Abstract
A docking procedure is described that allows the transmembrane surface of a G protein-coupled receptor (GPCR) to be swept rapidly for potential binding sites for cholesterol at the bilayer interfaces on the two sides of the membrane. The procedure matches 89% of the cholesterols resolved in published GPCR crystal structures, when cholesterols likely to be crystal packing artifacts are excluded. Docking poses are shown to form distinct clusters on the protein surface, the clusters corresponding to “greasy hollows” between protein ridges. Docking poses depend on the angle of tilt of the GPCR in the surrounding lipid bilayer. It is suggested that thermal motion could alter the optimal binding pose for a cholesterol molecule, with the range of binding poses within a cluster providing a guide to the range of thermal motions likely for a cholesterol within a binding site.
Introduction
G protein-coupled receptors (GPCRs) operate in membranes rich in cholesterol, the cholesterol content affecting the function of many GPCRs (1). These effects of cholesterol and other lipids could follow from changes in the mechanical properties of the membrane or from direct binding to specific sites on the protein (1, 2, 3, 4, 5, 6, 7, 8, 9). Arguing for the importance of direct binding is the observation that phospholipid structure affects GPCR function even in nanodisks or detergent micelles in which no bulk lipid bilayer is present (6, 10).
Most cholesterol molecules, when in a lipid bilayer, orient with their −OH groups close to the glycerol backbone region, with their hydrophobic rings and chains in the hydrophobic interior, the long axis of the molecule being more or less parallel to the bilayer normal (5, 11). These cholesterols will be referred to as interfacial cholesterols to distinguish them from the small proportion of cholesterols buried deep within the bilayer (12). Molecular dynamics (MD) simulations suggest that interfacial cholesterols interact extensively with the transmembrane (TM) surfaces of GPCRs, adopting an orientation much like that in a simple lipid bilayer (13, 14, 15, 16, 17, 18). As well as the large numbers of cholesterols interacting nonspecifically with the TM surface, these studies report a few hot-spots in which the probability of cholesterol contact is high, although even here, cholesterols are free to rotate about their long axes (17), suggesting that the sites are not deep energy wells into which a cholesterol falls but rather are energetically shallow sites able to accommodate cholesterols in a range of poses (19). A combined NMR and denaturation study on cholesterol binding to the β2 adrenergic receptor (β2AR) is also consistent with the presence of a small number of high-affinity sites together with a number of lower affinity sites (20).
The picture derived from x-ray crystallographic studies is rather different. The PDB database contained, at the time this manuscript was prepared, 51 GPCR structures of high resolution (3.5 Å or better) containing resolved cholesterols. The Orientations of Proteins in Membranes (OPM) database provides predicted locations for the lipid bilayer around a membrane protein (21), and most of the resolved cholesterols are located with their −OH groups close to the polar-hydrophobic interfaces of the predicted bilayers, as shown for β2AR in Fig. 1. The number of resolved cholesterols per structure is small, varying between one and four (see Tables 1, S1, and S2). The exact relationship between these cholesterols and the sites predicted by MD simulation is unclear; they could correspond to hot-spots of the kind suggested by the MD simulations but could also be the result of packing constraints in the crystal. There remains a need for a simple and rapid method for identifying potential binding sites for cholesterol on GPCRs, capable of distinguishing between high-affinity binding and weak background binding.
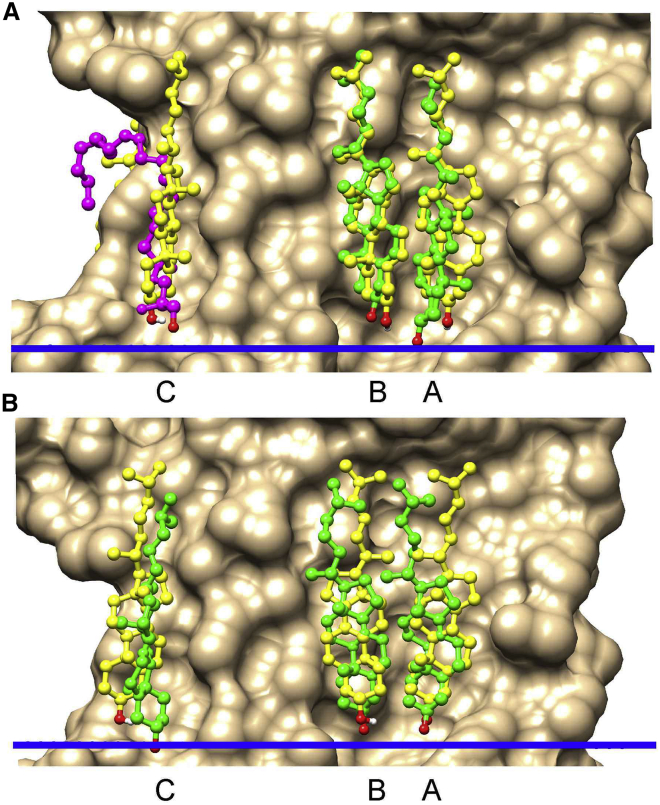
Figure 1.
Docking and crystallographic results for β2AR. (A) and (B) show surfaces on the IC side for structures 3D4S and 5D5A, respectively. Cholesterol molecules are shown in ball and stick mode: crystallographic structures, green and docked structures, yellow. Oxygen atoms are colored red. Sites are labeled as in Table 1. For the 3D4S structure, 1-oleoyl-R-glycerol (magenta) is observed at site C in the crystal structure. The IC side of the hydrophobic slab representing the bilayer core is shown by the blue bar. To see this figure in color, go online.
Table 1.
Cholesterol Binding to β2AR (IC Side)
| PDB | Space Group | Binding Sitea |
||
|---|---|---|---|---|
| A | B | C | ||
| 3d4s | P 212121 | CLR402 | CLR403 | OLC 406 |
| 2rh1 | C 1 2 1 | CLR412 | CLR414 | CLR413 |
| 3ny8 | P 212121 | CLR1201 | CLR1202 | OLC 1206 |
| 3ny9 | P 212121 | CLR1201 | CLR1202 | OLC 1204 |
| 3nya | P 212121 | CLR1201 | CLR1202 | – |
| 3pds | P 212121 | CLR1202 | – | – |
| 5d5a | C 1 2 1 | CLR1206 | CLR1208 | CLR1207 |
| 5x7d | P 212121 | CLR1204 | CLR1203b | – |
The nomenclature for bound cholesterol (CLR) molecules is that given in the respective PDB file. In some structures, as indicated, binding site C is occupied by a molecule of 1-oleoyl-R-glycerol (OLC). Entries in italics indicate that a docking pose is located at the site as indicated by an rmsd with the crystallographic CLR molecule of 4 Å or less or, in the case of a bound OLC molecule, where a docking pose overlaps with the OLC, as shown in Fig. 1 A.
A docking pose with an rmsd value of 4.3 Å with CLR1203 was observed at this site.
Attempts to define binding motifs for cholesterol on membrane proteins have, unfortunately, proved to be of limited value (22). CRAC, CARC, and CCM motifs have all been suggested, all involving a small number of apolar and aromatic residues, together with a basic residue that can hydrogen bond to the cholesterol −OH group. However, many resolved cholesterols are not bound at any of these motifs, and many examples of these motifs are not occupied by resolved cholesterols (1, 5, 22). In part, the problem arises because although hydrogen bonding of its −OH group is likely to be important in anchoring a cholesterol to the interface, analysis shows that 49% of the resolved cholesterols in GPCR structures are not hydrogen bonded to the GPCR. In a membrane, these non-hydrogen-bonded cholesterols are likely to hydrogen bond to water or to the glycerol backbones and headgroups of neighboring phospholipid molecules. Indeed, many cholesterols hydrogen bonded to a GPCR in a crystal may not be so bonded in a membrane because the surrounding lipid bilayer will provide a much larger number of potential hydrogen bond partners than a GPCR.
The usefulness of MD simulations in characterizing cholesterol binding to GPCRs is limited by the very long simulation times required to ensure system equilibration (17). Here, it is shown that the AutoDock Vina molecular docking tool (23) can be used to sweep the TM surfaces of GPCRs for potential cholesterol binding sites in just a few minutes using a conventional personal computer. The success of the docking approach depends on the creation of model interfaces on the extracellular (EC) and intracellular (IC) sides of the membrane to which a cholesterol −OH group can hydrogen bond, mimicking the hydrogen bond donors and acceptors present in the interface regions of a lipid bilayer. In a Vina docking run, a single cholesterol is docked onto the protein surface, generating a maximum of 20 docking poses. To ensure complete coverage of the surface, a set of five sequential docking runs is performed for each of the two monolayers making up the bilayer, as described in Methods. This generates a maximum of 200 docking poses, which are then processed to produce a meaningful set of potential binding sites, aiming to filter strong binding sites from weaker, background binding sites and from “false” sites. Two criteria are employed in the filtering, based on the observation that, in GPCR crystal structures, all resolved cholesterols are oriented close to the bilayer normal, and all make extensive contacts with the protein surface. The average tilt angle between the long axis of the cholesterol ring system and the normal to the EC and IC planes predicted by the OPM database is 14.4 ± 6.7° with a maximal tilt angle of 30°. The number of residues contacted by a resolved cholesterol varies from 8 to 13 with an average of 10.1 ± 1.5; these numbers exclude cholesterols at the packing interface in the A2AR crystal structure in which the number of contacts is only five (see further discussion below). The analysis also excludes cholesterol hemisuccinate molecules resolved in some structures because interaction of a hemisuccinate group with a bilayer surface will be different from that of a cholesterol −OH group. The criteria required for a binding site were therefore chosen to be a maximum tilt angle for a bound cholesterol of 30° and a minimum number of residue contacts of eight. In what follows, a pose will refer to a pose that has met these criteria, unless otherwise made clear.
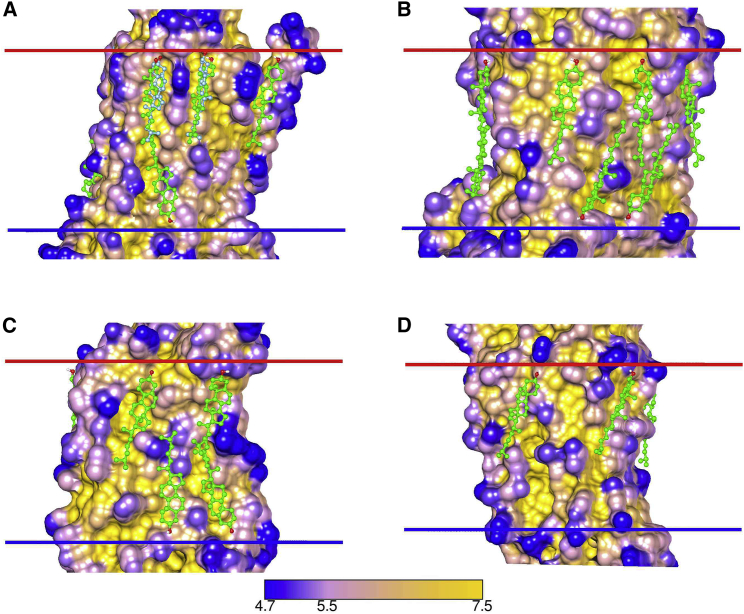
The test of a successful docking protocol is its ability to identify known, “true,” ligand binding sites, with only a small number of false positives. The 131 interfacial cholesterols resolved in GPCR structures provide a large database for testing. The approach adopted here had a success rate of 89% in identifying these cholesterols when cholesterols at packing interfaces in A2AR structures were excluded. The question of false positives is less clear. For example, docking studies with the available β2AR structures identified not only the three crystallographic binding sites (Table 1) but also an average of 3 ± 0.5 additional sites on the IC side with a further 4.9 ± 1.8 sites on the EC side (Table S7). It is not possible to say with certainty whether or not these additional sites correspond to “real” sites, a problem faced by any in silico approach. However, cholesterols were selected from the initial set of poses on the basis that they matched the characteristics of the crystallographically resolved cholesterols. Further, as shown in Fig. 2 for GPCRs of classes A, B, C, and F, all these poses are located in hollows between protein ridges, where crystallographically resolved cholesterols are also observed to bind. These “greasy hollows” are unlike the highly structurally specific binding sites characteristic of ligand (or drug) binding sites—binding of cholesterol is less structurally demanding. Last, as the cholesterol content of the membrane surrounding a GPCR is very high, even sites of low affinity will, in a membrane, be at least partly occupied by cholesterol.
Figure 2.
Surface view of GPCRs with docked cholesterol molecules for the following: (A) Class A, A2A receptor, 5IU4; (B) Class B, calcitonin-like receptor, 6E3Y; (C) Class C, mGlu5 receptor, 4OO9; and (D) Class F, smoothened receptor, 5L7D. Surfaces are colored by distance from a bulk solvent layer calculated using the Depth program (29) with distances (Å) given by the bottom scale. Docked cholesterols are colored green, and in (A), crystallographic cholesterols are colored blue. EC and IC interfaces are shown by the red and blue bars, respectively. To see this figure in color, go online.
Methods
Crystal structures for GPCRs with resolutions of 3.5 Å or better were identified on the OPM (http://opm.phar.umich.edu) and Protein Data Bank (PDB) (https://www.rcsb.org/pdb) databases. Structures were assigned to active, inactive, or active intermediate states based on the packing of the TM helices (Table S7). Docking was performed using AutoDock Vina (23) running under Chimera (24). Given the large number of GPCR structures to be searched, a protocol was designed that can run unattended through large sets of structures. Vina employs a search box to define the space around the protein to be searched. Here, the search was restricted to the TM region of the protein, and the EC and IC monolayers were searched separately. The required dimensions of the search box are the same for all monomeric GPCRs, but the coordinates defining the position of the box are not. Fortunately, the OPM database provides coordinates for membrane proteins all centered about the middle of the predicted hydrophobic slab representing the lipid bilayer (21), allowing the same search box to be used for all the structures.
The cholesterol ligand was prepared for docking with free rotation about the C-OH bond using AutoDock 4 (25) and with an H atom added to the cholesterol oxygen. Proteins were prepared using the DockPrep facility in Chimera, repairing incomplete side chains using the Dunbrack rotamer library. The AddH command was used to add hydrogens, with ionizable residues assumed to have protonation states applicable at a physiological pH, and files in the pdbqt (PDB, partial charge (q), and atom type (t)) format required by Vina were also generated using the DockPrep facility. Weighting factors for hydrogen bonds and hydrophobic effects were changed from the default values to −2.0 and −0.001, respectively, values appropriate for a membrane environment (12). Vina performs a number of individual sampling “runs,” the number being defined by the exhaustiveness parameter, each run starting from a random ligand conformation (26). In a study with flexible peptides as ligands, docking was found to have converged when the exhaustiveness parameter was set at 200 (26). Here, docking was also found to have converged with an exhaustiveness value of 200, and so this was the value adopted. Protein structure files downloaded from the OPM database contain sets of dummy atoms marking the interfaces on the two sides of the bilayer (21). These dummy atoms were converted into NH3 groups (see below) and combined with the protein coordinate pdbqt files using in-house Python code.
Docking poses obtained for individual GPCR structures were sorted into clusters using simple threshold clustering based on a root mean-square deviation (rmsd) between poses of less than 4 Å (27). Poses were rejected when a cholesterol contacted fewer than eight residues, or when the angle between the long axis of the cholesterol molecule and the bilayer normal was greater than 30°. The large search boxes used here meant that the maximum of 20 poses returned by a single Vina run was insufficient to cover all possible binding sites, and so five sequential runs were performed for each monolayer for each structure, with the poses selected from the first run (in pdbqt format) being combined with the pdbqt file used in that run to generate the pdbqt file for the second run and so on. Poses from sets of GPCR structures were clustered using the density-based spatial clustering of applications with noise method with a threshold rmsd value of 3.5 Å and a minimal number of neighbors of two (27).
Systematic residue numbers were obtained from the GPCR database (http://gpcrdb.org) (28). Residue depths from a theoretical bulk solvent layer around the protein were determined using the Depth server (http://cospi.iiserpune.ac.in/depth/htdocs/intro.html) with a solvent neighborhood radius of 3 Å and a minimal number of neighborhood solvent molecules of five (29). Residues within 4 Å of a bound cholesterol molecule were identified using protein and cholesterol molecules prepared as for docking.
A table of cholesterol dockings (Table S7) together with associated structure files in PDB format for downloading are available on the DeepCholesterol web site (https://deepcholesterol.soton.ac.uk).
Results and Discussion
A protocol for identifying interfacial cholesterol binding sites
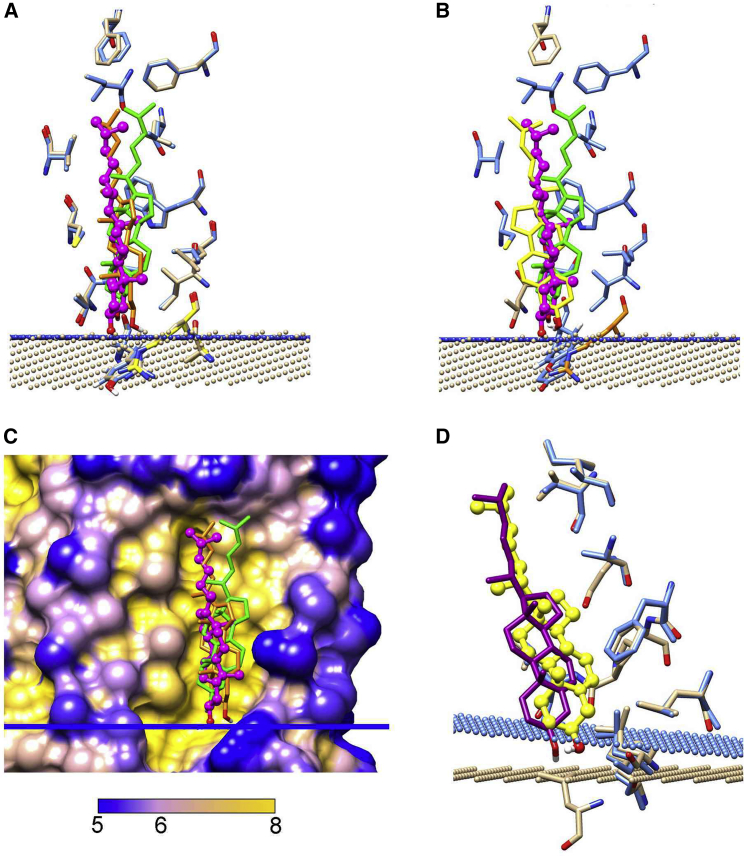
The protocol adopted for docking is illustrated in Fig. 3 for the IC side of 3D4S. The default version of Vina uses weighting factors for the hydrophobic effect and for the hydrogen bonding of −0.0351 and −0.587, respectively, values designed for an aqueous environment. These values are unsuitable for a hydrophobic environment and result in docking poses limited to a restricted area of the protein surface with many cholesterols docked with their −OH groups in what would be the interior of the membrane (Fig. 3 A). In a previous article, it was shown that the appropriate weighting factors for a membrane were −0.001 and −2.0, respectively (12). Using these parameters, docking poses are more widely spread over the TM surface but with many poses very unlike those observed crystallographically (Fig. 3 B). The final step was to introduce interfaces on the EC and IC sides to which the cholesterol −OH group could hydrogen bond (Fig. 3 C). Wiener and White (30) described the hydrocarbon/headgroup boundary of a lipid bilayer as “one of tumultuous chemical heterogeneity because of the thermal motion of the bilayer.” The exact nature of the model interfaces is therefore not important, just as long as they provide ample potential hydrogen bond donors and acceptors. Here, the interfaces were created by converting the dummy interface atoms in OPM structure files to NH3; all docked cholesterols now adopt orientations with their −OH groups close to the interface (Fig. 3 C).
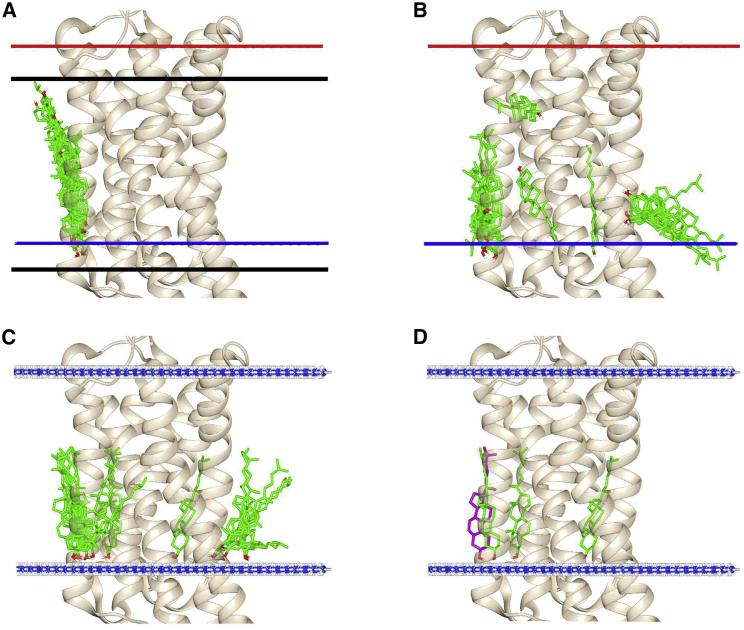
Figure 3.
The docking protocol. The figure shows docking on the IC side of 3D4S. Cholesterol docking poses are shown in green (A–C) or in green and magenta (D). In (A) and (B), the locations of the EC and IC interfacial planes predicted by OPM are shown as red and blue bars, respectively; in (C) and (D), the interfacial planes have been replaced by planes of NH3 molecules (ball and stick; N, blue; H, white). In (A), the top and bottom edges of the Vina search box are shown by black lines. (A) shows the results of a single docking run with default Vina binding parameters and (B) with binding parameters appropriate for a hydrophobic environment. (C) shows the results of a single docking run with modified binding parameters and the inclusion of interfacial planes of NH3. (D) shows the three binding poses selected from the first docking run (green), together with the additional binding pose selected from the second docking run (magenta). To see this figure in color, go online.
The docking run shown in Fig. 3 C returned 20 poses, the maximal possible number. Many of these poses are very similar, and so poses were sorted into clusters, in this case giving one cluster of eight poses, five of two poses, and two single poses. Poses were then filtered on the basis of tilt angle and the numbers of contact residues (see Methods), leading to the rejection of nine poses. For the remaining poses that were in clusters, the pose of highest energy in that cluster was chosen to represent the cluster. This resulted in a final selection of three poses (Fig. 3 D). To ensure complete coverage of the surface, four further sequential runs were performed (see Methods). In this example, the second run resulted in one more selected pose (Fig. 3 D) with runs three to five resulting in no further poses. For all GPCRs studied, the majority of selected poses were found in runs one and two, with a few in runs three and four and none in run five, showing that all poses that met the chosen criteria had been detected.
A comparison between crystallographic and docking results for β2AR
Crystal structures for β 2AR in the inactive state each contain between one and three resolved cholesterols, all on the IC side; docking studies successfully identified 16 of the 17 cholesterols contained in the eight available structures (Table 1). The cholesterols are located at distinct sites, two of which are occupied by cholesterol in all but one structure, the third site being only occupied by cholesterols in two structures but being occupied by 1-oleoyl-R-glycerol in three others. Docking to the 3D4S structure identified a total of four binding sites on the IC side (Table S3), two corresponding to the two cholesterol binding sites A and B reported for the 3D4S structure and one corresponding to site C occupied by 1-oleoyl-R-glycerol (Fig. 1 A; Table 1). In the 5D5A structure, all three sites A–C are occupied by cholesterol (Fig. 1 B), and docking identifies six sites on the IC side (Table S3), of which three correspond to the three crystallographic binding sites (Fig. 1 B; Table S3). The observation that a cholesterol docks to site C in 3D4S (Fig. 1 A) shows that docking is able to detect binding sites for cholesterol even when the site is not occupied by cholesterol in the crystal. In fact, a cholesterol was resolved at site C only in the two structures that crystallize in the C121 space group (Table 1); this space group results in a multilayered structure of symmetry-related dimers with site C at the protein-protein interface (31), suggesting that crystal packing could be an important factor in resolving cholesterol at this site. Agreement between the lists of residues making up the docking and crystallographic binding sites is very good (Table S3).
rmsd values between docked and crystallographically resolved cholesterols are higher than would be expected in a drug docking study, varying for the structures shown in Fig. 1 between 0.9 and 2.9 Å. These values are, however, comparable to rmsd values between corresponding resolved cholesterols in the 3D4S and 5D5A structures; after aligning the two structures, the rmsd values for cholesterols at sites A and B are 2.6 and 1.0 Å, respectively. The high value at site A is not due to a problem with protein alignment as the rmsd value between the residues that make up site A (residues 70–115 and 151–166; Table S3) is just 0.7 Å. Rather, the cholesterol at site A adopts a pose in 5D5A with its smooth A face against the protein surface, whereas in 3D4S, the cholesterol binds edge-on, even though the residues making up site A in the two structures are very similar, with 10 residues in common, the only difference being that Leu155 is also a part of site A in the 5D5A structure (Table S3). These results are consistent with the idea that cholesterol binding sites are able to accommodate cholesterols in a range of poses.
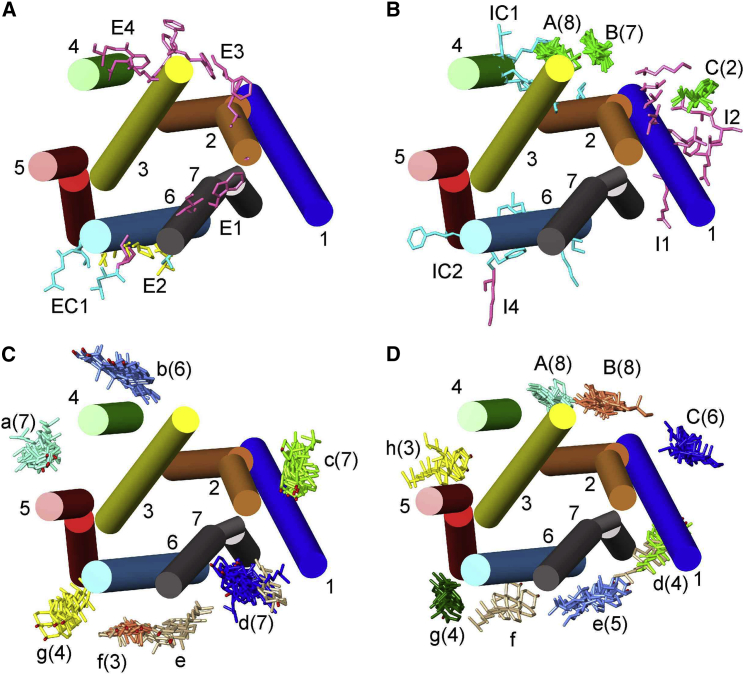
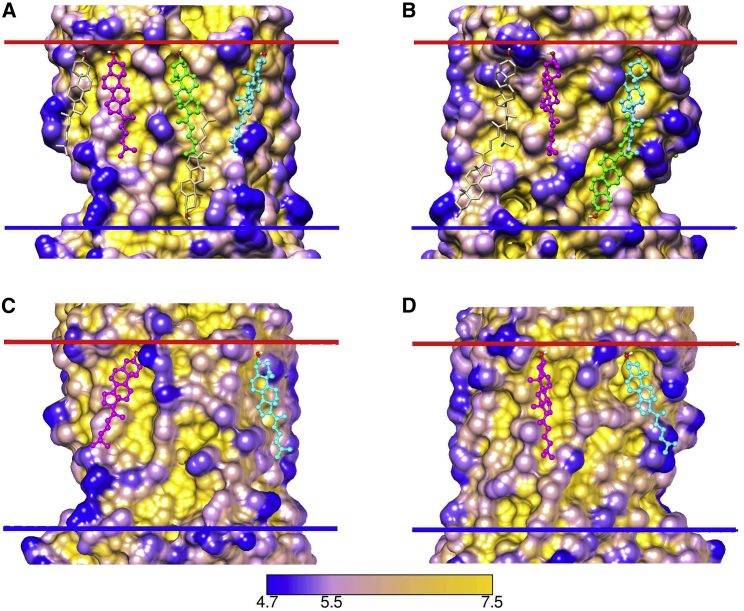
Docking results for all 8 β2AR structures containing resolved cholesterols are given in Table S7, and a cluster analysis of the poses is shown in Fig. 4, together with a comparison with the crystallographic results. In Fig. 4, C and D, clusters are shown in different colors with the number of poses in the cluster given in brackets; poses not in clusters are colored tan. The analysis identifies six clusters on the EC side and seven clusters on the IC side, three of which correspond to the crystallographic sites A–C; 87 and 93% of the poses on the EC and IC sides, respectively, are located in clusters. A spread of poses is observed in each cluster, together with differences in the numbers of poses in each cluster (Fig. 4). Because all eight β2AR structures are in the same, inactive state, with highly superimposable TM α-helices, any particular binding site would have been expected to be present in all eight structures. The observed variability in poses turns out to be due in large part to differences in the protein tilt angles predicted by OPM, these differences resulting in changes in the pose giving the most stable hydrogen bonding interaction with the interface, as illustrated in Fig. 5 for two β2AR structures, 3NY8 and 3NY9. Fig. 5 A shows that the pose at site A is different for 3NY8 (orange) and 3NY9 (green), despite the fact that the positions of the residues making up the binding site are superimposable in the two structures. The predicted membrane interfaces on the IC side are, however, different for 3NY8 (tan) and 3NY9 (blue). Fig. 5 B shows that if the interface for the 3NY9 structure is exchanged for that of the 3NY8 structure, then the pose (yellow) shifts and becomes very similar to that of the crystallographically resolved cholesterol (magenta) and to the pose for the 3NY8 structure in Fig. 5 A. Fig. 5 C shows the protein surface at site A with the crystallographically resolved cholesterol and the two poses located in a hollow on the IC side. A larger effect is seen at site d (Fig. 5 D), in which no pose is observed for 3NY9 until its interface is replaced by that of 3NY8. These results establish that differences in poses between different crystal structures of a protein in the same conformational state are not due to structural differences in the TM regions but, rather, to differences in predicted interface tilts.
Figure 4.
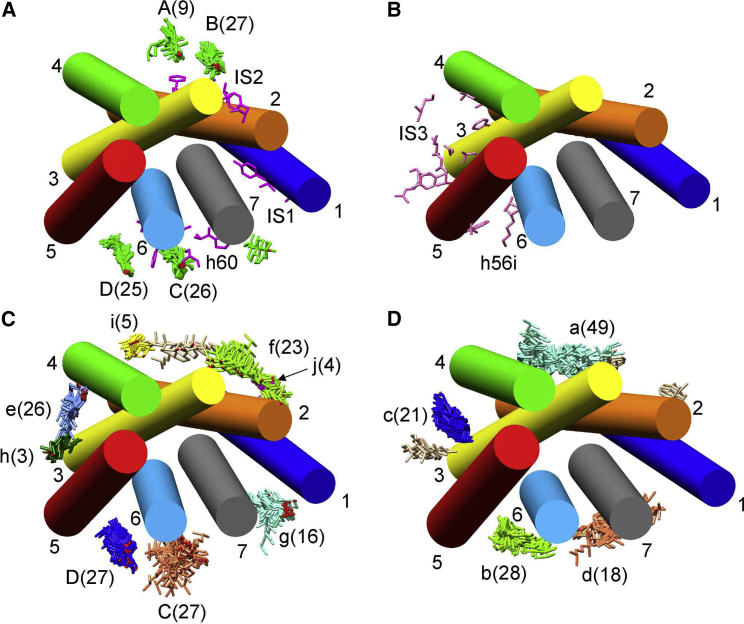
Cluster analysis of poses for β2AR, all views being from the EC side. The eight structures included in the analysis are listed in Table 1. All structures have been aligned to 3D4S, with TM α-helices shown and numbered. (A) and (B) show crystallographic and MD results, and (C) and (D) show docking results for the EC (A and C) and IC (B and D) monolayers. In (A), residues on the EC side identified in MD simulations as being close to bound cholesterols are shown in pink (E1–E4) (13, 14) or blue (EC1) (16) and in yellow when present in both sets of MD simulations. In (B), crystallographically resolved cholesterol molecules on the IC side are shown in green, located at sites A–C (Table 1); the number of cholesterols resolved at a given site is given in brackets. Residues identified in MD simulations are again shown in pink (I1, I2, I4) (13, 14) or blue (IC1, IC2) (16). (C) and (D) show clusters of poses on the EC and IC sides, respectively, colored by cluster, with single poses colored tan. The numbers in brackets give the number of poses in each cluster, and in (D), clusters corresponding to the three crystallographic sites A–C on the IC side are labeled in capitals with additional clusters in (C) and (D) in lower case. To see this figure in color, go online.
Figure 5.
Effect of interfacial layer tilt on docking. (A) and (B) show results at site A for 3NY8 and 3NY9 after aligning the 3NY8 structure to that of 3NY9. The IC interfaces for 3NY8 and 3NY9 are shown as tan and blue spheres, respectively, together with the crystallographically resolved cholesterol for 3NY9 (ball and stick, magenta). In (A), the pose (lines) is shown for 3NY8 (orange) and 3NY9 (green). Residues close to the docked cholesterol (within 4 Å) for 3NY9 are colored blue. Residues close to the docked cholesterol for 3NY8 are colored tan when they are also close to the docked cholesterol for 3NY9 and yellow when only close to the docked cholesterol for 3NY8. (B) shows the effect on poses when the IC interface for 3NY9 is exchanged for that of 3NY8. The crystallographically resolved cholesterol for 3NY9 is shown colored magenta. The pose for 3NY9 with the 3NY9 interface (as in (A)) is colored green and that for 3NY9 with the 3NY8 interface is colored yellow. Residues close to the docked cholesterol for 3NY9 with both the 3NY9 and the 3NY8 interfaces are colored blue. Two residues close to the docked cholesterol only for 3NY9 with the 3NY9 interface are colored tan; the single residue close to the docked cholesterol only for 3NY9 with the 3NY8 interface is colored orange. (C) shows the protein surface at site A for 3NY9 colored by surface depth with cholesterol molecules colored as in (A); the blue bar shows the position of the IC interface for 3NY9. (D) shows the effect of tilt on docking at site d (Fig. 4). The pose for 3NY8 is shown in yellow (ball and stick), but no pose is observed at this site for 3NY9. However, after aligning 3NY9 to 3NY8 and replacing the 3NY9 interface with that of 3NY8, a pose is observed (lines, magenta). Residues close to the docked cholesterol for 3NY8 are shown in tan and those close to the docked cholesterol for 3NY9 with the 3NY8 interface are shown in blue. To see this figure in color, go online.
The tilt of the protein relative to the two interfaces as reported by OPM is particularly sensitive to the distribution of charged residues in the interhelical loops close to the interfaces. For 3NY8 and 3NY9, residue differences close to the interfaces include, on the IC side, the side chain of Glu338, which adopts a different position in the two structures, and the side chain of Lys60, which is missing in 3NY9; on the EC side, the side chain of Lys97 adopts a different position in the two structures, and the side chains of Trp32 and Gln299 are missing in 3NY9. To what extent these predicted differences in protein tilt might be observed in a real membrane is unclear, but MD simulations also show differences in protein tilt between 3NY8 and 3NY9 (Memprotmd.bioch.ox.ac.uk) (32). If thermal motion of protein loops and of the lipid bilayer results in fluctuations in tilt, the optimal cholesterol binding pose could vary with time in the manner shown in Fig. 5. Cluster analysis (Fig. 4, C and D) could then provide information about the range of possible binding poses for cholesterol at any given site. If only a single crystallographic structure is available for a protein, equivalent information can be obtained by generating a set of tilted interfacial planes around the protein. As an example, the EC and IC interfacial planes around 3D4S were rotated by 5, 0, and −5° around the Chimera x and z axes to generate nine pairs of planes. Cholesterol docking to these nine structures (Fig. S1) results in a distribution of docking clusters very similar to that shown in Fig. 4, C and D.
A comparison between crystallographic and docking results for other GPCRs
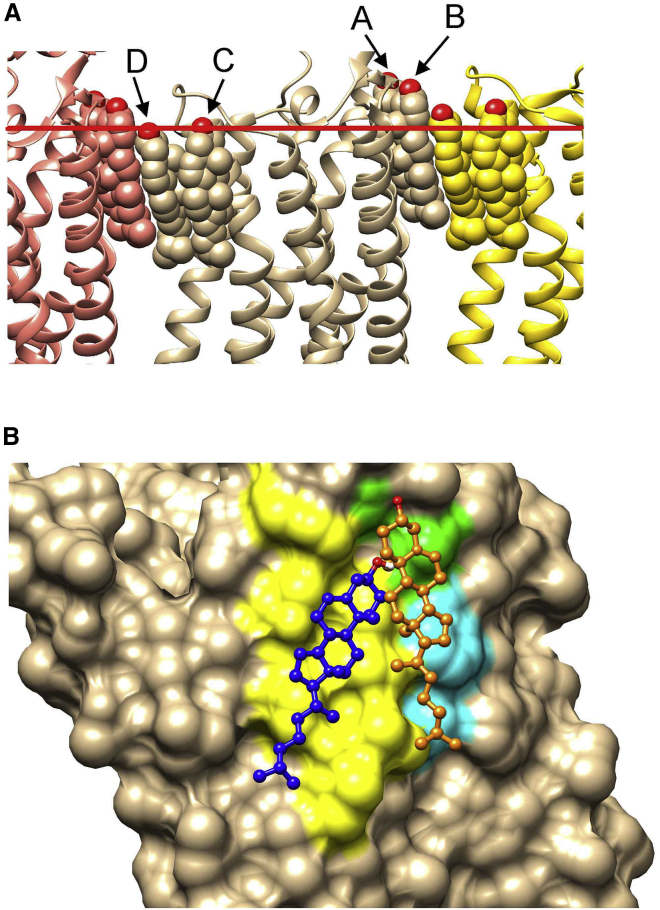
The PDB database contains 27 A2AR structures in an inactive state containing resolved cholesterols at four sites on the EC side (Table S1). Docking results for these structures are shown in Fig. 6 and Table S1. Of the 51 cholesterols observed at sites C plus D (Fig. 6 A), all but one are reproduced by poses, but of the 27 cholesterols observed at site B, only one is matched by a pose, and none of the nine cholesterols at site A have matches (Table S1). All the structures show type 1 crystal packing with each A2AR molecule surrounded by four symmetry mates within membrane-like layers (33). In this arrangement, every A2AR molecule forms one parallel and one antiparallel dimer with two of its neighbors, both involving direct protein-protein interactions. The remaining two neighbors pack to form rows of dimers, packing being mediated solely by cholesterols, the dimer structure being unusual in that packing results in a zig-zag rather than a planar structure, with each A2AR displaced from its two neighbors (Fig. 7 A). It is also noticeable in Fig. 7 A that cholesterols occupying sites A and B at the packing interface are located with their −OH groups well above the interface plane predicted by OPM, unlike the cholesterols at sites C and D. An analysis of all resolved cholesterols bound to GPCRs in the PDB database shows that 74% have their OH groups located within 2.5 Å of an interface. Of the 26% of cholesterols with OH groups further than 2.5 Å from an interface, 81% come from A2AR, all from cholesterols at sites A and B; excluding these molecules, the proportion of cholesterols with OH groups within 2.5 Å of an interface becomes 94%. Site A is also unusual in that cholesterols at this site make contact with just five residues, compared with the average of 10.1 ± 1.5 contacts at other sites. Finally, thermal B factors for cholesterols at site A are unusually large (circa 95 for the 5MZP structure shown in Fig. 7 A); only 9 of the 26 structures show a resolved cholesterol at site A, whereas all but two show cholesterols at sites B–D (Table S1). These observations suggest that the dimer arrangement observed in the crystal might not be observed in a membrane; although A2AR forms oligomers in membrane-like environments, this is dependent on the presence of the 95 residue-long C-terminal sequence, which is removed from the construct used in crystallographic studies (34). If A2AR adopted a pose in a bilayer like that observed in the crystal, there would need to be a thickening of the bilayer close to sites A and B to satisfy the hydrophobic requirements of the cholesterols at these sites, but MD simulations (32) provide no evidence for any such distortion. The simplest explanation for the docking results is that sites A and B correspond to crystal packing “artifacts” and that only sites C and D will be observed in a membrane. As described below, this is consistent with MD simulations of cholesterol binding.
Figure 6.
Cluster analysis of poses for A2AR, all views being from the EC side. The 27 structures included in the analysis are listed in Table S1. All structures have been aligned to 4EIY. (A) and (B) show crystallographic and MD results, and (C) and (D) show poses for the EC (A and C) and IC (B and D) monolayers. In (A), resolved cholesterols on the EC side are shown in green, located at sites A–D as given in Table S1; the number of cholesterols at a given site is given in brackets. Residues identified in MD simulations as being close to bound cholesterols are shown in pink on the EC (IS1, IS2, h60) and IC (IS3, h56i) sides (17, 36). (C) and (D) show clusters of poses on the EC and IC sides, respectively, colored by cluster, with single poses colored tan. The numbers in brackets give the number of poses in each cluster, and in (C), the clusters corresponding to the crystallographic sites C and D on the EC side are labeled in capitals with additional clusters in lower case. To see this figure in color, go online.
Figure 7.
(A) The packing interface for A2AR crystallized in the C2221 space group. An A2AR monomer in the 5MZP structure is shown (ribbons; tan), together with the two neighboring molecules that pack to give a layer structure (ribbons; orange and yellow). A2AR monomers are separated by cholesterols (spheres) packed at the protein-protein interfaces; the four cholesterols per monomer resolved in this structure are colored by the monomer to which they belong, and binding sites A–D are labeled as in Table S1. The position of the interface on the EC side as predicted by OPM is shown by the red bar. (B) The surface of an A2AR monomer is shown on the EC side of the membrane for the 4EIY structure, with the crystallographic cholesterol molecule at site B (Table S1) shown in orange (ball and stick). The pose in cluster f (Fig. 6 C) is shown in blue (ball and stick). Residues identified as being both part of cholesterol binding site IS2 predicted in MD simulations (36) and as being adjacent to the pose (blue) are shown in yellow. Residues that are only part of site IS2 are shown in light blue, and residues that are only adjacent to the pose are shown in green. To see this figure in color, go online.
A cluster analysis of the poses on the EC side of A2AR assigns the 134 poses to eight clusters with three poses not assigned to a cluster (Fig. 6 C). Three of the clusters contain 26 or 27 poses each, including the two clusters C and D that correspond to binding sites in the crystallographic structures. Cluster f is a rather diffuse cluster involving interactions with TM2 and 3, adjacent to the crystallographic binding site B. On the IC side, 120 poses are assigned to four large clusters (Fig. 6 D). Docking results for inactive A2AR structures crystallized without cholesterol (Fig. S3, A and B) are very similar to those with cholesterol, except that for structures without cholesterol, the proportion of poses in many of the clusters is less. This effect arises from a difference in the tilt of the interface planes predicted by OPM for structures with cholesterol, which crystallize in the C2221 crystal form, and those without cholesterol, which crystallize in other forms. All the A2AR structures in the C2221 form have a fully resolved N-terminus with Pro2 or Ile3 at the predicted EC interface. However, the most N-terminal region of TM1 is not resolved in structures in other crystal forms, structures starting at ca Ser6. This has a marked effect on the tilt of the EC interfacial plane predicted by OPM as OPM locates the most N-terminal resolved residue at the interface. In the C2221 crystal form, the N-terminal Pro1 makes contact with helix H8 in an adjacent crystal plane, stabilizing the N-terminal region in a fully extended, resolvable form; in the other crystal forms, there are no such adjacent H8 regions. Shortening the N-terminal chain in a C2221 structure and recalculating the OPM plane results in a tilted plane as observed for the structures in crystal forms other than C2221. Structures with a short N-terminal region show distortion of the surrounding bilayer in MD simulations (32). It is a moot point what the structure of the full N-terminal region will be in a membrane because the interaction stabilizing an extended N-terminus in the crystal will be absent.
Results for the remaining GPCRs containing resolved cholesterols are listed in Table S2; 16 of the 23 resolved cholesterols are reproduced in the docking studies. The dimeric metabotropic glutamate receptor structure shows six resolved cholesterols, all located at or close to the dimer interface. Four of these cholesterols are matched in the docking studies (Table S2); the other two cholesterols (CLR1902 and 1904) make only small numbers of residue contacts (4, 5, 6), and no poses match these two cholesterols (Table S2).
Is cross-docking a problem?
AutoDock Vina employs rigid protein structures with no allowance for ligand-induced conformational changes. This may not be a problem in “self-docking” studies in which a ligand is docked into a structure from which that same ligand has been first extracted. However, it could be a problem in “cross-docking” studies in which a ligand is docked into a structure that was not determined in the presence of the ligand under study (35). For the particular case of docking cholesterol onto the TM region of a membrane protein, cross-docking is less likely to be a problem because the energy well in which a cholesterol binds is not deep, and considerable movement within the site is possible (Fig. 2). This is confirmed by the analysis of the docking results for A2AR at sites C and D on the EC side (Fig. 6). Because activation of GPCRs results in little change in TM helix packing on the EC side, it is possible to analyze together the inactive, active intermediate, and active state structures of A2AR. The PDB database contains 17 structures for A2AR with no bound cholesterols, crystallized from detergent either with or without cholesterol, from detergent containing cholesterol hemisuccinate, or from a cubic phase containing cholesterol. Of these, all 17 show a pose at site D and 14 show a pose at site C (Table S4), providing no evidence for a cross-docking problem.
Eight structures exist for β2AR crystallized from a cholesterol-containing cubic phase, showing up to three cholesterol binding sites each (Table 1), but only in two cases is site C occupied by cholesterol; in three structures, it is occupied by 1-oleoyl-R-glycerol, and in three other structures, the site is unoccupied. However, poses are observed at site C for the three structures where site C is occupied by 1-oleoyl-R-glycerol (Fig. 1; Table 1) and in one structure (3NYA) where site C is empty (Table S7). In the remaining two crystal structures with an empty site C (3PDS and 5X7D), no pose was observed at the site (Table S7). However, these two structures show very different tilt angles in the membrane to the other β2AR structures and show no pose at site B (Table 1), so that the lack of docking at site C could be due to the unusual tilt angle. Thus, β2AR structures also provide no evidence that cross-docking is a problem.
A comparison between MD simulations and docking results
The agreement between MD simulations and docking is generally good, with the docking approach detecting more of the crystallographically determined binding sites for cholesterol than the MD approach. MD simulations of A2AR by the Lyman group (17, 36) identified five potential binding sites for cholesterol (Fig. 6; Table S5). One of these (h60) corresponds to site C in the crystal structure, but the other three crystallographic sites for cholesterol were not identified in the MD simulations. Importantly, however, residues identified near binding sites in the MD simulations match well to residues close to poses (Fig. 7 B; Table S5). In particular, both the MD and docking studies detect a cholesterol binding site on TM2/3 at the crystal packing interface close to crystallographic site B (Fig. 7 B), although neither sites A nor B were detected in either the MD or docking studies. The fact that both sites A and B were absent from both the MD and the docking studies supports the suggestion that these sites could be crystal packing artifacts.
Agreement between MD simulations (13, 14, 16, 37) and docking results is also good for β2AR (Fig. 4). The MD simulations of Cang et al. (13, 14) detect a site (I2) corresponding to crystallographic binding site C on the IC side, in good agreement with the docking results. The MD simulations of Manna et al. (16) detect a site (IC1) corresponding to crystallographic binding site A on the IC side, also in good agreement with the docking results. On the EC side, Cang et al. (13, 14) and Manna et al. (16) detect a binding site (E2 and EC1, respectively) that matches residues in docking clusters f and g (Fig. 4 C). MD simulations by Mahmood et al. (37) identified two binding sites for cholesterol on β2AR, one on the EC side and one on the IC side. Neither correspond to a crystallographic binding site, but the six residues identified as part of the binding site on the IC side (Val126, Val129, Val206, Pro211, Ile214) are all present in cluster h (Fig. 4 D; Table S7).
MD simulations by Cang et al. (14) for β1AR (2VT4) identified six residues on TM6 involved in an interaction with cholesterol, of which four are part of a docking site on the IC side, and one is part of a docking site on the EC side (Table S7). Cang et al. (14) also identified an occupied CCM motif on β1AR (Lys159, Cys163, Trp166), part of one of the docking sites on the IC side (Table S7).
A comparison between the docking results for rhodopsin and opsin and the results of MD simulations is presented in Table S6. MD simulations were interpreted in terms of groups of residues that showed preferential interaction with cholesterol; these groups contained a spread of residues too wide to correspond to a single binding site (38, 39, 40) but generally map well onto the docking clusters (Fig. S2; Table S6). MD groups A1, C1, and D1 contain residues from TM1/7 on the IC side, close to the palmitoyl chains attached to Cys322 and Cys323, and in simulations, cholesterol was observed binding between the palmitoyl chains and the protein surface (40). No equivalent docking poses were observed (Table S6), presumably because the docking procedure does not allow the marked deviation in palmitoyl chain conformation from that in the crystal required to allow the chain to wrap around a bound cholesterol. However, group C1 contains five additional residues not present in groups A1 and D1, and these residues are close to docking clusters d and f (Fig. S2, A and B). Five of the eight residues in group A2 are close to cluster d (Fig. S2 B), and four of the five residues in group A3 are close to cluster a (Fig. S2 A). Of the five residues in group B1 and the three in group B2, all but one are close to clusters b, c, and e (Fig. S2, A and B). Group C2 contains nine residues of which eight are close to clusters g and h (Fig. S2 B), and group C3 contains just Ile263, which is close to cluster i on the EC side (Fig. S2 A). Seven of the nine residues in opsin in group D2 are close to cluster j on the IC side (Fig. S2 F), but the three residues on TM6 in group D3 are not close to any of the identified clusters for opsin (Table S6).
A coarse grain MD simulation of the smoothened receptor using the 5L7D structure identified one major binding site for cholesterol in the TM region, involving residues 276, 279, 283, 286, 312, 313, 316, 317, and 320 (41). This agrees well with the residues identified for the most energetically favorable binding pose (275, 279, 311, 312, 313, 316, 317, and 320; Table S7).
A comparison between inactive and active conformations of GPCRs
GPCRs cycle between one or more inactive (basal) states, observed in the absence of ligands or in the presence of inverse agonists or antagonists, and an active state requiring both a bound agonist and a bound G protein or equivalent; for some GPCRs, the binding of just an agonist results in an active intermediate state (42). Binding of inverse agonists or antagonists results in changes around the ligand binding site but does not affect packing of the TM helices. In contrast, activation of a GPCR leads to changes in helix packing on the IC side with an outward movement of TM5 and 6 and an inward movement of TM7; helix packing in the active intermediate state is intermediate between those in the inactive and active states (42, 43). The largest number of GPCR active state structures exists for rhodopsin. Activation of rhodopsin leads to changes in surface topology and changes in the shapes and sizes of some sites where cholesterol binds (Fig. 8). On the EC side of the inactive state is cluster a, which contains poses from 9 of the 11 available inactive structures, but no poses are present in this region on any of the eight available active state structures (Fig S2 A; Table S7); shapes of the hollows containing cluster a change on activation (Fig. 8, A and C). Similarly, on the IC side cluster d, which contains poses from all 11 available inactive structures, disappears on activation (Fig. S2 B; Table S7), activation again leading to changes in the shape of the binding region (Fig. 8, B and D). The loss of binding sites is quite localized, neighboring binding sites being unaffected (Figs. 8 and S2). The observed disappearance of the two binding clusters on activation is statistically highly significant, with p values of 0.0004 and <0.00001 for clusters a and d, respectively. Opsin has the same TM helix packing as activated rhodopsin and so, as expected, cholesterol binding to opsin is very similar to that to activated rhodopsin (Fig. S2; Table S7).
Figure 8.
Cholesterol binding to inactive (1U19) and active (2X72) forms of rhodopsin. Shown are depth-colored surface plots for inactive (A and B) and active forms (C and D) with the (A and C) and (B and D) views being related by a 180° rotation. (A) shows a pose on the EC side (green, ball and stick), part of cluster a (Fig. S2; Table S5), absent in (C); poses that are parts of cluster b (magenta, ball and stick; Fig. S2) and adjacent to TM4 (cyan, ball and stick) on the EC side are seen at equivalent positions in (A) and (C). (B) shows a pose on the IC side (green, ball and stick), part of cluster d (Fig. S2; Table S5), absent in (D); poses that are parts of clusters f (cyan, ball and stick) and i (magenta, ball and stick) on the EC side (Fig. S2) are found at equivalent positions in (B) and (D). Poses not specifically mentioned are shown colored tan. To see this figure in color, go online.
Inactive, active intermediate, and active structures are available for A2AR (Fig. S3). Only two structures are available for the active state but docking clusters for cholesterol are very similar for the active and active intermediate states. Conversion from the inactive to the active intermediate state leads to the loss of a docking cluster on the EC side, in the vicinity of TM3/4, that contains seven poses from the nine inactive structures (p value for the change, 0.0019). Activation also leads to the appearance of a new cluster, between TM4 and 5, containing four poses from the seven active intermediate structures (p value for the change, 0.029). On the IC side, formation of the active intermediate state leads to the loss of the cluster, in the vicinity of TM7, that contains five poses from the nine inactive structures (p value for the change, 0.017).
Conclusions
GPCRs in mammalian cells are surrounded by membranes rich in cholesterol. Effects of cholesterol on GPCRs include an increase in thermal stability, a reduced number of energetically favorable receptor conformations, and changes in ligand affinity (1). Crystallographic studies identify just a small number of cholesterol binding sites on the TM surfaces of GPCRs, whereas MD simulations suggest that cholesterol molecules can interact with most of the TM surface. An analysis of GPCR crystal structures by Gimpl (1) showed that cholesterol binding was not restricted to any particular binding motif, so that a search for binding sites based on motif is of limited value. Similarly, the practicability of an MD-based search for binding sites is restricted by the long simulation times required to ensure system equilibration (17). Here, a docking protocol is described that allows the TM surface of a GPCR to be swept for potential binding sites for cholesterol in a few minutes on a personal computer. Docking was performed using AutoDock Vina, and docking poses were filtered to select those with characteristics (number of contacts and binding angle) that matched those of cholesterol molecules in GPCR crystal structures. The docking approach identified 89% of the cholesterols resolved in GPCR structures when cholesterols likely to be crystal packing artifacts were excluded. Agreement between the docking approach and MD studies was good, with the docking approach identifying more crystallographic sites than the MD approach.
Docking results for sets of crystallographic structures of the same conformational state of a protein varied between structures but with the docking poses falling into a number of distinct clusters. For example, for the eight available structures of β2AR in the inactive state, the average numbers of clusters on the EC and IC sides were 4.9 ± 1.7 and 5.3 ± 1.1, respectively (Fig. 4). Coloring the β2AR surface by depth shows that the clusters are located in hollows between surface ridges (Fig. S4). This confirms the results of MD simulations (4, 17) that show that cholesterol binding sites are not deep energy wells into which a cholesterol falls to occupy a single, well-defined conformational state but, rather, are “greasy hollows” within which a cholesterol can adopt a range of binding poses. The most favorable binding pose for a cholesterol in one of these hollows depends on the angle of tilt of the GPCR in the surrounding lipid bilayer. The angle of tilt, as calculated by the OPM database (44), is sensitive to the distribution of charged residues close to the bilayer surface so that the optimal cholesterol binding pose is likely to be affected by thermal motion, both of the protein and of the lipid bilayer. If this is the case, then the range of cholesterol binding poses at a given site, as shown in Fig. S4, will represent the likely range of thermal motions for a cholesterol molecule at the site.
Extension of these results to a real biological membrane is complicated by the possible existence of membrane regions enriched in cholesterol and by competition between phospholipids and cholesterol for binding to the protein surface. Effects of cholesterol on GPCR function could follow from binding between TM helices if binding between the helices restricted their relative motion; this would not require binding to be highly specific. The observation of different patterns of cholesterol binding for inactive and active conformations of GPCRs (Figs. 8, S2, and S3) suggests that the presence of cholesterol could affect the energetics of conformational changes on the GPCRs. Finally, it has been suggested that the presence of cholesterol could affect the packing of GPCRs into oligomeric structures (45).
Acknowledgments
The School of Biological Sciences, University of Southampton is thanked for computing facilities.
Editor: Michael Grabe.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.03.025.
Supporting Material
References
- 1.Gimpl G. Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids. 2016;199:61–73. doi: 10.1016/j.chemphyslip.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Simmonds A.C., East J.M., Lee A.G. Annular and non-annular binding sites on the (Ca2+ + Mg2+)-ATPase. Biochim. Biophys. Acta. 1982;693:398–406. doi: 10.1016/0005-2736(82)90447-3. [DOI] [PubMed] [Google Scholar]
- 3.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Lee A.G. Biological membranes: the importance of molecular detail. Trends Biochem. Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Song Y., Kenworthy A.K., Sanders C.R. Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci. 2014;23:1–22. doi: 10.1002/pro.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawaliby R., Trubbia C., Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fantini J., Di Scala C., Barrantes F.J. A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes. Sci. Rep. 2016;6:21907. doi: 10.1038/srep21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M.F. Soft matter in lipid–protein interactions. Annu. Rev. Biophys. 2017;46:379–410. doi: 10.1146/annurev-biophys-070816-033843. [DOI] [PubMed] [Google Scholar]
- 9.Salas-Estrada L.A., Leioatts N., Grossfield A. Lipids alter rhodopsin function via ligand-like and solvent-like interactions. Biophys. J. 2018;114:355–367. doi: 10.1016/j.bpj.2017.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inagaki S., Ghirlando R., Grisshammer R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khelashvili G., Harries D. How sterol tilt regulates properties and organization of lipid membranes and membrane insertions. Chem. Phys. Lipids. 2013;169:113–123. doi: 10.1016/j.chemphyslip.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee A.G. A database of predicted binding sites for cholesterol on membrane proteins, deep in the membrane. Biophys. J. 2018;115:522–532. doi: 10.1016/j.bpj.2018.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cang X., Du Y., Jiang H. Mapping the functional binding sites of cholesterol in β2-adrenergic receptor by long-time molecular dynamics simulations. J. Phys. Chem. B. 2013;117:1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- 14.Cang X., Yang L., Jiang H. Cholesterol-β1 AR interaction versus cholesterol-β2 AR interaction. Proteins. 2014;82:760–770. doi: 10.1002/prot.24456. [DOI] [PubMed] [Google Scholar]
- 15.Prasanna X., Chattopadhyay A., Sengupta D. Cholesterol modulates the dimer interface of the β2-adrenergic receptor via cholesterol occupancy sites. Biophys. J. 2014;106:1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manna M., Niemelä M., Vattulainen I. Mechanism of allosteric regulation of β2-adrenergic receptor by cholesterol. eLife. 2016;5:e18432. doi: 10.7554/eLife.18432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouviere E., Arnarez C., Lyman E. Identification of two new cholesterol interaction sites on the A2A adenosine receptor. Biophys. J. 2017;113:2415–2424. doi: 10.1016/j.bpj.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guixà-González R., Albasanz J.L., Selent J. Membrane cholesterol access into a G-protein-coupled receptor. Nat. Commun. 2017;8:14505. doi: 10.1038/ncomms14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A.G. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 20.Gater D.L., Saurel O., Milon A. Two classes of cholesterol binding sites for the β2AR revealed by thermostability and NMR. Biophys. J. 2014;107:2305–2312. doi: 10.1016/j.bpj.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomize A.L., Pogozheva I.D., Mosberg H.I. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaipuria G., Ukmar-Godec T., Zweckstetter M. Challenges and approaches to understand cholesterol-binding impact on membrane protein function: an NMR view. Cell. Mol. Life Sci. 2018;75:2137–2151. doi: 10.1007/s00018-018-2789-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Morris G.M., Huey R., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rentzsch R., Renard B.Y. Docking small peptides remains a great challenge: an assessment using AutoDock Vina. Brief. Bioinform. 2015;16:1045–1056. doi: 10.1093/bib/bbv008. [DOI] [PubMed] [Google Scholar]
- 27.Stevens T.J., Boucher W. Cambridge University Press; Cambridge, UK: 2015. Python Programming for Biology. [Google Scholar]
- 28.Pándy-Szekeres G., Munk C., Gloriam D.E. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan K.P., Nguyen T.B., Madhusudhan M.S. Depth: a web server to compute depth, cavity sizes, detect potential small-molecule ligand-binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 2013;41:W314–W321. doi: 10.1093/nar/gkt503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiener M.C., White S.H. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys. J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherezov V., Rosenbaum D.M., Stevens R.C. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stansfeld P.J., Goose J.E., Sansom M.S.P. MemProtMD: automated insertion of membrane protein structures into explicit lipid membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W., Chun E., Stevens R.C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonenbach N.S., Rieth M.D., O’Malley M.A. Adenosine A2a receptors form distinct oligomers in protein detergent complexes. FEBS Lett. 2016;590:3295–3306. doi: 10.1002/1873-3468.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland J.J., Nandigam R.K., Vieth M. Lessons in molecular recognition. 2. Assessing and improving cross-docking accuracy. J. Chem. Inf. Model. 2007;47:2293–2302. doi: 10.1021/ci700253h. [DOI] [PubMed] [Google Scholar]
- 36.Lee J.Y., Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J. Am. Chem. Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmood I., Liu X., Hoshino T. Influence of lipid composition on the structural stability of g-protein coupled receptor. Chem. Pharm. Bull. (Tokyo) 2013;61:426–437. doi: 10.1248/cpb.c12-01059. [DOI] [PubMed] [Google Scholar]
- 38.Grossfield A., Feller S.E., Pitman M.C. A role for direct interactions in the modulation of rhodopsin by ω-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khelashvili G., Grossfield A., Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with rhodopsin identified from microsecond length molecular dynamics simulations. Proteins. 2009;76:403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horn J.N., Kao T.C., Grossfield A. Coarse-grained molecular dynamics provides insight into the interactions of lipids and cholesterol with rhodopsin. In: Filizola M., editor. G Protein-Coupled Receptors - Modeling and Simulation. Springer; 2014. pp. 75–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedger G., Koldsø H., Sansom M.S.P. Cholesterol interaction sites on the transmembrane domain of the hedgehog signal transducer and class F G protein-coupled receptor smoothened. Structure. 2019;27:549–559.e2. doi: 10.1016/j.str.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weis W.I., Kobilka B.K. The molecular basis of G protein–coupled receptor activation. Annu. Rev. Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wacker D., Fenalti G., Stevens R.C. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J. Am. Chem. Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 45.Prasanna X., Sengupta D., Chattopadhyay A. Cholesterol-dependent conformational plasticity in GPCR dimers. Sci. Rep. 2016;6:31858. doi: 10.1038/srep31858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.