited in GenBank under accession no. CP038791 (BioProject no. PRJNA530748).
ACKNOWLEDGMENTS
We thank Alexandra Collaud and Alexandra Rossano for technical assistance.
This study was financed by grant no. 407240_167083 from the Swiss National Science Foundation within the Antimicrobial Resistance National Research Program NRP72 to Mireille Meylan and Vincent Perreten.
REFERENCES
- 1.Sköld O. 2009. Sulfonamides and trimethoprim, p 259–269. In Mayers DL. (ed), Antimicrobial drug resistance, vol 1 Humana Press, New York, NY. [Google Scholar]
- 2.Zinner SH, Mayer KH. 2015. Sulfonamides and trimethoprim, p 410–418.e412. In Bennett JE, Dolin R, Blaser MJ (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 8th ed, vol 1 Elsevier Health Sciences, London, United Kingdom. [Google Scholar]
- 3.Papich MG. 2016. Trimethoprim + sulfamethoxazole, p 820–822. In Papich MG. (ed), Saunders handbook of veterinary drugs: small and large animals, 4th ed Elsevier, St. Louis, MO. [Google Scholar]
- 4.Christaki E. 2017. Folate inhibitors, p 1280–1284.e1281. In Cohen J, Powderly WG, Opal SM (ed), Infectious diseases, 4th ed, vol 2 Elsevier, St Louis, MO. [Google Scholar]
- 5.Federal Office of Public Health and Federal Food Safety and Veterinary Office. 2018. Swiss Antibiotic Resistance Report 2018. Usage of Antibiotics and Occurrence of Antibiotic Resistance in Bacteria from Humans and Animals in Switzerland. November 2018. FOPH publication no. 2018-OEG-87. http://anresis.ch/index.php/anresisch-data.html.
- 6.Huovinen P, Sundström L, Swedberg G, Sköld O. 1995. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother 39:279–289. doi: 10.1128/AAC.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sköld O. 2001. Resistance to trimethoprim and sulfonamides. Vet Res 32:261–273. doi: 10.1051/vetres:2001123. [DOI] [PubMed] [Google Scholar]
- 8.Perreten V, Boerlin P. 2003. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother 47:1169–1172. doi: 10.1128/AAC.47.3.1169-1172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts MC, Schwarz S, Aarts HJ. 2012. Erratum. Acquired antibiotic resistance genes: an overview. Front Microbiol 3:384. doi: 10.3389/fmicb.2012.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tagg KA, Francois Watkins L, Moore MD, Bennett C, Joung YJ, Chen JC, Folster JP. 2019. Novel trimethoprim resistance gene dfrA34 identified in Salmonella Heidelberg in the USA. J Antimicrob Chemother 74:38–41. doi: 10.1093/jac/dky373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hausherr A. 2018. Resistance to antibiotics and quaternary ammonium compounds of Escherichia coli from calves at the beginning of the fattening period in Switzerland. DVM thesis Vetsuisse Faculty, University of Bern, Bern, Switzerland. [Google Scholar]
- 12.Donà V, Perreten V. 2018. Comparative genomics of the first and complete genome of “Actinobacillus porcitonsillarum” supports the novel species hypothesis. Int J Genomics 2018:5261719. doi: 10.1155/2018/5261719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kehrenberg C, Schwarz S. 2005. dfrA20, a novel trimethoprim resistance gene from Pasteurella multocida. Antimicrob Agents Chemother 49:414–417. doi: 10.1128/AAC.49.1.414-417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toleman MA, Bennett PM, Bennett DMC, Jones RN, Walsh TR. 2007. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis 13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 17.Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev 35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 18.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 20.Donà V, Bernasconi OJ, Pires J, Collaud A, Overesch G, Ramette A, Perreten V, Endimiani A. 2017. Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolates from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob Agents Chemother 61:e01245-17. doi: 10.1128/AAC.01245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; twenty-seventh informational CLSI supplement M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of sequenced bacteria containing dfrA35 found in the NCBI GenBank database (accessed 30 November 2018). Download Table S1, PDF file, 0.03 MB (31.6KB, pdf) .
Copyright © 2019 Wüthrich et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.