Abstract
Noncoding RNAs (ncRNAs) such as microRNAs (miRNAs) and long ncRNAs (lncRNAs) have been shown to function as pivotal regulators in the carcinogenesis of renal cell carcinoma (RCC). However, the functions and underlying mechanisms of most ncRNAs in RCC are still elusive, and the crosstalks of different layers of ncRNAs are seldom reported. Here we showed that miR-124 and maternally expressed gene 3 (MEG3) were both significantly reduced in RCC, and combined expression of miR-124 and MEG3 emerged as an independent prognostic factor in our RCC cohort. Overexpression of miR-124 or MEG3 inhibited cell proliferation, migration, and invasion in vitro, and restrained tumor growth in vivo. EZH2 knockdown induced the epigenetic silencing of miR-124 and MEG3 expression by H3K27me3. Besides, miR-124 directly targeted the TET1 transcript, and then the interaction resulted in the upregulation of MEG3. Furthermore, we demonstrated that MEG3 induced p53 protein accumulation, whereas p53 was a positive transcriptional regulator of the miR-124. In addition, tumor-suppressive PTPN11 was identified as a direct target of miR-124, as well as the MEG3- and p53-regulated gene. Our study identifies three crosstalks between miR-124 and MEG3, which provide a plausible link for these two ncRNAs in RCC. Both ncRNAs exert important antitumor effects in RCC pathogenesis and might serve as prognostic biomarkers and molecular therapeutic targets.
Keywords: miR-124, MEG3, methylation, p53, renal cell carcinoma, EZH2, PTPN11, noncoding RNA
Introduction
Renal cell carcinoma (RCC) accounts for approximately 3% of all adult cancers,1 and poor survival is manifested in RCC patients, especially for those with metastasis.2 Increasing evidence indicates that miRNAs and long noncoding RNAs (lncRNAs) play vital roles in the regulation of various cellular processes. In our previous study, we identified that a robust panel of miRNA signatures could distinguish RCC from normal kidney tissues, and we also constructed a five-miRNA-based classifier as a reliable predictive tool for cancer-specific survival in RCC patients. miR-124, one member of the five-miRNA-based classifier, was identified as a significantly downregulated and prognostic miRNA in RCC.3 In another study that first drew the lncRNA expression profile in RCC, we identified that one nominated lncRNA, maternally expressed gene 3 (MEG3), was downregulated in RCC.4 In this study, we performed the lncRNA promoter methylation methylated DNA immunoprecipitation (MeDIP)-chip of human RCC tissue samples, and combined analysis of lncRNA promoter methylation MeDIP-chip and lncRNA microarray expression profiling indicated that MEG3 was both downregulated and hypermethylated in RCC.
miR-124 and MEG3 function as tumor suppressor genes involved in tumorigenesis; their overexpression could inhibit cell proliferation, induce cell growth arrest, and promote cell apoptosis in various cancers. Although previous studies indicated that CpG island methylation contributed to downregulation of miR-124 expression and promoted tumor progression in several tumors,5, 6 the definite roles of miR-124 in RCC remain elusive at present. Besides, hypermethylation of promoter of the MEG3 gene has been identified to contribute to the loss of MEG3 expression in tumors, whereas little is known about the MEG3 expression pattern and its biological functions in RCC carcinogenesis. Besides, whether there exists some regulatory relations or signal crosstalks between miR-124 and MEG3 in renal cancer is also puzzling.
In this study, we demonstrated that both miR-124 and MEG3 expression were downregulated, and their combined expression pattern had the potential to facilitate survival prediction in RCC. Besides, a series of in vitro and in vivo experiments determined the tumor-suppressive roles of miR-124 and MEG3. Furthermore, we identified three crosstalks between miR-124 and MEG3, which explained their reciprocal regulations. In addition, PTPN11 was identified as a crucial downstream gene involved in the miR-124/MEG3 regulatory network. This study helps broaden our knowledge of the expression pattern, clinical significance, physiopathologic functions, and reciprocal regulations of two noncoding RNAs, miR-124 and MEG3.
Results
miR-124 and MEG3 Expressions Were Significantly Reduced in RCC
A total of 525 clear cell RCC (ccRCC) miRNA expression profiles (level 3 data) were obtained from The Cancer Genome Atlas (TCGA). Defining 1.5-fold difference as the cutoff level, a significant difference in expression was observed in 143 of 1,046 detected human miRNAs between RCC and adjacent normal tissues, including 62 downregulated and 81 upregulated miRNAs (Figure S1A).
Our previous study described lncRNAs microarray profiles in six pairs of ccRCC and corresponding adjacent nontumorous tissues.4 Using 1.5-fold expression difference as a cutoff level, 59 downregulated and 74 upregulated lncRNAs were identified between RCC and adjacent normal tissues (Figure S1B). In addition, we performed lncRNA promoter methylation MeDIP-chip analysis with three paired RCC and adjacent normal tissues to identify differentially methylated lncRNAs. This assay turned out to identify 3,837 lncRNAs that were significantly differentially methylated. Finally, combined analysis of lncRNA promoter methylation MeDIP-chip and lncRNA microarray expression profiling revealed that MEG3 was both downregulated and hypermethylated in RCC (Figure S1C).
To identify noncoding RNAs (ncRNAs) with altered expression in RCC, we performed a series of microarray analysis regarding ncRNA expression. Among all downregulated ncRNAs, miR-124 (Figure S1A) and MEG3 (Figure S1B) were the top two decreased (Figure S1C).
miR-124 and MEG3 Expression Were Positively Correlated and Associated with Overall Survival in RCC Patients
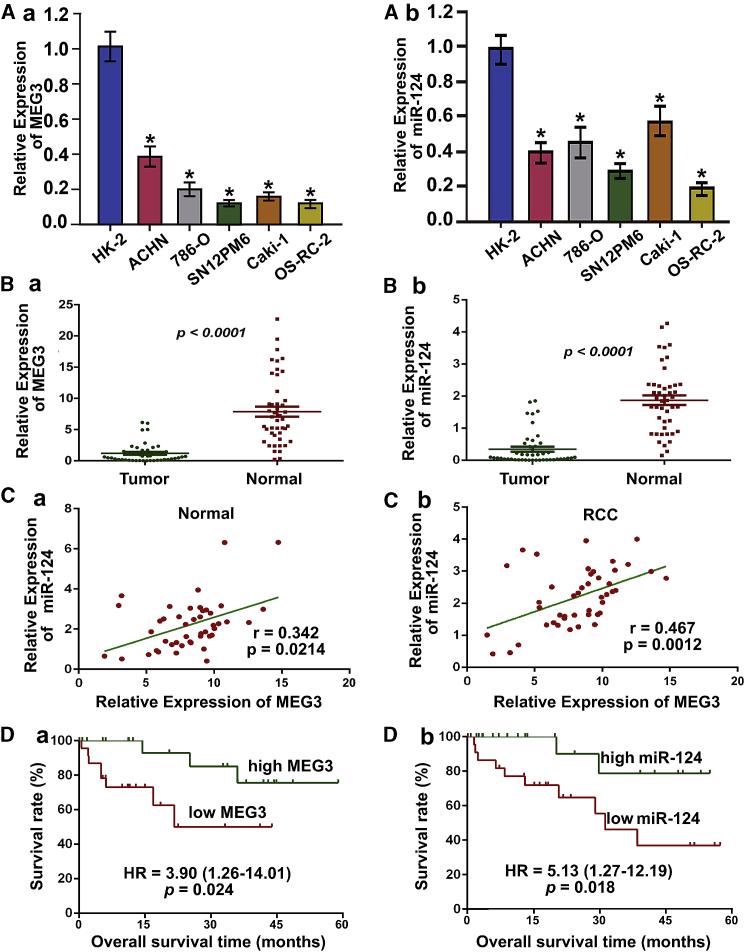
Via qRT-PCR, we then verified the expression levels of miR-124 and MEG3 in five RCC cell lines in comparison with the expression in HK2 (Figure 1A). Next, we assessed the expression of miR-124 and MEG3 in a further 45 pairs of RCC and adjacent normal tissues (Table S4) to validate the microarray analysis findings (Figure 1B). Statistically, the expression levels of miR-124 and MEG3 were significantly reduced in RCC. Moreover, the relationships between miR-124, MEG3 expression, and the clinicopathologic factors of RCC were evaluated, and we found that miR-124 and MEG3 expression were significantly correlated with tumor stage, grade, and lymph node metastasis (p < 0.05) (Table S5). Because miR-124 and MEG3 were both significantly reduced and shown to be correlated with RCC progression, we further examined their expression relationship using the Pearson correlation coefficient analysis. The expression of miR-124 was positively correlated with MEG3 expression in both RCC and adjacent normal tissues (Figure 1C; Figure S4A). These data indicated that miR-124 and MEG3 expression were remarkably decreased in RCC cell lines and tissues, and were positively correlated.
Figure 1.
Decreased miR-124 and MEG3 Expressions Were Positively Correlated and Associated with Poor Overall Survival in RCC Patients
(A) miR-124 and MEG3 expressions were reduced in RCC cell lines including ACHN and 786-O, compared with primary normal human renal epithelial cells (HK2). Data were presented as mean ± SD (**p < 0.01). (B) Both miR-124 and MEG3 expressions were reduced in human RCC compared with adjacent nonmalignant tissues. Data were presented as mean ± SD (**p < 0.01). (C) Positive correlation between miR-124 and MEG3 expression levels in RCC and matched nonmalignant tissues. Statistical analysis was performed using Pearson correlation coefficient analysis, with r and p values as indicated. (D) Kaplan-Meier curves with log rank tests showed that patients with high miR-124 and MEG3 expression survived significantly longer than those with low miR-124 and MEG3 expression (p < 0.001).
To investigate the potential prognostic significance of these two non-coding RNAs in the carcinogenesis and progression of RCC, we analyzed the correlation of miR-124 or MEG3 expression and overall survival (OS) of patients with RCC. With the median fold change (T/N) in miR-124 or MEG3 expression chosen as the cutoff value, patients with higher miR-124 or MEG3 expression survived statistically significantly longer than those with lower expression levels (Figure 1D). Moreover, the kidney renal clear cell carcinoma (KIRC) dataset also identified a similar significant association between miR-124 or MEG3 expression and OS in RCC (Figures S2 and S3; Tables S2 and S3; p < 0.05). Furthermore, a multivariate Cox regression analysis was conducted to evaluate whether miR-124 and MEG3 were independent prognostic factors for RCC, which indicated that high expression levels of miR-124 and MEG3 in RCC were associated with favorable prognosis for OS (hazard ratio [HR] = 0.61, 95% confidence interval [CI]: 0.46–0.82; p = 0.001; Figure S4B), independent of other clinicopathologic covariates (Table S6). These results suggested that miR-124 and MEG3 might be used as independent prognostic factors for RCC.
Overexpression of miR-124 and MEG3 Inhibit Cell Proliferation and Tumor Growth in RCC
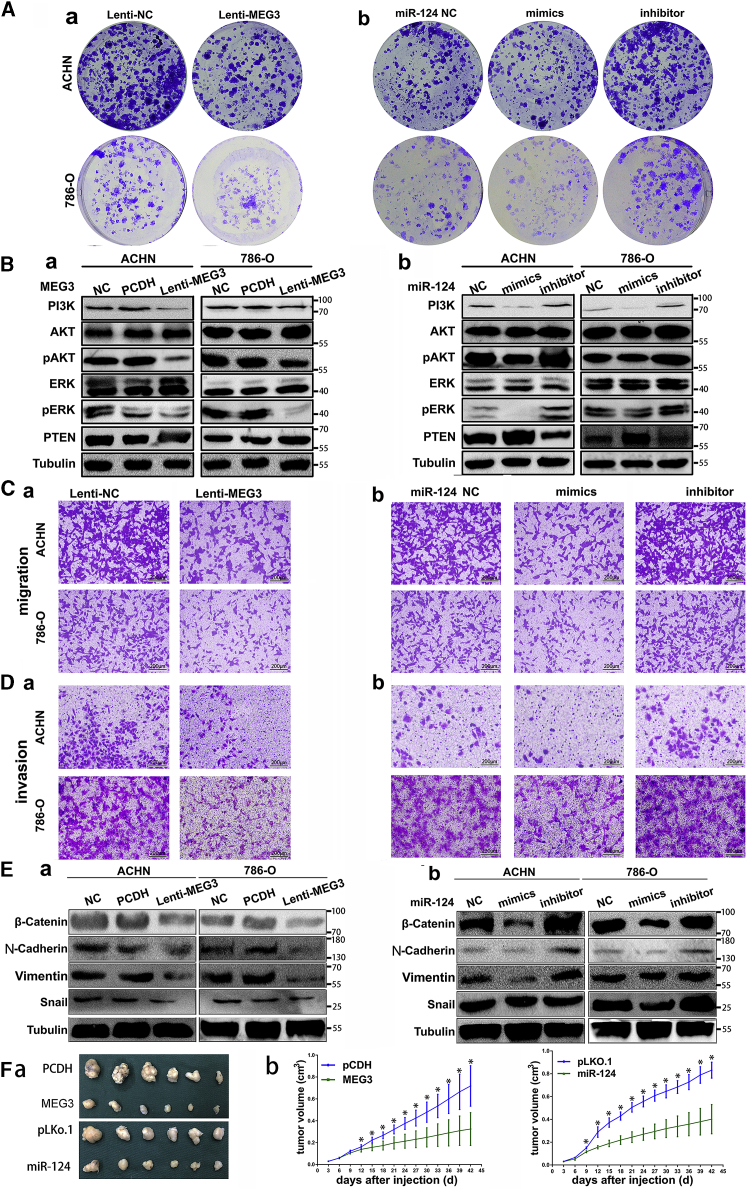
To examine the effects of miR-124 and MEG3 on RCC cell proliferation, we transfected MEG3, miR-124 mimics, or miR-124 inhibitor into ACHN and 786-O cells, which changed the expression levels of MEG3 or miR-124 in cells. Using MTS and colony formation assays, we demonstrated that ectopic expression of MEG3 and miR-124 significantly inhibited RCC cell growth (Figure 2A; Figure S5). Conversely, knockdown of miR-124 with inhibitors promoted the cell proliferation.
Figure 2.
Overexpression of miR-124 and MEG3 Could Inhibit Cell Proliferation, Migration, Invasion, and Tumor Growth
(A) Representative photographs of the colony formation assay using RCC cells following lenti-MEG3, miR-124 mimics, and inhibitors transfection. *p < 0.05 compared with NC groups. (B) Western blot analysis of the proliferation-related proteins expression after transfecting with lenti-MEG3 (a), miR-124 mimics, and inhibitors (b). Tubulin served as an internal control. *p < 0.05 compared with NC. (C and D) Migration (C) and invasion assay (D) for RCC cells. Representative photographs were taken at ×200 magnification; number of migrated cells was quantified in 10 random images from each treatment group. (E) Western blot analysis of the EMT-related proteins expression after transfecting with lenti-MEG3 (a), miR-124 mimics, and inhibitors (b). Tubulin served as an internal control. *p < 0.05 compared with NC. (F) Xenograft tumor experiments of ACHN cells that stably expressed MEG3 or miR-124. Representative figures of resected tumors (a) and tumor volume growth data with time (b) were shown. *p < 0.05.
We further explored whether miR-124 and MEG3 restoration or downregulation modulated the expression of downstream proliferation-related proteins in RCC cells. In ACHN and 786-O cells, downregulation of phosphatidylinositol 3-kinase (PI3K), pAKT, and pERK protein levels and induction of PTEN expression were exhibited after miR-124 and MEG3 overexpression, whereas the reverse tendency was shown after miR-124 inhibitor transfection (Figure 2B). Besides, ACHN cells that stably expressed MEG3 or miR-124 generated smaller xenograft tumors in nude mice (Figure 2Fa) and exhibited slower tumor growth rates in vivo (Figure 2Fb). These findings indicated that miR-124 and MEG3 overexpression inhibited RCC cell proliferation and tumor growth by modulating the expression of proliferation-related proteins.
Overexpression of miR-124 and MEG3 Could Suppress Invasion and Migration
To assess the impacts on cell migration and invasion, ACHN and 786-O cells were infected with lenti-MEG3 or lenti-NC and miR-124 mimics, inhibitors, or negative control (NC). Compared with the NC, overexpression of miR-124 and MEG3 significantly inhibited cell migration (Figure 2C) and invasion (Figure 2D) of ACHN and 786-O cells.
Further, epithelial-mesenchymal transition (EMT)-related proteins were detected after miR-124 and MEG3 restoration or downregulation. As shown in Figure 2E and Figure S6, N-cadherin, β-catenin, Vimentin, Snail, and MMP-2/7/9 were downregulated in ACHN and 786-O cells after miR-124 and MEG3 overexpression, whereas the reverse tendency was shown after miR-124 inhibitor transfection. Taken together, these findings stated that miR-124 and MEG3 inhibited RCC cell invasion and migration, owing to the suppression of EMT-related proteins.
EZH2-Mediated Silencing of miR-124 and MEG3
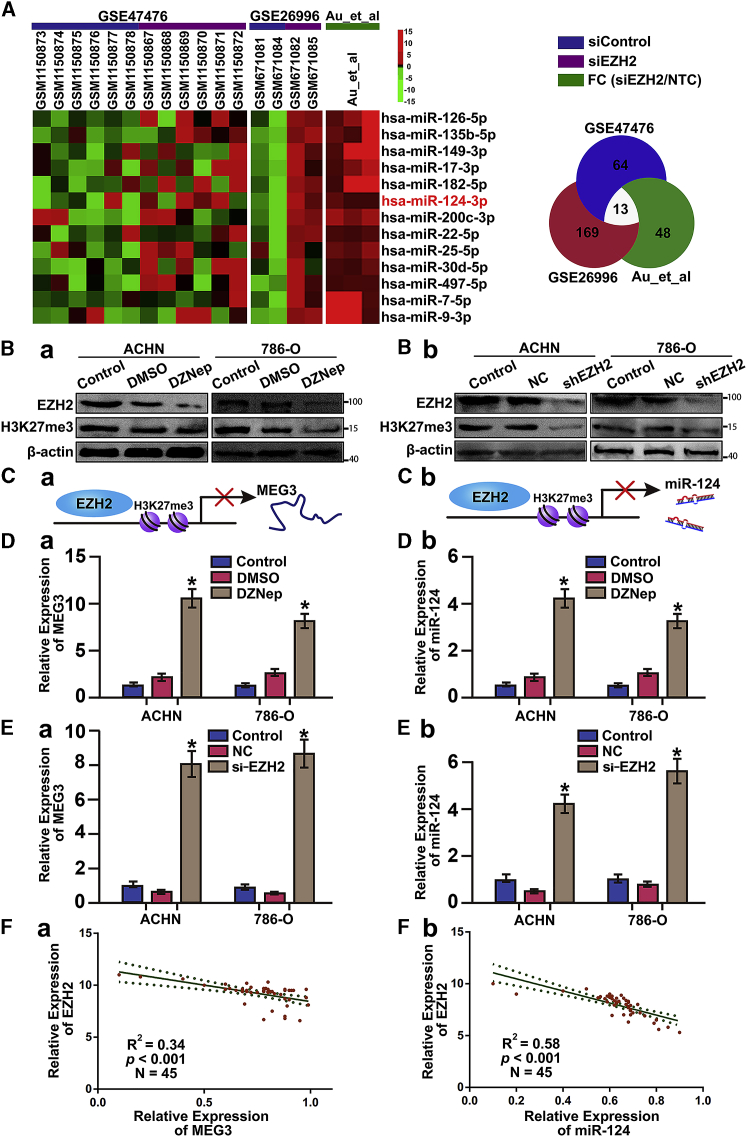
EZH2, a histone-lysine N-methyltransferase enzyme, is highly expressed in many cancers and mediates transcriptional repression of target genes. We searched the database of GEO and Gene Cloud of Biotechnology Information (GCBI) for the gene expression profiles, and selected six available GEO datasets including 16 chips (GEO: GSE47476 and GSE26996) and one publication that evaluated the downstream events following EZH2 silencing (Figure 3A). Using hierarchical clustering and heatmap analysis, we identified 13 EZH2-silenced miRNAs including miR-124, whose expression was significantly restored after EZH2 abrogation (Figure 3A).
Figure 3.
EZH2-Mediated Silencing of miR-124 and MEG3
(A) Hierarchical clustering and heatmap analysis of GSE47476, GSE26996, and one publication from GEO and GCBI database; 13 EZH2-mediated silence miRNAs were identified. (B) DZNep and siRNA to knock down EZH2, which could also inhibit the trimethylation of H3K27 and reduces EZH2 expression. (C) The model of EZH2 induced the epigenetic silencing of miR-124 and MEG3 expression by H3K27me3. (D) Silencing EZH2 using DZNep induced miR-124 and MEG3 expression. (E) Silencing EZH2 using EZH2 siRNA induced miR-124 and MEG3 expression. Data were presented as mean ± SD (*p < 0.05). (F) Pearson correlation coefficient analysis of the expression of miR-124 and MEG3 both were negatively correlated with EZH2 expression in RCC tissues.
We hypothesized that EZH2 induced the epigenetic silencing of miR-124 and MEG3 expression by H3K27 trimethylation (H3K27me3) (Figure 3C). To confirm the repressive effects of EZH2 on transcription of miR-124 and MEG3, we treated RCC cells with DZNep or small interfering RNA (siRNA) for EZH2, which resulted in reduction of EZH2 expression and inhibition of H3K27me3 (Figure 3B). By qPCR, induction of miR-124 and MEG3 expression was detected (Figure 3D). Besides, miR-124 and MEG3 were significantly upregulated in EZH2 siRNA-transfected ACHN and 786-O cells when compared with siRNA control (Figure 3E).
Given that miR-124 and MEG3 were both significantly downregulated and correlated with EZH2, we further examined their expression relationship using the Pearson correlation coefficient analysis. The expression of miR-124 and MEG3 both were negatively correlated with EZH2 expression in RCC tissues (Figure 3F).
Overexpression of miR-124 Decreased Expression of TET1 by Directly Targeting the 3ʹ UTR
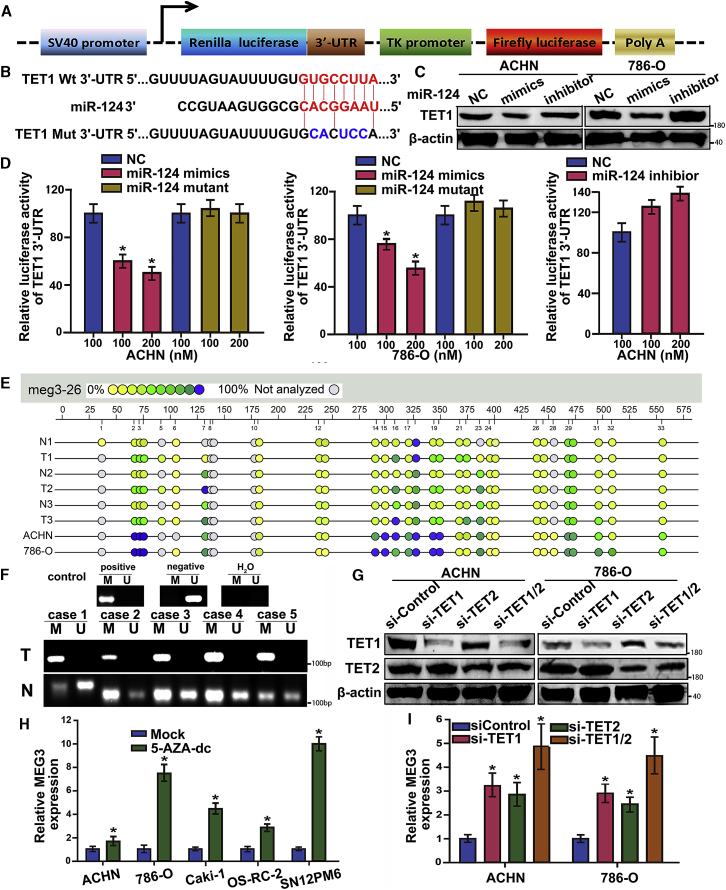
Ten-eleven translocation 1 (TET1) plays important roles in the DNA demethylation process, and bioinformatics analysis showed that TET1 contains the conserved putative miR-124 target sites in its 3′ UTR (Figure 4B). To validate whether TET1 was a bona fide target of miR-124, we constructed luciferase reporter plasmids containing either wild-type or mutated miR-124 seed sequence in the 3′ UTR of TET1 (Figure 4A). Following introduction of wild-type or mutant reporters, we found that ectopic expression of miR-124 suppressed the luciferase activity of the reporter containing wild-type 3′ UTR (Figure 4D), but not the reporter containing the mutated miR-124 binding site. In addition, we examined the effects of overexpression or silencing of miR-124 on the expression of TET1 protein by western blot. TET1 protein expression was dramatically decreased or increased in RCC cells transfected with miR-124 mimics or inhibitors, respectively (Figure 4B).
Figure 4.
miR-124 Regulates MEG3 by Directly Targeting TET1
(A) Schematic diagram of our constructed luciferase reporter plasmids. (B) The seed sequence of miR-124 matches the 3′ UTR of TET1. Mutations of miR-124 introduced into the 3′ UTR of TET1 (bottom). (C) Western blot analysis of the protein levels of TET1 in the ACHN and 786-O cells transfected with miR-124 mimics and inhibitors. β-Actin served as internal control. (D) Luciferase assay. Luciferase reporters harboring putative target sites in the 3′ UTRs of TET1 were co-transfected with small RNA molecules in ACHN and 786-O cells. Relative luciferase activity was plotted as the mean ± SEM of three independent experiments. *p < 0.05. (E) Aberrant methylation in MEG3 promoter determined by Sequenom MassARRAY analysis in three matched RCC and adjacent normal tissues, as well as two cell lines ACHN and 786-O. Epigram shows profile of unit-specific methylation of CpG sites in the MEG3 promoter region; open and filled circles denote unmethylated and methylated CpG sites, respectively. (F) Using methylation-specific PCR, average methylation levels of each CpG unit were measured in five matched cancer-adjacent normal renal tissue and renal cancer tissue. (G) Western blot analysis of the protein levels of TET1 in the ACHN and 786-O cells transfected with shRNA. β-Actin served as an internal control. (H) qRT-PCR demonstrated MEG3 transcript in five renal cancer cell lines after treatment with 5-aza-2-deoxycytidine compared with mock-treated cells. (I) Using qRT-PCR, we analyzed the MEG3 level after transfection with siRNA targeting TET1/2 in the ACHN and 786-O cells.
MEG3 Expression Was Epigenetically Regulated by TET1
Methylation within the promoter region has been associated with downregulation of gene expression, whereas bioinformatics analysis revealed the promoter region of MEG3 contained typical CpG islands. To explore the epigenetic mechanism for the downregulation of MEG3 expression in RCC, we examined MEG3 promoter methylation levels by Sequenom MassARRAY. Compared with normal tissues, MassARRAY assay revealed higher levels of methylation at multiple tested sites in three RCC tumor tissues and two RCC cell lines (Figure 4E). Furthermore, we evaluated promoter methylation status of MEG3 in RCC tissues by performing methylation-specific PCR, and found that the MEG3 promoter was hypermethylated in five RCC tissues (Figure 4F). To examine the role of aberrant methylation in the loss of MEG3 expression, we further evaluated the effect of the demethylation agent 5-Aza-2-deoxycytidine (5-Aza-DC) on MEG3 expression. The findings demonstrated that MEG3 expression increased after incubation with 5-Aza-DC in both ACHN and 786-O cells (Figure 4H). To further elucidate the methylation-dependent mechanism of MEG3 alteration, we silenced TET1 in ACHN and 786-O via RNAi, which dramatically increased the levels of MEG3 expression (Figure 4I).
Activation of p53 by MEG3 in RCC
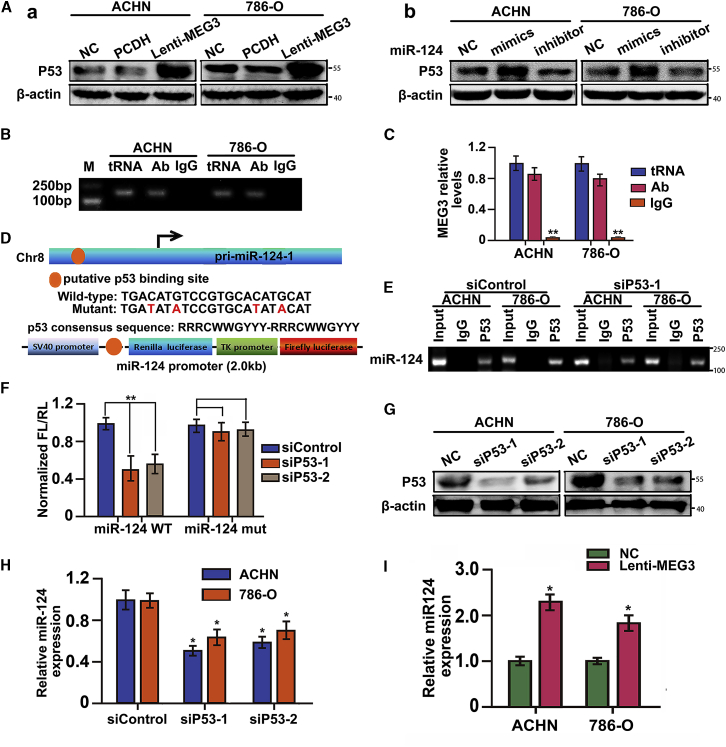
Apart from being a regulator, p53 also functions as a downstream effector of some molecules such as MEG3. To understand the molecular mechanism by which MEG3 inhibited RCC cell proliferation and EMT, we examined whether MEG3 could affect p53 expression and function. Thus, we transfected either the overexpression constructs for MEG3 or miR-124 mimics or inhibitor into RCC cells. Western blot found that cells transfected with MEG3 or miR-124 mimics both had significant increases in p53 protein levels (Figure 5A). To explore whether the induction of p53 in MEG3-transfected cells was functionally active or inactive, we performed RNA immunoprecipitation (RIP) with an antibody against p53 from cellular extracts. We found significant enrichment of MEG3 with the p53 antibody (Figures 5B and 5C) compared with the immunoglobulin G (IgG) control antibody. These data indicated that the p53 protein accumulated by transfection of MEG3 was functionally active.
Figure 5.
miR-124 Regulation by MEG3 via p53
(A) Western blot analysis of the protein levels of p53 in the ACHN and 786-O cells transfected with lenti-MEG3 or miR-124 mimics and inhibitors. β-Actin served as an internal control. (B) RIP was performed using the p53 antibody to immunoprecipitate MEG3 and a primer to detect MEG3 (left, ACHN cells; right, 786-O cells). Ab, p53 antibody; IgG, an antibody control; M, Marker; tRNA, total RNA, as positive control. (C) MEG3 relative level was measured after the p53 antibody immunoprecipitating MEG3. **p < 0.01. (D) A schematic of the luciferase vectors with miR-124 promoter fragments and sequence information for the p53 binding site mutations. Sequence alignment of predicated putative p53 binding site in the promoter of miR-124. Schematic diagram of our constructed miR-124 luciferase reporter plasmids. (E) ChIP assay were performed using the p53 antibody to immunoprecipitate the promoters of miR-124 promoters in wild-type p53 or p53 knockdown cells. (F) Luciferase assay using the DNA sequence of miR-124 promoters with wild-type (WT) or mutant (mut) p53-binding sites under p53 knockdown. (G) Inhibition of p53 protein expression by siRNAs to p53 (sip53-1 and sip53-2). ACHN and 786-O cells were transfected with sip53 or siRNA control as indicated. After 48 h, p53 and internal control β-actin were detected by western blot. (H) Expression levels of miR-124 in ACHN and 786-O cells harboring wild-type p53, and p53 knockdown using siRNA. (I) Expression levels of miR-124 in ACHN and 786-O cells after transfection with lenti-MEG3.
p53 Upregulates the Expression of the miR-124
Bioinformatics analysis indicated that the promoter of miR-124 clusters contains p53 binding sites (Figure 5D); then we sought to study the transcriptional regulatory effects of p53 on miR-124. Using siRNA, p53 was knocked down both in ACHN and 786-O cells, as proved by western blot (Figure 5G). We further analyzed direct interaction of p53 and p53-binding sequences on miR-124 promoter by chromatin immunoprecipitation (ChIP). ChIP analysis revealed that p53 directly bonded to the predicted p53-binding sequences on miR-124 promoter (Figure 5E). Besides, we also employed a luciferase reporter assay to verify whether the p53 binding sites in the miR-124 promoter could modulate miR-124 transcription. We found that p53 knockdown significantly reduced the transcriptional activity of the miR-124 promoter with wild-type p53 binding sites. However, this reduction of miR-124 transcriptional activity after p53 knockdown was abrogated in miR-124 luciferase reporter with mutant binding sites (Figure 5F), which further implied that p53 affected miR-124 expression via transcriptional regulation. As shown by real-time RT-PCR, miR-124 was downregulated via transfection with p53 siRNAs (Figure 5H). Furthermore, we detected the transactivation of miR-124 by the MEG3 using qRT-PCR (Figure 5I). Taken together, these results indicated that MEG3 was a positive transcriptional regulator of the miR-124 in RCC cells, probably through the MEG3-p53-miR-124 axis.
miR-124 and MEG3 Inhibit Cell Proliferation and EMT by Suppressing PTPN11 Expression
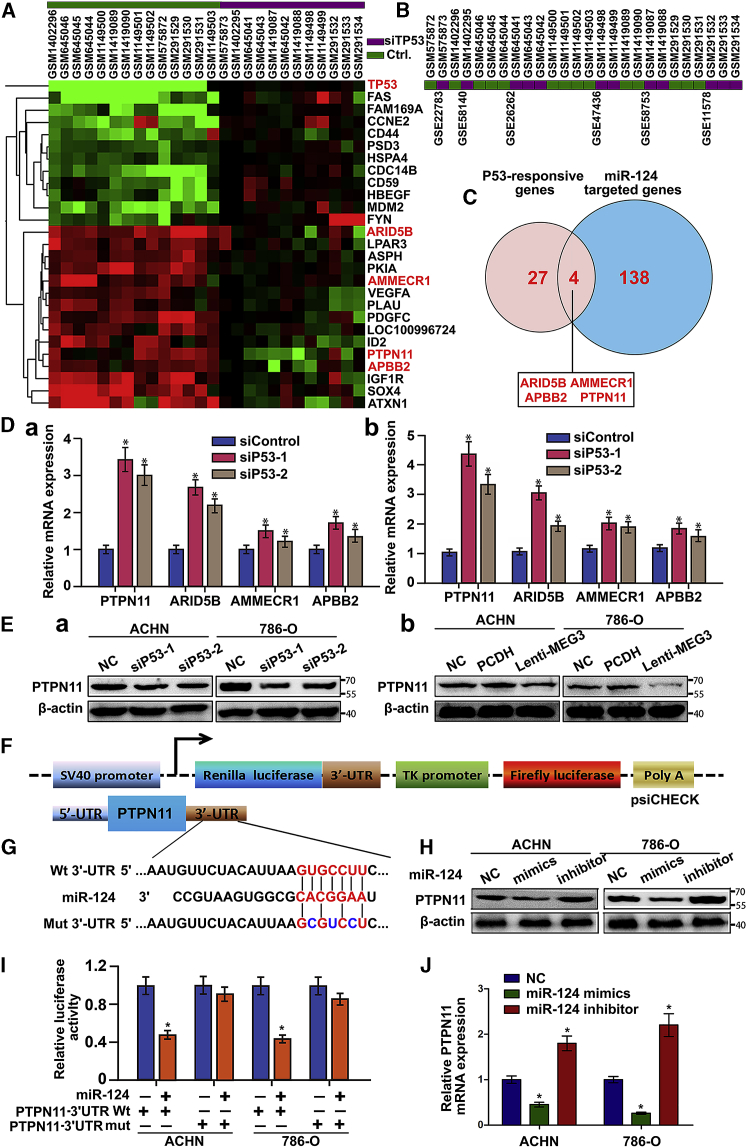
Because three crosstalks between miR-124 and MEG3 have been established, we further explored how miR-124 and MEG3 could inhibit cell proliferation and EMT. We searched the database of GEO and GCBI for the gene expression profiles and then selected six available GEO datasets that compared the two groups of si-p53 and si-Control (Figure 6B). Making use of hierarchical clustering and heatmap analysis, we identified 27 p53-responsive genes, of which 15 genes were significantly upregulated after p53 siRNA transfection (Figure 6A). Bioinformatics analysis of miR-124-targeted genes showed that the 3′ UTR of 138 genes had the predictive binding sites. To identify common downstream pathways of miR-124 and MEG3, we further made a combined analysis of p53-responsive genes and miR-124-targeted genes, which picked out four eligible genes (ARID5B, AMMECR1, APBB2, and PTPN11) to mediate the tumor-suppressive functions of both miR-124 and MEG3 (Figure 6C). To validate the microarray results, we silenced p53 with siRNAs, which resulted in significant upregulation of PTPN11 mRNA (Figure 6D) and protein expression (Figure 6Ea). Besides, western blot also proved that PTPN11 protein was downregulated by overexpression of MEG3 (Figure 6Eb).
Figure 6.
miR-124 and MEG3 Inhibit Cell Proliferation and Metastasis by Suppressing PTPN11 Expression
(A) Hierarchical clustering and heatmap analysis of p53-responsive genes after si-p53 was transfected. (B) Color bars showed the database of GEO and GCBI we searched for the gene expression profiles and selected six available GEO (GSE22783, GSE58140, GSE26262, GSE47436, GSE58763, and GSE11578) datasets including 26 chips, which compared the two groups of si-p53 and si-Control. (C) Schematic diagram showed a combined analysis of si-p53-responsive genes and miR-124-targeted genes. (D) Using qRT-PCR, we found that the expressions of ARID5B, AMMECR1, APBB2, and PTPN11 were significantly increased when we knocked down p53 with siRNA in ACHN (Da) and 786-O (Db) cells. Data were presented as mean ± SD (*p < 0.05). (E) Western blot analysis of the protein levels of PTPN11 in the ACHN and 786-O cells transfected with p53 siRNAs (Ea) or lenti-MEG3 (Eb). β-Actin served as an internal control. (F) Schematic diagram of our constructed luciferase reporter plasmids. (G) The seed sequence of miR-124 matches the 3′ UTR of PTPN11. Mutations sequence in the 3′ UTR of PTPN11 (bottom). (H) Western blot analysis of the protein levels of PTPN11 in RCC cells transfected with miR-124 mimics and inhibitors. β-Actin served as internal control. (I) Luciferase reporters harboring putative target sites in the 3′ UTR of PTPN11 were co-transfected with indicated small RNA molecules in ACHN and 786-O cells. Relative luciferase activity was plotted as the mean ± SEM of three independent experiments. *p < 0.05. (J) Relative expression of PTPN11 was measured by qRT-PCR in RCC cells after transfection with miR-124 mimics or inhibitors.
Next, sequence bioinformatics analyses revealed predicated putative miR-124 binding sites in the 3′ UTR of PTPN11 mRNA (Figure 6G). Therefore, we constructed the psiCHECK-2 plasmid to contain either the wild-type or mutated miR-124 binding sequences in the 3′ UTR of PTPN11 (Figure 6F), and performed the luciferase reporter assays. Overexpression of miR-124 decreased the relative luciferase activity of reporter harboring wild-type PTPN11 3′ UTR, whereas this inhibitory effect turned out to be abrogated with luciferase reporter containing mutant 3′ UTR (Figure 6I). Consistently, western blot showed that protein expression of PTPN11 was strikingly decreased or increased by transfecting miR-124 mimics or inhibitor, respectively (Figure 6H). Moreover, ectopic expression of PTPN11 abrogated the inhibitory effects of MEG3 or miR-124 on RCC cell proliferation, migration, and invasion (Figures S7 and S8). Together, these results indicated that PTPN11 was the direct target of miR-124, as well as the MEG3- and p53-regulated genes in RCC cells.
Discussion
In the present study, through searching the KIRC TCGA miRNA profiling and the combined analysis of RCC lncRNA expression and methylation profiles, we initially identified that two ncRNAs, miR-124 and MEG3, were significantly downregulated, and simultaneously MEG3 was hypermethylated. Validation in our samples and cell lines also validated their downregulation, positive correlation, as well as prognostic values for OS. Given that overexpression of miR-124 and MEG3 in RCC cells could suppress cell proliferation, invasion, migration, and tumor growth, we demonstrated that miR-124 and MEG3 could function as potential tumor suppressors in RCC. To elucidate the interactive regulation of these two ncRNAs, we proposed three crosstalks between miR-124 and MEG3, which may provide a plausible link between the positive correlation of miR-124 and MEG3. First, EZH2 mediated epigenetic silencing of miR-124 and MEG3. Second, miR-124 induced MEG3 expression by directly targeting the epigenetic regulator TET1. Third, MEG3 upregulated miR-124 via p53 promoter activation. Following the establishment of three crosstalks between miR-124 and MEG3, we further probed into the common downstream pathway of miR-124 and MEG3. Therefore, we conducted combined analysis of p53-responsive genes and miR-124-targeted genes, identifying PTPN11 as a crucial gene that might mediate the tumor-suppressive functions of miR-124 and MEG3. Finally, we proposed a signaling model for the crosstalk between miR-124 and MEG3, which regulated cell proliferation and EMT of RCC through PTPN11 (Figure S9).
To begin with, we conducted a comprehensive combined analysis of miRNA and lncRNA expression in RCC patient samples. We found that miR-124 and MEG3 were two of the most significantly reduced ncRNAs in RCC. Their expression levels were significantly correlated with TNM stages, grades, tumor size, and metastasis. Moreover, patients with lower levels of miR-124 or MEG3 expression had significantly poorer prognosis. For miR-124, our data were similar to the findings in other cancers,5, 6, 7, 8, 9, 10 in which miR-124 expression was found to be reduced. Besides, our survival analyses showed that miR-124 expression was an independent prognostic factor for OS in RCC, and our logistic regression analysis discovered significant correlation between its expression and TNM stage, tumor grade, and lymph node metastasis. These results were consistent with the findings in other tumor types.8, 9, 10 As an imprinted gene, MEG3 has been found to be decreased in various types of malignancies; it has also been recognized as a reliable biomarker to predict the survival in patients with various cancers.11 Our study strengthened these conclusions by proving that MEG3 was downregulated in RCC by hypermethylation, and it possessed the potential for survival prediction. These results indicated that miR-124 and MEG3 might serve as prognostic biomarkers in RCC.
Our in vitro and in vivo experiments showed that both miR-124 and MEG3 suppressed cell proliferation, migration, invasion, and tumor growth, implying their potential roles of tumor suppressors. Consistent with our results, increasing evidence has demonstrated miR-124 was a typical tumor suppressor. Wang et al.5 reported that decreased miR-124 expression was highly methylated and suppressed pancreatic cancer progression and metastasis by targeting Rac1. Taniguchi et al.10 found that the miR-124 inhibited colorectal cancer growth through modulating PTB1/PKM1/PKM2 feedback cascade. Previous studies also supported our data that ectopic expression of MEG3 markedly inhibited growth of cancer cells.12 Besides, the study by Chen et al.13 also indicated that MEG3 inhibited cell proliferation and induced apoptosis, partially via the activation of the endoplasmic reticulum stress and p53 pathway. Additional data from Zhuo et al.14 further demonstrated MEG3, acting as a potential biomarker in predicting the prognosis of hepatocarcinoma, was regulated by UHRF1 via recruiting DNMT1 and regulated p53 expression. Consistent with these findings, our study provided novel evidence for the biological and clinical significance of MEG3 as a tumor suppressor by inhibiting cell proliferation and invasion for the first time, providing an additional molecular therapeutic target for RCC.
Although we found out that these two ncRNAs were significantly deregulated in RCC and could act as tumor suppressors, there are no reports that show any associations between them presently. To explore possible interaction or reciprocal regulation of these two ncRNAs, we further found three crosstalks between miR-124 and MEG3, which may provide a plausible link between the positive correlation of miR-124 and MEG3. EZH2, a histone-lysine N-methyltransferase enzyme, can interact with other components such as EED and SUZ12 to form a PRC2 complex that contributes to transcriptional repression of target genes. We proved silencing EZH2 through DZNep or RNAi dramatically increased the levels of miR-124 and MEG3 expression. In accordance with our results, a previous study reported that inhibition of EZH2 by DZNep affected the expression of miR-124 in vivo.15 Interestingly, Xu et al.16 provided evidence that EZH2 was also a target gene of miR-124, and miR-124-EZH2 interaction could regulate the choice between neuronal and astrocyte differentiation. Interestingly, others have reported evidence that MEG3 interacts with EZH2 in embryonic stem cells.17 Currently, little is known about the mechanism by which EZH2 represses the expression of miR-124 and MEG3. We hypothesize that EZH2-mediated H3K27me3 epigenetic silence may contribute to ncRNA transcriptional repression.
Methylation within the promoter region has been associated with loss of MEG3 expression in various cancers.18 A negative correlation between MEG3 expression and DNA demethylation regulator TET1 has been reported previously, raising the possibility that MEG3 might be regulated by TET1-associated epigenetic silencing. Moreover, bioinformatics analysis showed that the TET1 3′ UTR had the predictive binding site of miR-124. Therefore, we hypothesized that reduced miR-124 expression could potentially mediate MEG3 silencing through the modulation of TET1 activity. We next undertook to examine the impact of interaction of miR-124 and TET1 on MEG3 expression in RCC cells. Previous studies had found that miR-29a could regulate the expression of MEG3 through the modulation of DNMT3B activity in hepatocellular cancer.18 Yan et al.19 found that the suppression of miR-148a might contribute to the downregulation of MEG3 in gastric cancer by modulation of DNMT1. Interestingly, others have reported some miRNAs directly targeted TET1, including miR-22, miR-191, and miR-26.20, 21, 22 Moreover, some lncRNAs were also reported to be downregulated by TET-dependent promoter hypermethylation, including MEG3, POU3F3, HOXD-AS1, and ANRIL.23, 24, 25, 26 Our results indicated that miR-124 reduced the expression of its direct target TET1, which was involved in epigenetic regulation of MEG3 expression. Therefore, we expanded the regulatory network comprising TETs, miRNAs, and hypermethylated lncRNAs by establishing a novel interactive paradigm. These findings enriched our understanding of how miRNAs interact with and regulate lncRNAs.
Previous experiments had demonstrated that p53 was a target of MEG3, and that its protein level could be significantly increased through the overexpression of MEG3 in cancer cells.27 MEG3 could also induce p53 expression via an indirect mechanism, because MEG3 could suppress MDM2 expression and attenuate the inhibitory effect of MDM2 on p53.27 Our present study found that re-expression of MEG3 directly induced p53 accumulation through direct binding with p53. As a tumor suppressor, p53 plays pivotal roles in tumor suppression and mediates the functions of many other tumor suppressors. We proposed the tumor-suppressive effects of MEG3 on RCC might be partly mediated by activation of p53. Liu et al. proposed that miR-124 was differentially regulated by wild-type or mutant p53 under photodynamic therapy in colorectal cancer, which implied that miR-124 was a plausible p53-targeted miRNA.28 As the p53 transcriptional targets, these miRNAs have been implicated in targeting genes that are involved in various p53 signaling pathways, contributing to critical and extensive function of p53. Kidney cancer TCGA data showed that miR-124 expression was positively correlated with p53. Furthermore, knocking down p53 with siRNA could silence miR-124 expression, whereas ectopic expression of MEG3 activated p53 and stimulated miR-124 transcription activation. Therefore, as a p53-responsive miRNA, miR-124 could be induced by MEG3 via p53 accumulation.
Recent studies demonstrate that crosstalk among competitive endogenous RNAs (ceRNAs) and miRNAs represents a novel layer of gene regulation and plays important roles in cancer progression. Here we further reviewed recent findings regarding the critical role of ncRNAs and created the miRNA-lncRNA regulatory networks. First, for the miRNAs and lncRNAs with opposite expression pattern, ceRNA theory and miRNAs-translation factors (TFs)-lncRNAs model are mostly investigated to explain their relationship. However, when they are consistently expressed, especially jointly downregulated, EZH2-mediated epigenetic silencing, miRNAs-DNMTs-lncRNAs and lncRNAs-TFs-miRNAs may represent as the potential underlying mechanisms. In the present study, miR-124 and MEG3 both significantly silenced in RCC, and their expression was positively correlated. We further identified three crosstalks between miR-124 and MEG3; these underlying mechanisms provided valuable examples to facilitate our understanding of ncRNA-regulatory networks in tumors. These findings also provided insights into how miRNAs interact with and regulate lncRNAs.
Since three crosstalks between miR-124 and MEG3 have been established, we further identified how miR-124 and MEG3 inhibited cell proliferation and EMT. To find the common downstream pathway of miR-124 and MEG3, we made a combined analysis of si-p53-responsive genes and miR-124-targeted genes, screening out four promising genes (ARID5B, AMMECR1, APBB21, and PTPN11). Therefore, we hypothesized that these four genes may mediate the tumor-suppressive function of miR-124 and MEG3. Subsequent experiments indicated that PTPN11 was the direct target of miR-124, as well as the MEG3- and p53-regulated gene in RCC cells. Although PTPN11 mutation was initially discovered to be involved in the development of Noonan syndrome, it has become apparent that PTPN11 is also involved in cell proliferation, invasion, migration, and tumor growth.29 Consistent with our results, others reported that miR-489 and miR-1213 could directly target the 3′ UTR of PTPN11, resulting in inhibition of tumor progression.30, 31 Thus, it may be reasonable that MEG3 and miR-124 exerted their tumor-suppressive functions through downregulating PTPN11.
In summary, we detected the downregulation pattern of miR-124 and MEG3 in RCC, and confirmed their tumor-suppressive effects in vitro and in vivo. Our present study highlights the interplays between these two classes of ncRNAs, miRNAs and lncRNAs, and provides three crosstalks of interaction between miR-124 and MEG3. We also identified that PTPN11 was the direct target of miR-124, as well as the MEG3- and p53-regulated gene, and could mediate their tumor-suppressive functions. This study extends our knowledge of miR-124 and MEG3 in the pathology and progression of RCC, and emphasizes the importance of ncRNAs such as miR-124 and MEG3 as crucial regulators in tumor biology.
Materials and Methods
RCC miRNA and lncRNA Expression Profilings
Gene expression and miRNA-seq data (level 3) of KIRC were downloaded from TCGA (https://tcga-data.nci.nih.gov/docs/publications/). lncRNA expression profiling was obtained from our previous study in which we compared six pairs of RCC and corresponding nontumorous tissue. lncRNA promoter methylation MeDIP-chip analysis (Ouyi, Shanghai, China) was also performed on three paired RCC and adjacent normal tissues to identify differentially methylated lncRNAs.
Cell Culture and Transfection
Human renal cancer cell lines (ACHN, 786-O, SN12PM6), normal renal epithelial cell line (HK-2), and human 293T cells were maintained in DMEM (high-glucose) containing 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere of 5% CO2, whereas Caki-1 and OS-RC-2 were cultured in RPMI-1640 medium. Indicated overexpression plasmids were transiently transfected into the cells using FuGENE HD transfection reagent (Roche, Switzerland), with empty vector as control. miR-124 mimics and inhibitor, siRNAs of specific genes, and NC were respectively transfected into cells by X-tremeGENE Transfection Reagent (Roche, Switzerland).
Colony Formation Assay
Exponentially growing treated ACHN and 786-O cells were digested and then seeded at 1,000 cells per well in six-well plates. The cells continued to be cultured for 2 weeks, and culture medium was changed every 3 days. After staining cells with 0.5% crystal violet solution for 20 min, the numbers of cell colonies were calculated and analyzed.
In Vitro Migration and Invasion Assay
About 1 × 105 ACHN cells or 1 × 104 786-O cells were suspended and plated in the upper chambers of 24-well Transwell plates (Corning) in FBS-free medium. Complete medium (10% FBS) was deposited in the lower chambers to serve as a chemo-attractant. After 12 h for cells to pass through the membrane, cells remaining on the upper filter were removed, whereas cells that passed through the Transwell filter were stained by 0.5% crystal violet for 20 min. Images were taken of five random optical fields (200×) on each filter, and cell numbers were then counted.
To evaluate cell invasion, we coated Transwell membranes with Matrigel (BD Biosciences) prior to plating indicated cells. After 24 h for ACHN and 12 h for 786-O, crystal violet staining and cell counting were performed as above.
Animal Experiments
A total of 5 × 106 treated ACHN cells were injected subcutaneously into the right flank of 4-week-old BALB/c nude mice, tumor growth of mice was monitored every 3 days, and mice were sacrificed 6 weeks after inoculation. Tumor volume was calculated using the formula, 0.5ab2, where “a” and “b” meant long diameter and short diameter, respectively. All animal experiments were approved by the Committee of Animal Experimental Ethics, Huazhong University of Science and Technology.
Methylation Status Analysis and Demethylation Assay
Genomic DNA was isolated with QIAamp DNA Mini Kit (QIAGEN), and bisulfite modification of the genomic DNA was carried out using an EpiTect Bisulfite Kit (QIAGEN) according to the manufacturer’s instructions. Quantitative methylation analysis of the promoter of MEG3 coding gene was performed by the Sequenom MassARRAY platform (Ouyi, Shanghai, China) according to the manufacturer’s protocol. To determine whether MEG3 expression can be induced after demethylation, we treated the five RCC cell lines with 10 μM DNA demethylation drug 5-Aza-DC. The methylation-specific PCR primers were listed in Table S1.
Dual-Luciferase Reporter Assay
3′ UTRs of target gene with the wild-type or mutated binding site of hsa-miR-124 were inserted into psi-CHECK2 luciferase reporter vector and validated by sequencing. ACHN and 786-O cells were seeded in a 24-well plate until 60%–80% confluence and then were co-transfected with wild-type or mutated constructs and miR-124 mimics or control. The dual-luciferase activities were measured, whereas Renilla luciferase signal was normalized to the firefly luciferase signal.
RIP and Chromatin Immunoprecipitation Assay
To validate the association of MEG3 and p53, we performed RIP using Magna RIP Kit (Millipore) according to the manufacturer’s instructions. To determine whether p53 could directly activate the miR-124 promoter, chromatin immunoprecipitation was conducted through the EZ-Magna ChIP kit (Millipore). All procedures were performed following the manufacturer’s instructions. All used primers were listed in Table S1.
Statistical Analysis
The data were presented as the mean ± SD. Differences among groups were determined by a two-way ANOVA followed by a post hoc Tukey’s test. Comparisons between two groups were performed with an unpaired Student’s t test. Survival curve was plotted using the Kaplan-Meier method and compared by the log rank test. A value of p < 0.05 was considered significant.
Author Contributions
H.Z. and K.T. contributed equally to the work. H.Z., K.T., and H.X. designed the study. K.T., H.Z., H. Liu, and L.Y. performed cellular function experiments and provided the clinical samples analysis. K.T., J.Z., K.C., and H. Li did the data analysis. J.H. and W.G. contributed to language editing. K.T. drafted the manuscript. K.T., H.Z., and H. Liu prepared all of the tables and figures. H.Z. and Z.Y. conceived the study. The final manuscript was reviewed and approved by all of the authors.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (grants 81670645, 81602236, and 81874089), the National Major Scientific and Technological Special Project for Significant New Drugs Development (2012ZX09303018), the Chenguang Program of Wuhan Science and Technology Bureau (grant 2015070404010199), and the Graduates’ Innovation Fund, Huazhong University of Science and Technology (grant 5003540056).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.04.005.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.MacLennan S., Imamura M., Lapitan M.C., Omar M.I., Lam T.B.L., Hilvano-Cabungcal A.M., Royle P., Stewart F., MacLennan G., MacLennan S.J., UCAN Systematic Review Reference Group. EAU Renal Cancer Guideline Panel Systematic review of perioperative and quality-of-life outcomes following surgical management of localised renal cancer. Eur. Urol. 2012;62:1097–1117. doi: 10.1016/j.eururo.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 3.Tang K., Xu H. Prognostic value of meta-signature miRNAs in renal cell carcinoma: an integrated miRNA expression profiling analysis. Sci. Rep. 2015;5:10272. doi: 10.1038/srep10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu G., Yao W., Wang J., Ma X., Xiao W., Li H., Xia D., Yang Y., Deng K., Xiao H. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS ONE. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P., Chen L., Zhang J., Chen H., Fan J., Wang K., Luo J., Chen Z., Meng Z., Liu L. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33:514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 6.Shi X.B., Ma A.H., Xue L., Li M., Nguyen H.G., Yang J.C., Tepper C.G., Gandour-Edwards R., Evans C.P., Kung H.J., deVere White R.W. miR-124 and Androgen Receptor Signaling Inhibitors Repress Prostate Cancer Growth by Downregulating Androgen Receptor Splice Variants, EZH2, and Src. Cancer Res. 2015;75:5309–5317. doi: 10.1158/0008-5472.CAN-14-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei J., Wang F., Kong L.Y., Xu S., Doucette T., Ferguson S.D., Yang Y., McEnery K., Jethwa K., Gjyshi O. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73:3913–3926. doi: 10.1158/0008-5472.CAN-12-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y.X., Wang L., Liu W.J., Zhang H.T., Xue J.H., Zhang Z.W., Gao C.J. MiR-124-3p/B4GALT1 axis plays an important role in SOCS3-regulated growth and chemo-sensitivity of CML. J. Hematol. Oncol. 2016;9:69. doi: 10.1186/s13045-016-0300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zu L., Xue Y., Wang J., Fu Y., Wang X., Xiao G., Hao M., Sun X., Wang Y., Fu G., Wang J. The feedback loop between miR-124 and TGF-β pathway plays a significant role in non-small cell lung cancer metastasis. Carcinogenesis. 2016;37:333–343. doi: 10.1093/carcin/bgw011. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., Ito Y., Otsuki Y., Uno B., Uchiyama K., Akao Y. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Lyu Y., Lou J., Yang Y., Feng J., Hao Y., Huang S., Yin L., Xu J., Huang D., Ma B. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017;31:2543–2551. doi: 10.1038/leu.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C., Huang C., Wang J., Huang H., Li J., Xie Q., Liu Y., Zhu J., Li Y., Zhang D. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1α translation. Oncogene. 2017;36:3878–3889. doi: 10.1038/onc.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R.P., Huang Z.L., Liu L.X., Xiang M.Q., Li G.P., Feng J.L., Liu B., Wu L.F. Involvement of endoplasmic reticulum stress and p53 in lncRNA MEG3-induced human hepatoma HepG2 cell apoptosis. Oncol. Rep. 2016;36:1649–1657. doi: 10.3892/or.2016.4919. [DOI] [PubMed] [Google Scholar]
- 14.Zhuo H., Tang J., Lin Z., Jiang R., Zhang X., Ji J., Wang P., Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol. Carcinog. 2016;55:209–219. doi: 10.1002/mc.22270. [DOI] [PubMed] [Google Scholar]
- 15.Clarke M.A., Luhn P., Gage J.C., Bodelon C., Dunn S.T., Walker J., Zuna R., Hewitt S., Killian J.K., Yan L. Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int. J. Cancer. 2017;141:701–710. doi: 10.1002/ijc.30781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L., Dai W., Li J., He L., Wang F., Xia Y., Chen K., Li S., Liu T., Lu J. Methylation-regulated miR-124-1 suppresses tumorigenesis in hepatocellular carcinoma by targeting CASC3. Oncotarget. 2016;7:26027–26041. doi: 10.18632/oncotarget.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braconi C., Kogure T., Valeri N., Huang N., Nuovo G., Costinean S., Negrini M., Miotto E., Croce C.M., Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J., Guo X., Xia J., Shan T., Gu C., Liang Z., Zhao W., Jin S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med. Oncol. 2014;31:879. doi: 10.1007/s12032-014-0879-6. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X., Hu C., Arnovitz S., Bugno J., Yu M., Zuo Z., Chen P., Huang H., Ulrich B., Gurbuxani S. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016;7:11452. doi: 10.1038/ncomms11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Zhou Z.Q., Yang Z.R., Tong D.N., Guan J., Shi B.J., Nie J., Ding X.T., Li B., Zhou G.W., Zhang Z.Y. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66:136–151. doi: 10.1002/hep.29116. [DOI] [PubMed] [Google Scholar]
- 22.Huang H., Jiang X., Wang J., Li Y., Song C.X., Chen P., Li S., Gurbuxani S., Arnovitz S., Wang Y. Identification of MLL-fusion/MYC┤miR-26┤TET1 signaling circuit in MLL-rearranged leukemia. Cancer Lett. 2016;372:157–165. doi: 10.1016/j.canlet.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao H., Duan M., Lin L., Wu C., Fu X., Wang H., Guo L., Chen W., Huang L., Liu D. TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget. 2017;8:18337–18347. doi: 10.18632/oncotarget.15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Zheng J., Deng J., You Y., Wu H., Li N., Lu J., Zhou Y. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714–1726.e5. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Zhuang C., Liu Y., Chen M., Chen Y., Chen Z., He A., Lin J., Zhan Y., Liu L. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J. Exp. Clin. Cancer Res. 2016;35:99. doi: 10.1186/s13046-016-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng W., Wang J., Zhang J., Cai J., Bai Z., Zhang Z. TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life. 2016;68:355–364. doi: 10.1002/iub.1490. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 28.Liu K., Chen W., Lei S., Xiong L., Zhao H., Liang D., Lei Z., Zhou N., Yao H., Liang Y. Wild-type and mutant p53 differentially modulate miR-124/iASPP feedback following photodynamic therapy in human colon cancer cell line. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.477. e3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Zhang F., Niu R. Functions of Shp2 in cancer. J. Cell. Mol. Med. 2015;19:2075–2083. doi: 10.1111/jcmm.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikkawa N., Hanazawa T., Fujimura L., Nohata N., Suzuki H., Chazono H., Sakurai D., Horiguchi S., Okamoto Y., Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br. J. Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J., Huang X., Tan W., Yu D., Du Z., Chang J., Wei L., Han Y., Wang C., Che X. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 2016;48:747–757. doi: 10.1038/ng.3568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.