Abstract
We report the x-ray crystal structure of intact, full-length human immunoglobulin (IgG4) at 1.8 Å resolution. The data for IgG4 (S228P), an antibody targeting the natriuretic peptide receptor A, show a previously unrecognized type of Fab-Fc orientation with a distorted λ-shape in which one Fab-arm is oriented toward the Fc portion. Detailed structural analysis by x-ray crystallography and molecular simulations suggest that this is one of several conformations coexisting in a dynamic equilibrium state. These results were confirmed by small angle x-ray scattering in solution. Furthermore, electron microscopy supported these findings by preserving molecule classes of different conformations. This study fosters our understanding of IgG4 in particular and our appreciation of antibody flexibility in general. Moreover, we give insights into potential biological implications, specifically for the interaction of human anti-natriuretic peptide receptor A IgG4 with the neonatal Fc receptor, Fcγ receptors, and complement-activating C1q by considering conformational flexibility.
Introduction
Antibodies, also known as immunoglobulins (Ig), share a unique three-dimensional structure, which is instrumental for both specific interaction with its target molecule and its inherent conformational stability in physiological environments. Antibodies generally exhibit a common domain architecture and overall assembly. They are composed of constant and variable domains, which both differ in their degree of sequence similarity based on their natural abundance. Consequently, the human antibody repertoire is classified in five different functional classes (IgA, IgE, IgD, IgG, IgM), and isotypes (e.g., IgG1, IgG2, IgG4). Each of these exhibit unique structural features (e.g., disulfide scrambling in IgG2; lack of effector function; Fab-arm exchange (FAE) in IgG4; secretory components in IgGA; multimerization by a cysteine incorporated in an 18 amino acid extension at the heavy chain carboxy terminus in IgM). In addition, their Fc glycan moieties play crucial roles in biological function, in maintaining solubility, and in facilitating cellular processing of IgG in antibody-dependent cellular cytotoxicity, antibody-dependent cellular phagocytosis, and complement-dependent cytotoxicity (1, 2, 3, 4).
In general, Igs possess conformational diversity and high inherent flexibility, contributing to the lack of high-resolution structural information for intact full-length Igs. To date, only five full-length IgG structures were deposited in the Protein Data Bank (PDB): one IgG2a antibody (PDB: 1IGT (5)), three IgG1 antibodies (PDB: 1HZH (6), 1IGY (7), 1MCO (8)), and one IgG4 antibody (PDB: 5DK3 (9)).
In this study, we report the high-resolution x-ray crystal structure of a therapeutically relevant, intact full-length anti-NPRA IgG4-humanized monoclonal antibody (mAb) at 1.8 Å resolution. The anti-NPRA antibody specifically targets the natriuretic peptide receptor A (NPRA) extracellular domain bound to its natural cyclic natriuretic peptide ligands (10). NPRA is a homodimeric membrane guanylyl cyclase expressed in a variety of tissues, including kidney, heart, vasculature, brain, and lung. The natural cyclic peptide ligands for this receptor include atrial natriuretic peptide (ANP) and B-type natriuretic peptide (BNP), which are produced by the heart in response to stress and are elevated in patients with heart failure (10). The anti-NPRA mAb described in this study potentiates ANP- and BNP-dependent cyclic guanosine monophosphate production in NPRA-presenting human embryonic kidney cells (Fig. S1) and appears to stabilize the receptor-peptide complex. Therefore, its potentiating role could allow for therapeutic treatment. The anti-NPRA antibody is a member of the human IgG4 subclass. Among all human IgG subclasses, IgG4 are the least-represented subclass in serum. IgG4 antibodies display a number of unique functional and biological properties (11, 12, 13, 14, 15) and thus are of particular interest. It is noninflammatory, does not activate the complement, and can undergo FAE, in which heavy chains can dissociate and reassociate in vivo to generate bispecific but functionally monovalent antibodies. In the case of the anti-NPRA IgG4, we specifically altered the sequence by incorporating an S228P amino acid substitution within the hinge region to revert the natural occurring human IgG4 wild-type sequence to that of human IgG1 to stabilize the molecule against FAE.
Here, we performed a comprehensive structural analysis of anti-NPRA. The crystal structure revealed that Fab and Fc portions adopt the common canonical conformation. However, they differ substantially in their relative Fab-Fc orientation from all other crystal structures reported so far. To investigate anti-NPRA structural diversity in solution, we performed small angle x-ray scattering (SAXS) measurements to determine its preferential conformation in solution. In addition, we carried out molecular dynamics (MD) simulations and transmission electron microscopy (TEM) in combination with two-dimensional (2D) image class analysis to gain further understanding. Hence, we report a detailed and comprehensive structural study, including the varied solution conformations exhibited by anti-NPRA, and provide new insights into how solution conformation might affect biological function to its natural ligands as well as to the neonatal Fc receptor (FcRn), the Fcγ receptor (FcγR), and complement-activating component (C1q).
Methods
Antibody generation
Functional anti-NPRA antibodies were discovered using a human Fab phage display library (HuCAL GOLD; MorphoSys, Planegg, Germany) (16). The resulting anti-NPRA Fab was grafted on an IgG4 (S228P) framework. This mutation was specifically introduced to prevent FAE.
Anti-NPRA crystallization and data collection
The investigated mAb of subtype IgG4 was produced by mammalian cell culture technology (13, 17) using Chinese hamster ovary cells. The material was purified according to procedures described in the literature (18). Protein monomer content was >99% (based on high-performance size exclusion chromatography). Protein concentration was determined by ultraviolet/visible spectroscopy at 280 nm (19). The sample purity and integrity was determined by high-performance size exclusion chromatography, native and non-native sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and hydrophobic interaction chromatography (20).
Initial crystal hits were found with Wizard-I-Screen (Emerald BioAgriculture, Butte, Montana) under a condition containing 20% polyethylene glycol (PEG 8000), 0.2 M calcium acetate, and 0.1 M imidazole (pH 8.0). This condition was refined using a systematic grid varying pH and PEG concentration. The best crystals grew within 10 days at a protein concentration of 14.5 mg/mL with 5% PEG 8000, 0.2 M calcium acetate, and 0.1 M imidazole (pH 7.5). Crystals were cryoprotected with 30% glycerol and flash cooled in liquid nitrogen. The crystallographic experiments were performed on the X06SA beam line at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). Data were processed with MOSFLM (21), XDS (22), SCALA, and POINTLESS (23) within autoPROC (24).
Structure solution and refinement
The structure was determined by molecular replacement with PHASER (25) using PDB: 1HZH (6) as a start model and refined with autoBUSTER (24, 26). For molecular replacement with PHASER (25), the start model was divided into Fc-, CH1-VL-, and VH-VL-parts. Model building was done with COOT (27). Side chains with alternative conformations were modeled, and solvent molecules were added. The quality of the final structure was assessed with MolProbity (28).
Structure interpretation
The visualization and analysis of the structure and the comparison to other complete antibody structures was performed with PyMol (Schrödinger, New York, NY) (29). The elbow angles of the two Fabs in each structure were calculated, as were the rotation angle between FabHL and FabKM, the angle between the long axes of FabHL and FabKM, the angle between the Fab-dyad and the Fc-symmetry axis, the angles between the major axis of either FabHL or FabKM and Fc, and the radius of gyration. These calculations were performed with Python scripts adopted to Rupp et al. (30). The secondary structure elements were calculated from the atomic coordinates with the program ksdssp (31).
SAXS
SAXS data were collected at beamline BM29 (European Synchrotron Radiation Facility, Grenoble, France) at 0.99 Å wavelength, 2.8 m camera distance, and with a 0.0039–0.49 Å−1 scattering vector. The measurements used the automated sample exchange unit. A concentration series was carried out to eliminate any concentration dependence starting with a high concentration of 5 mg/mL. Initial analysis was performed using Scatter software (http://Bioisis.net). Crystal structures 1IGT, 1IGY, and anti-NPRA were used in Scatter for Kratky analysis. Ab initio shape determination was performed using DAMMIF (32) where 20 runs were merged. Rigid body modeling used the program SASREF (33), a simulated annealing protocol that avoids steric clashes. The FabHL or FabKM and Fc domains were extracted from the PDB model into three separate domains with the Fc fixed in position. The H and K chain linker regions were also extracted and split into three sections with the middle section containing the Cys-Pro-Pro-Cys region. A contact of 5 Å was constrained between each cysteine in the linker to mimic the disulphide bonds, enabling the linkers to act as hinges. Several independent SASREF runs were completed.
TEM
Carbon-coated 400 mesh electron microscopy grids (QUANTIFOIL Micro Tools, Großlöbichau, Germany) were hydrophilized by electric glow discharging for 30 s at a low pressure in air. Then 20 μL of anti-NPRA solution (10 μg/mL) was adsorbed onto the hydrophilic grids for 1 min. The grids were washed twice on drops of distilled water and stained on a drop of 2% uranyl acetate in distilled water. Samples were analyzed in a Zeiss EM900 electron microscope (Carl Zeiss, Oberkochen, Germany) at a calibrated magnification of 54,500×. Images were recorded on 8.3 × 10.2 cm Kodak 4489 Electron Microscope Film (Kodak, Rochester, NY). The 29 negatives were digitized at a resolution of 2400 dpi using an Epson Perfection 4990 Photo scanner (Seiko Epson, Nagano, Japan), resulting in a pixel size of 2.13 Å.
The complete single-particle analysis was performed using RELION 1.4 (34, 35). 671 particles were picked manually from six micrographs and 2D classified into 10 classes in three rounds. Non-antibody particles were discarded after each round. Three class averages from the last round were selected as references for automatic particle picking (36). A total of 4663 particles were selected from the original six micrographs and 2D classified into 50 classes in two rounds, resulting in 2867 particles. A total of 18,257 particles were selected from the remaining 22 micrographs, which were combined with the first data set. This data set was then sorted and 2D classified in two rounds, resulting in 4136 particles. For the final classification into 40 classes, frequencies were limited to 10 Å in the alignment steps.
Computational methods
MD simulations were run with GROMACS (version 5) (37), using the Amber99sb-ILDN protein force field (38, 39), Glycam06j (40) force field for the glycan, and TIP3P water model (41). To ensure a neutral overall charge of the simulation systems, 150 mM sodium chloride was added. Periodic boundary conditions were used, and long-range Coulomb interactions were treated with the particle-mesh Ewald method (42). Temperature and pressure were kept constant at 298 K and one bar, respectively. All simulations are based on our 1.8 Å resolution x-ray crystal structure. The missing residues in the hinge region and the loop in Cγ1 were added with MODELLER (43). Simulations were carried out with two different starting structures. First, two 500 ns MD simulations were initiated from the λ-shaped (x-ray) conformer. Second, two 500 ns MD simulations were initiated from an (idealized) Y-shaped conformation, which was constructed by fitting the individual mAb domains from our λ-shaped x-ray structure to the corresponding domains of the IgG1 model of Padlan (44). From these two trajectories, another ten simulations (each 400 ns) were initiated using different random seeds to generate starting velocities, according to a Maxwell-Boltzmann distribution at 298 K: Five simulations were started from a snapshot taken after 100 ns, another set of five simulations were started from a snapshot taken after 500 ns. The total sizes of the simulation systems were ∼571,000 and 585,000 atoms for the λ- and Y-shaped conformers, respectively. The free energy difference between the λ-shaped and Y-shaped conformers was calculated from the conformational enthalpy, entropy, and hydration free energy of the two conformers, ΔG = Gλ − GY = ΔHc − TΔSc + ΔGhyd. The conformational enthalpies, Hc, were approximated by the average potential energies, <Ec>, as obtained from the force field. For the configurational entropies, Sc, the quasiharmonic approximation of Schlitter (45) was used. Hydration free energies, ΔGhyd, were calculated using the three-dimensional reference interaction site model (3D-RISM) integral equation theory (45, 46, 47, 48, 49, 50, 51, 52). For further details, see Supporting Materials and Methods.
Results
X-ray crystal structure
We determined the high-resolution structure of full-length anti-NRPA IgG4 mAb at a resolution of 1.8 Å. The anti-NPRA mAb crystallized in space group P21, with one copy of the full- length anti-NPRA molecule per asymmetric unit, corresponding to a Matthew’s coefficient of 2.95 and a solvent content of 58.3%. Stereochemical analysis reveals an overall good structure quality. 98% of all residues are in allowed regions of the Ramachandran plot. An overall high data set quality is reflected in Rmeas and Rmerge (5.41 and 4.5%, respectively, Table 1). The majority of amino acid residues are visible in the electron density map. However, some residues in the hinge region of heavy chain H (residues H:219–236 following the Kabat numbering convention) and in the hinge region of heavy chain K (residues K:218–236) were not modeled because of missing or weak electron density. Interestingly, the last residue traceable in the electron density of both heavy chains and which also precedes the hinge region (Ser217) adopts a different stereochemical orientation and thus might be the beginning of a break in symmetry that is necessary to connect the Fabs to the Fc. In addition, the loop amino acid residues in the constant domain of Cγ1 (residues H:133–135 and K:133–137) are disordered and hence not modeled because of missing or weak electron density. These residues are highly solvent exposed and not stabilized by crystal packing.
Table 1.
Crystallographic Data Collection and Model Refinement
| Data Collection Statistics | |
|---|---|
| Wavelength (Å) | 0.91 |
| Resolution range (Å)a | 40.0–1.8 (1.864–1.799) |
| Space group | P 21 |
| Unit cell dimensions a, b, c (Å)/α, β, γ (°) | 65.78, 160.51, 87.30/90, 110.72, 90 |
| No. of reflections (total/unique) | 15,552/52,613 |
| Multiplicity | 3.4 |
| Completeness (%)a | 99.5 (99.8) |
| I/σa | 16.6 (2.3) |
| Rmeas(%) | 5.41 |
| Rmerge(%)a,b | 4.5 (54.8) |
| Wilson B-factor | 25.27 |
| Model refinement statistics | |
| Resolution range(Å)a | 40.0–1.8 (1.85–1.80) |
| Rwork/Rfree(%)c | 18.3 (25.8)/20.6 (27.1) |
| Number of reflections (total/unique) | 155,361/11,511 |
| Number of atoms | 11,617 |
| RMS (bonds) (Å) | 0.011 |
| RMS (angles) (°) | 1.66 |
| Ramachandran favored (%) | 98 |
| Ramachandran outliers (%) | 0.31 |
| Clashscore | 0.89 |
Values in parentheses correspond to the outer resolution shell, reflecting the highest atomic resolution.
Rmerge = (∑|Ihkl − ‹I›|)/(∑Ihkl), where the average intensity ‹I› is taken over all symmetry-equivalent measurements and Ihkl is the measured intensity for any given reflection.
Rwork = Σ|Fo − Fc|= Σ |Fc|, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is the R factor calculated from 5% of reflections not included in the refinement.
A carbohydrate is linked to Asn297 in both anti-NPRA heavy chain Cγ2 domains (Fig. 1 B). The observed electron density allowed for building a biantennary carbohydrate structure. Both carbohydrates feature a fucose-branch attached to the first N-acetylglucosamine that is covalently bound to Asn297. For both α (1, 2, 3, 4, 5, 6) branches, a mannose and a N-acetylglucosamine component were modeled, whereas only the mannose component for the α (1, 2, 3) branch could be resolved. Crystallographic data and statistics as well as model refinement statistics are listed in Table 1.
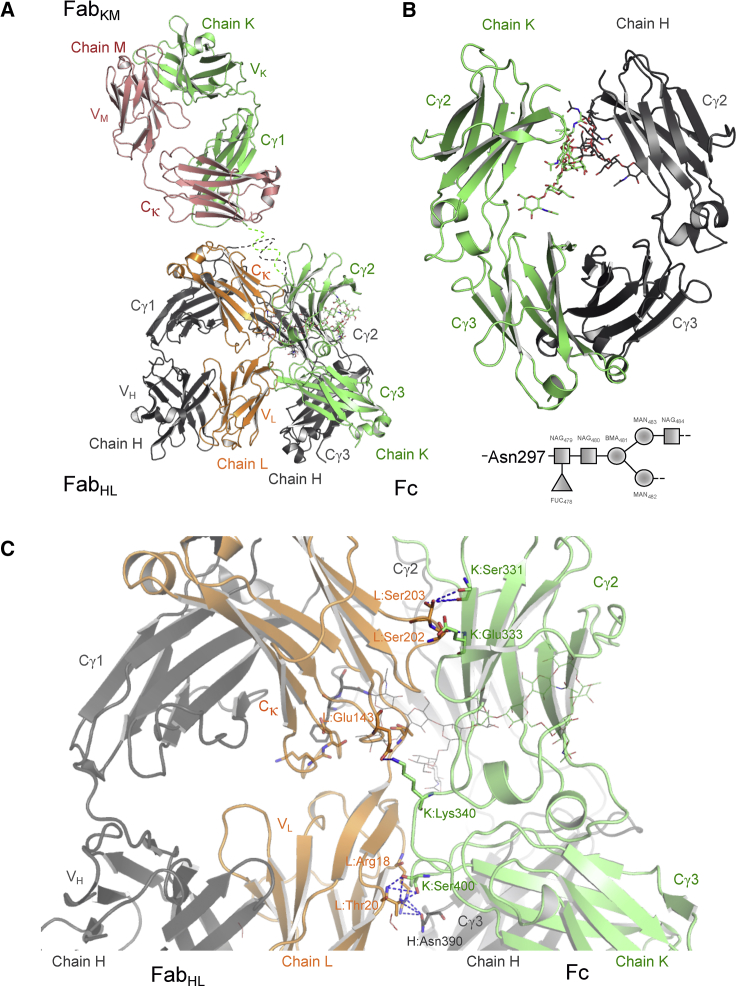
Figure 1.
Structural features of therapeutic anti-NPRA IgG4 antibody. (A) The crystal structure of anti-NPRA shows the unique/unusual λ-shaped conformation. The heavy chains K and H are highlighted in green and gray, respectively, and the two light chains L and M in orange and rose, respectively. Labels correspond either to domain or chain identifications. The glycan moieties are shown as sticks colored by atom type. The same color scheme and annotations are used in all figures. (B) Shown is the overall topology of the anti-NPRA Fc fragment depicting the conformational orientation of the internal carbohydrate moieties covalently linked to Asn297 in the Cγ2 domain in both heavy chains. Bottom: schematic drawing depicts the biantennary sequential arrangement of glycan elements. (C) Shown is a close-up view of the FabHL-Fc interface in the same orientation as in (A) showing a selection of the amino acids involved in polar contacts (dotted lines) at the interface. To see this figure in color, go online.
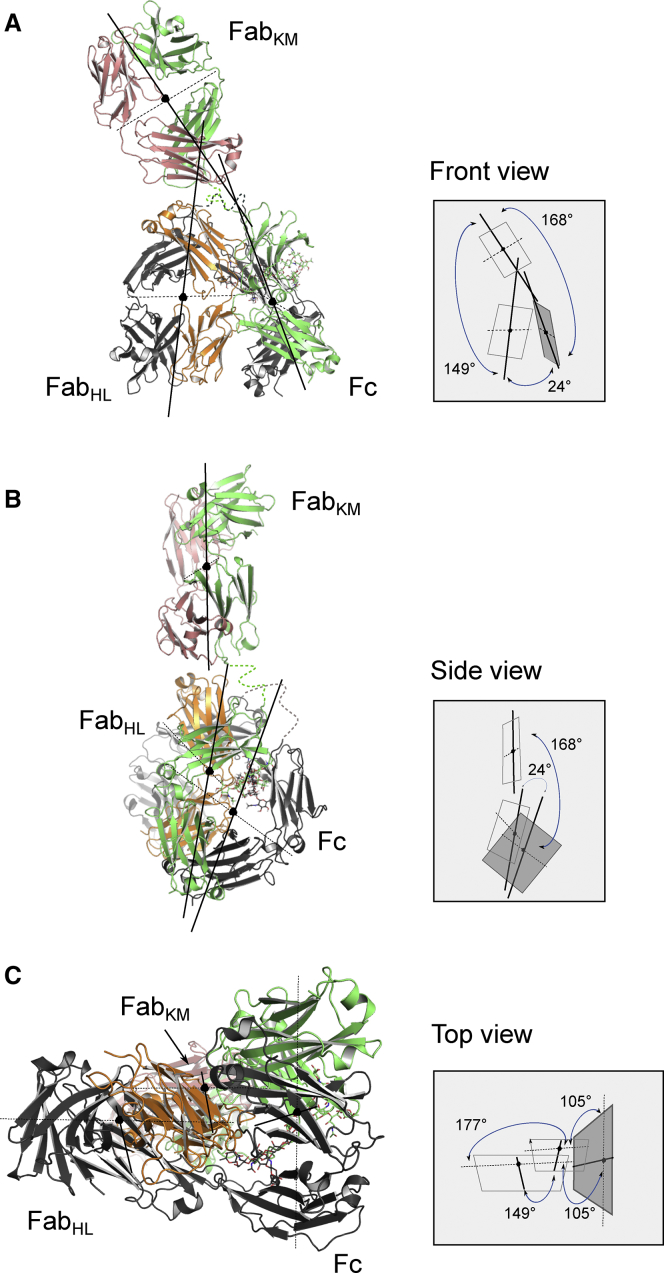
The anti-NPRA antibody domain assembly adopt the typical Ig fold characterized by an antiparallel β-sheet sandwich architecture. The superposition of isolated individual Fabs of anti-NPRA show that its overall structure is well preserved, which is reflected in the root mean-square deviation (RMSD) of 0.54 Å for main chain atoms. This indicates only slight conformational differences between both Fabs. Surprisingly, the domain architecture does not adopt a commonly displayed Y-shaped conformation. Instead, anti-NPRA adopts a rather unusual λ-shaped conformation with one Fab (FabHL) oriented toward the Fc-part (Figs. 1 A and 2). This can be quantified either by the angles of each individual Fab portion relative to the Fc or by an angle between the variable and constant domains (Fig. 2; Table 2; (5, 6, 7, 8, 9, 30)). The latter is referred to as the elbow angle and defined as the angle between the pseudo-twofold axes relating VL to VH and CL to CH1 (30) (Fig. 2). Therefore, the Fab elbow angle is a useful descriptor of the overall topology of the Fab. The anti-NPRA antibody exhibits elbow angles of 138° and 134° for its two Fab parts (FabHL and FabKM). The difference between these angles is comparable to x-ray structures of IgG1 b12 (PDB: 1HZH (6)) and IgG4 pembrolizumab (PDB: 5DK3 (9)).This suggests that there is no constraint regarding individual Fab conformation and orientation due to crystal contacts. The 177° rotation angle between the individual Fab portions (FabHL and FabKM) is nearly identical to all currently published full-length antibody structures.
Figure 2.
Asymmetry of anti-NPRA IgG4. Shown is the conformational arrangement of anti-NPRA emphasizing the Fab orientation relative to the Fc fragment. The crystal structure is shown on the left, and a schematic illustration of the relative domain positions and orientations is shown on the right. Major, minor, and depth axes of each Fab and the Fc are indicated by solid and dotted lines, respectively. The major and minor axes of each domain define the planes that are schematically shown on the right. The plane describing the Fc is highlighted in dark gray, and planes symbolizing the Fabs are transparent. The angles between the center of mass (black bullet) of the three functional parts (FabHL, FabKM, and Fc) are annotated according to Table 2. (A) Shown is the front view of anti-NPRA showing that FabHL is packed against the Fc portion, with one Cγ2 domain of the Fc in contact with the Cκ domain of FabHL. (B) Shown is a side view of anti-NPRA illustrating that FabHL and Fc are situated in a line such that FabHL is almost completely covered by the Fc. (C) Shown is the top view of anti-NPRA illustrating the rotation of FabHL relative to Fc. FabKM extends into the background underneath the FabHL-Fc interaction region. To see this figure in color, go online.
Table 2.
Comparison of Geometrical Parameters for Published Complete Antibody Structures
| Antibody | Subclass | PDB Entry | Elbow Angle FabHL/FabKM | Rotation Angle FabHL to FabKM | Angle between Major Axis of Fabs | Angle between Fab-Dyad and Fc | Angle between Fc and FabHL/FabKM | Radius of Gyration/Å |
|---|---|---|---|---|---|---|---|---|
| b12 | human IgG1 | 1hzh | 174°/168° (a) | 176° | 143° (a) | 174° | 109°/105° | 52.3 |
| Mab 61.1.3 | murine IgG1κ | 1igy | 155°/155° (b) | 180° (b) | 115° (b) | 107° (b) | 78°/123° (b) | 46.9 |
| Mab231 | murine IgG2aκ | 1igt | 159°/143° (c) | 176° | 168° | 128 | 65°/115° (c) | 53.8 |
| Mcg | human IgG1 | 1mco | 118°/118° (d) | 176° | 168° | 179 | 84°/84° | 46.8 |
| Pembrolizumab | IgG4 S228P | 5dk3 | 137°/167° (e) | 177° | 105° | 139 | 90°/161° | 46.4 |
| Anti-NPRA | IgG4 S228P | 6gfe | 138°/134° | 177° | 149° | 105 | 24°/168° | 48.6 |
Values for a(6), b(7), c(5), d(8), e(9), and anti-NPRA were calculated from deposited x-ray crystal structures according to Rupp et al. (30).
The most striking feature of the structure is the angle between the individual Fab portions and their relative orientation with respect to the Fc domain (Fig. 2). Consequently, anti-NPRA adopts a rather unusual λ-shaped conformation. We further assessed this phenomenon to gain insights into 1) anti-NPRA’s conformational diversity, 2) its potential impact under physiological conditions, and 3) its putative implications for biological function and for potential therapeutic treatment.
Fc-Fab interface
The FabHL is oriented toward the Fc portion, forming a large interface. This interface was analyzed in more detail using the PISA (Proteins, Interfaces, Structures, and Assemblies) software (53) (Fig. 1 C; Table 3). Because of the incomplete electron density, the polypeptide chain in the hinge region could not be completely assigned. In the crystals, several Fab copies within the unit cell surround the Fc-part. In principle, there are several options to build the complete antibody. However, two crystallographic independent Fab molecules are considerably closer to the Fc than the remaining copies. For the anti-NPRA full-length structure reported in this study, the distances between the last residue located in Cγ1 (H:Lys218, and K:Ser217) and the first residue in Cγ2 (H/K:Gly237) are 31 and 38 Å, respectively. In contrast, the two next closest Fab molecules are 53 and 58 Å away. Both are oriented in a way that it is impossible to form a functional full-length antibody. Moreover, a comparison to other reported complete antibody structures (PDB: 5DK3, 1HZH, 1IGT, and 1IGY) reveals that Fab and Fc are usually 36 Å apart. This agrees with the average length of the hinge region calculated for the residue range of 217–237. However, two possible assignments remain. The interaction of the Fc and FabHL can either be cis or trans, in the sense that they belong to the same or two different heavy chains. Although our data do not allow for a unique assignment in this regard, we assumed a cis interaction. Considering the cis interaction, all contributions to the FabHL-Fc interface are due to light chain residues (Fig. 1 C; Table 3). There is no contribution of Cγ1 domain. The interaction surface of the light chain L with the heavy chain K comprises an area of 448 Å2. In contrast, the interaction surface of light chain L and heavy chain H is considerably smaller (264 Å2). The VL domain interacts with both Cγ3 domains. All residues involved originate from strands A, A′, B, and E of VL (nomenclature according to IMGT Collier de Perles (54, 55); Gelfand and Kister (56)). Strand E is in contact with the AB-loop of the cis Cγ3, whereas L:Arg18 at the beginning of strand B forms a hydrogen bond to H:Asn390 at the N-terminal end of strand D in the cis Cγ3. The trans Cγ3 contributes with residues from the DE-loop, which contact the surface generated by strands A, A′, and B of VL. K:Ser400 (Cγ3) forms two hydrogen bonds to L:Thr20, one of them involving the backbone nitrogen and carbonyl atoms and the other one the two side chain oxygens. The elbow region of VL contacts the carbohydrate chain attached to H:Asn297 of the cis Cγ3. The fucose branching from the first N-acetyl-glucosamine forms a hydrogen bond to L:Thr109. In close proximity to this interaction, H:Phe296 forms a π-π interaction with the peptide bond of L:Lys169 and L:Asp170 from the DE-loop of Cγ1. Residues of the FG-loop of Cγ2 interact with strand G of Cγ2, in which K:Ser331 forms a hydrogen bond with L:Ser203 and K:Glu333 with L:Ser202 (Fig. 1 C; Table 3).
Table 3.
Analysis of the Interaction Interface of FabHL and Fc
| FabHL | Fc Portion | Distance/Å | Interaction |
|---|---|---|---|
| L:Ser 203 [OG] | K:Ser 331 [OG] | 3.8 | Hydrogen bond |
| L:Thr 20 [OG1] | K:Ser 400 [OG] | 2.7 | Hydrogen bond |
| L:Thr 20 [N] | K:Ser 400 [O ] | 2.9 | Hydrogen bond |
| L:Ser 202 [O] | K:Glu 333 [N ] | 2.7 | Hydrogen bond |
| L:Glu 143 [OE2] | K:Lys 340 [NZ] | 3.7 | Hydrogen bond |
| L:Glu 143 [OE2] | K:Lys 340 [NZ] | 3.7 | Salt bridge |
| L:Arg 18 [NE] | H:Asn 390 [OD1] | 3.3 | Hydrogen bond |
| L:Lys 169 | H:Phe 296 | 4.5 | π-π interaction |
| L:Asp 170 | H:Phe 296 | 4.5 | π-π interaction |
Data generated using PISA software (53).
FG-loop conformation of the Cγ2 domain
The FG-loops (H/K:Asn325–Ser331) of heavy chains H and K are structurally not well ordered as evident from the weaker electron density in these regions. However, the polypeptide chain could be traced in both chains. In heavy chain H, the loop adopts the same conformation as in the crystal structure of IgG1 (e.g., PDB: 1HZH (6)). Thus, H:Leu328 interacts via van der Waals contacts with H:Pro238 located in the lower hinge region. In heavy chain K, the FG-loop protrudes outward by 7 Å for K:Gly327 and 10 Å for K:Leu328 when compared to heavy chain H. This has also been observed for the recently determined structure of the Fc-part of pembrolizumab (9). In contrast to the findings of Scapin et al. (9), the FG-loop in our structure adopts an “out” conformation in both heavy chains. A comparison of anti-NPRA and the structure described by Davies et al. (11) revealed that the conformational space of the FG-loop in our structure is not restricted by crystal contacts. These findings suggest that the FG-loop in IgG4 possesses a greater flexibility compared to IgG1 and can adopt at least two different conformations. This can be attributed to the two amino acid exchanges from IgG1 to IgG4 in the heavy chain, namely Ala327Gly and Pro329Ser, both of which lead to higher conformational flexibility.
Cγ3-Cγ3 interaction characteristic for IgG4
The heavy chain residue Arg409 is characteristic for the human IgG4 Cγ3 domain (rather than the Lys409 found in human IgG1, IgG2, and IgG3 isotypes). The residue has recently been found important in FAE (11, 57). Arg409 in the anti-NPRA x-ray crystal structure adopts two different conformations in each of both heavy chains. Conformation one is identical to the conformation of Arg409 in heavy chain A of the published x-ray crystal structure (PDB: 4C54 (11)) of IgG4-Fc and is assumed by 70% of heavy chain H and 40% in chain K. Conformation two, which had been modeled in the published structure of IgG4-Fc (PDB: 4B53 (57)), is assumed by 30% of heavy chain H and 60% of heavy chain K. A comparison of the interactions between Arg409 and Asp399 revealed that they form bidentate intermolecular interactions in conformation one and a monodentate intermolecular interaction in conformation two. This indicates that conformation one possess a more stable Cγ3-Cγ3 interface. Another residue that differs between IgG1 and IgG4 Cγ3-Cγ3 interfaces is Glu356 (Asp356 in IgG1). In IgG1, residue Asp356 contributes to a salt bridge to Lys439. Because glutamate residues confer more conformational flexibility than aspartate and Glu356 at this position generally adopts different conformations in IgG4 crystal structures, we cannot assign specific interactions to Lys439. However, Glu356 and Lys439 are in close proximity (4 Å).
SAXS
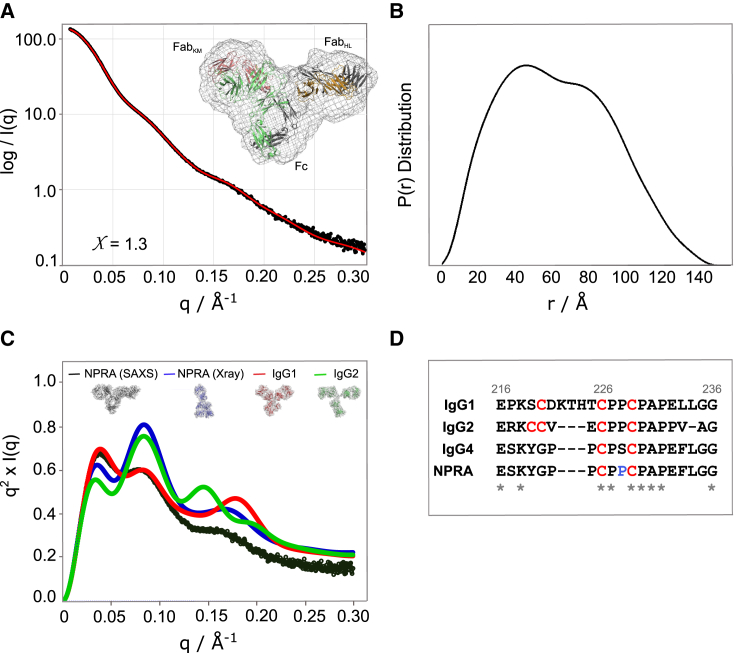
The SAXS analysis obtained the anti-NPRA mAb in solution as a globular molecule with a radius of gyration of 48 Å (±0.46) and a maximal diameter of 147 Å. These findings correspond well to the calculated radius of gyration of the x-ray crystal structure of 48.6 Å (Table 2). The bimodal P(r) curve was consistent with a T-shape or a Y-shaped molecule, which is in alignment with previously reported data. Three visible peaks in the Kratky plot indicate that the molecule has multiple domains connected to flexible linkers (58). The bimodal P(r) curve was also consistent with a T-shape or a Y-shaped molecule, which is in alignment with previously reported data (59, 60, 61) (Fig. 3 B). Three visible peaks in the dimensionless Kratky plot (Fig. 3 C) indicate that the molecule has multiple domains connected to flexible linkers (58). When compared to the anti-NPRA crystal structure (blue), the experimentally derived solution structure (black) shows a distinct conformational change in the second peak. This is not unexpected when comparing a solution with crystal structures. However, when both these curves are further compared to crystal structures of IgG1 (red) and IgG2 (green), it is apparent that the crystal structure adopts a conformation more similar to the Y-shaped IgG1, whereas the solution structure is more similar to the T-shaped IgG2 (Fig. 3 C). To identify a preferred conformation, rigid body fitting of the anti-NPRA antibody to the SAXS data was performed. A T-shaped model with a χ2 value of 1.3, indicating a good fit of the model to the experimental data, was found, which was also a visually good fit to the averaged DAMMIF model (Fig. 3, A and D). The independent refinements showed large changes in the positions of the Fab/Fc domains with the T-shaped and λ conformers predominating but the Y-shaped model also occurring (Fig. 4 C). This further supports the notion of the flexibility of individual domains in solution.
Figure 3.
Solution scattering data. (A) Scattering intensity plot is shown. Log SAXS intensity versus scattering vector, q, is shown. Plotted range represents the positive only data within the specified q range. Best fit between the calculated scattering curve from the best ensemble (red) and the experimental data (black) is shown. Rigid body modeling of anti-NRPA is shown. Shown is the best fitting conformer superimposed into ab initio bead model, an average of 20 runs. (B) The pair-distance P(r) distribution function is shown. Maximal dimension, dmax, is the largest non-negative value that supports a smooth distribution function. (C) The Kratky plot is shown. The first peak position for globular particles is dependent on the size. Anti-NPRA solution scattering data (black) and crystal structure (blue) are superimposed to crystal structures of IgG1 (PDB: 1IGY) (red) and IgG2 (PDB: 1IGT) (green) representing conformer ensembles of T-like, λ, Y, and T conformations, respectively. (D) A comparison of the primary sequence of hinge regions in three IgG subclasses is shown. Sequence conservation across IgG subclasses are indicated by an asterisk. To see this figure in color, go online.
Figure 4.
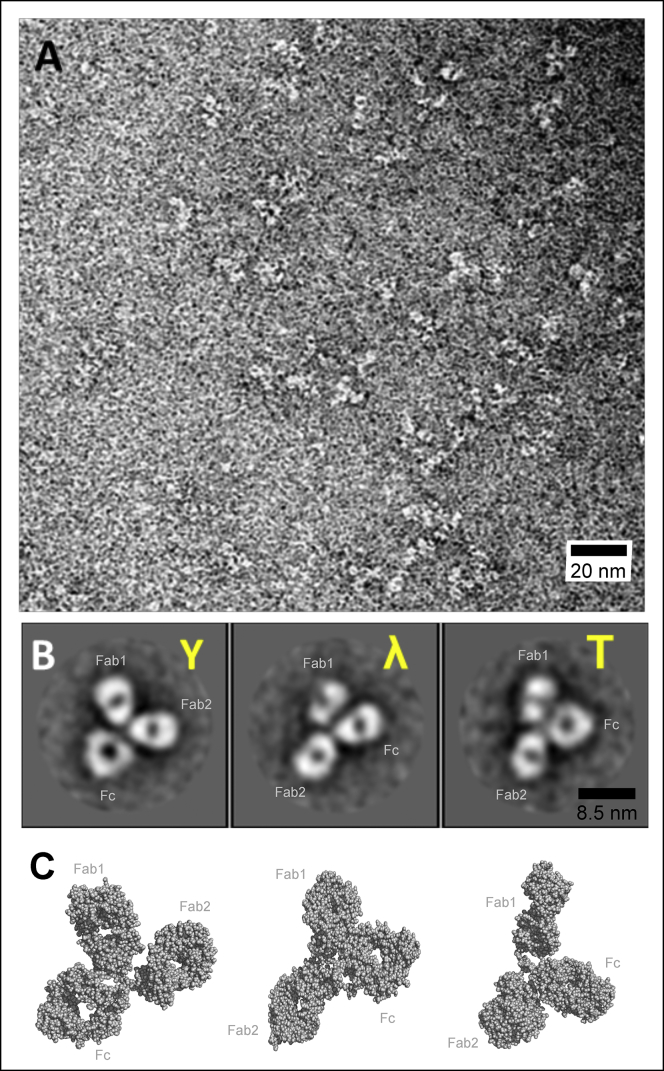
High-resolution images of anti-NPRA by negative-stain TEM. (A) Survey view of anti-NPRA with an increasing zoom level is shown. Shown are the contours of anti-NPRA IgG4 molecules feature different shapes that overall assemble in three globular circular or oval domains (2× Fab, 1× Fc). (B) 3 representative high-resolution views of selected anti-NPRA molecules demonstrate varying conformations and orientations. The scale bar in (A) corresponds to 20 nm, and the scale bar in (B) to 8.5 nm. (C) Shown is the visualization of conformer flexibility in solution by representative conformer ensembles resulting from SAXS data at similar orientations as in (B). To see this figure in color, go online.
Assessing the conformational space by TEM and 2D image analysis
The conformational space of the NPRA antibody dissolved in water was further investigated by 2D image analysis of negative-stain TEM images (Fig. 4 A). Single-particle images were semiautomatically picked from the micrographs. Several rounds of 2D classification and particle sorting resulted in a final data set comprising 4136 particles. Notably, reference images that were used for the semiautomatic particle selection did not distinguish between Y and λ conformers, and the 2D-classification method was reference free. 2D classification after the last round revealed that most antibody particles demonstrate a Y- or T-shaped conformation, whereas three class averages representing ∼10% of the data set showed antibody conformers that are similar to the λ-shaped crystal structure (Fig. 4, A and B). Despite the limited resolution of the TEM images, they support the notion of a dynamic conformational equilibrium and, hence, corroborate the findings of our x-ray structure determination, SAXS analysis, and MD simulations (see next paragraph).
MD simulations reveal conformational diversity
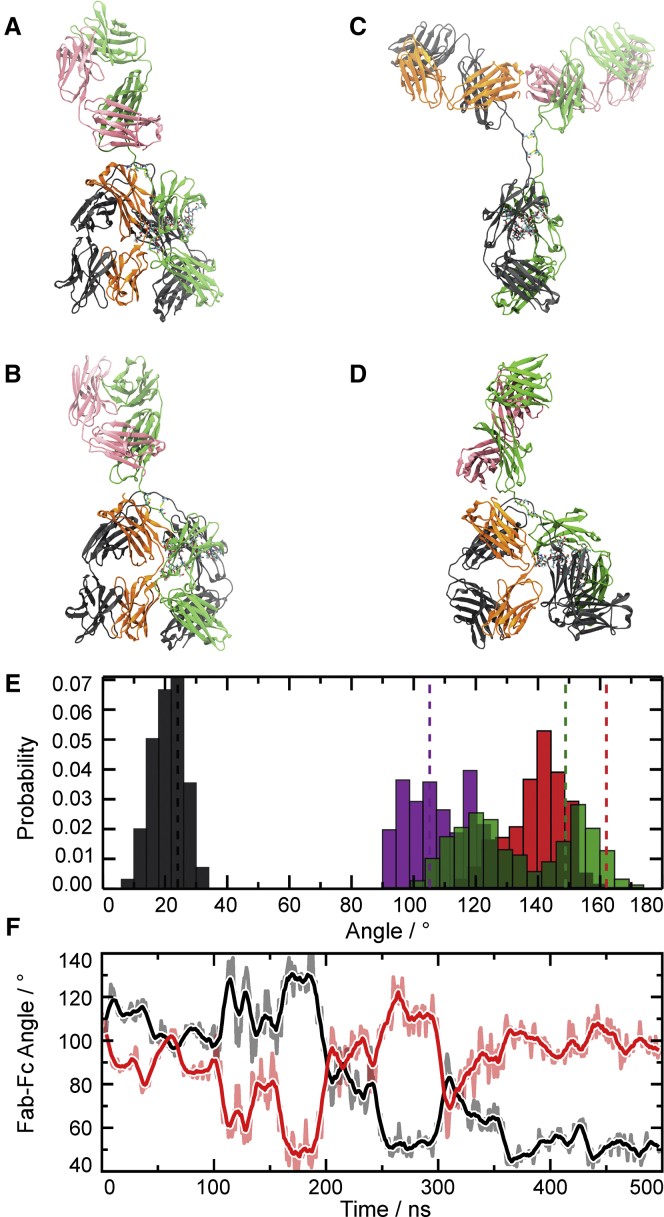
To more closely investigate the structural dynamics and energetic stability of anti-NPRA IgG4 in aqueous solution (as opposed to the crystal lattice), we carried out all-atom MD simulations. Two different structures were simulated (Fig. 5, A and C): the λ-shaped conformer present in the x-ray crystal structure and an (idealized) Y-shaped conformer that was obtained by fitting the individual domains of IgG4 onto the corresponding domains in the IgG1 structure of Padlan (44). The individual antibody domains are structurally very stable in the simulations (Cα-RMSD from the starting structure 2–4 Å, see Fig. S2). For the λ conformer, the Fab-Fc interface seen in the x-ray crystal structure is stable on the 500 ns timescale of the MD simulation. The only large-amplitude motion observed is that of the FabKM domain, which is not involved in the interface and can thus move freely relative to the other parts of the molecule (Fig. 5 E).
Figure 5.
MD Simulations of anti-NPRA. (A) X-ray crystal structure of λ-shaped conformation of anti-NPRA used as starting structure for the MD simulations is shown. (B) Shown is the structure of the λ conformer after 500 ns of MD simulation started from the x-ray structure. (C) Y-shaped conformation of anti-NPRA used as an alternative starting structure for MD is shown. (D) Shown is the structure after Y-to-λ conformational transition observed during MD simulation. (E) Shown is the distribution of angles (cf. Table 2) obtained from two 500 ns simulations of the λ-shaped conformer. The FabHL-Fc and FabKM-Fc angles are shown in black and red, respectively; the angle between the Fab-dyad and Fc is shown in magenta, and the angle between the two Fab domains is shown in green. The dashed lines depict the corresponding angle in the x-ray crystal structure. (F) Shown is the time course of the FabHL-Fc and FabKM-Fc angles (black and red curves, respectively) during representative MD simulation of Y-to-λ transition. To see this figure in color, go online.
Likewise, in the majority of the MD simulations started from the Y conformer, no large-scale domain rearrangements are observed. The antibody is slightly more compact and adopts a more T-shaped (rather than Y-shaped) conformation. Interestingly, in two of the ten MD simulations initiated from the Y-shaped conformation, large-scale rearrangements of the domains toward a λ-like conformer are observed (Fig. 5, D and F). The FabHL and FabKM domains are very mobile with respect to the Fc domain (Fig. 5 F), and after 500 ns of MD simulation, FabHL is in contact with Fc (Fig. 5 D). Although the global shape of the MD λ structure is similar to the x-ray structure, the relative orientations of the domains differ (compare Figs. 5, B and D). We speculate that this difference might be due to the limited timescale of the MD simulations, which only enable the formation of a first encounter complex on the way toward a λ structure that more closely matches the x-ray structure. To this end, the MD conformer (Fig. 5 D) would need to undergo further structural rearrangements, including a rotation of the individual domains with respect to each other, which are expected to happen on very long timescales that are inaccessible to MD simulations. The simulations nonetheless indicate that anti-NPRA adopts a dynamic equilibrium between different stable conformers in solution, including λ- and Y-shaped conformations.
To establish which of these conformers is preferred in solution, we calculated the free energy difference between the two conformers, ΔG = Gλ − GY, by combining our all-atom MD simulations with 3D-RISM theory (see Methods; Supporting Materials and Methods; Fig. S3). The free energy difference is estimated from the conformational enthalpies, configurational entropies, and hydration free energies according to ΔG = ΔHc − TΔSc + ΔGhyd. Our results show that because of the tight packing of the Fab to the Fc domain, the conformational enthalpy of the λ-shaped conformer is lower (more negative) than that of the Y-shaped conformer by ΔHc = −521 ± 50 kcal/mol; this contribution thus strongly favors the λ-shaped structure. Interestingly, however, this enthalpy difference is more than offset by the unfavorable free energy of hydration of the λ-shaped conformer, which differs from the Y conformer’s by ΔGhyd = +626 ± 33 kcal/mol. Because configurational entropy differences between the two conformers are comparably small (−TΔSc = −19 ± 12 kcal/mol), our simulations indicate that the Y-shaped conformer is preferred in solution, by ΔG = −86 ± 61 kcal/mol. Given the physical approximations and large statistical uncertainties, this result should be interpreted with care and only at a qualitative level. We conducted a number of control calculations under varying conditions, however, and these confirm this finding (see Supporting Materials and Methods). Our simulations hence provide atom-level insights into the molecular driving forces behind the dynamic conformational equilibrium, including an explanation for the observed presence of the λ-shaped conformer in the crystal structure. The lower solubility (less favorable hydration free energy) of the λ conformer primes it for crystallization. Upon crystal formation, the λ conformer is removed from the dynamic equilibrium between λ and Y conformers in solution, and according to Le Chatelier’s principle, the system will readjust itself by shifting the equilibrium toward the λ-shaped conformer.
Discussion
We report the full-length anti-NPRA x-ray crystal structure at 1.8 Å resolution. The anti-NPRA IgG4 is the sixth x-ray crystal structure of a complete IgG and the second complete IgG4 structure. Crystal structures of individual parts of IgG4 were previously reported (e.g., for the Fc portion (4C54 (11), 4C55 (11), 1ADQ (62)), the Cγ3 domain (PDB: 4B53 (57)) and a Fab (PDB: 1BBJ (63))). The crystal structures of the isolated Cγ3 domain (PDB: 4B53 (57)) and the IgG4 Fc portion (PDB: 4C54 and 4C55) can be superimposed on the Cγ3 domains of anti-NPRA with RMSD of 0.30 Å and 0.14 Å, respectively (Fig. S4; Table 4). In agreement with the finding of Davies et al. (11, 57), Arg409 plays a crucial role for the stability of the Cγ3-Cγ3 interface and for FAE, a phenomenon unique to IgG4. Arg409 can adopt two different conformations, producing a less well-defined hydrogen bonding network as compared to IgG1 structures. The exchange of Glu356 (IgG4) for Asp356 (IgG1) adds another residue with increased flexibility, most likely facilitating susceptibility for half-molecule loss. This contributes to previous findings that IgG4 molecules can naturally evolve as monovalent half molecules or bivalent full molecules with mono- or bispecificity in serum (11, 12, 64, 65). The latter depends on whether the interchain disulfide bonds are formed between identical or different IgG4 half molecules (66). The generation of mono- or bispecific IgG4 molecules was shown to depend on the dynamic equilibrium between inter- and intrachain disulfide bonds of the Cys-Pro-Ser-Cys motif within the hinge region, driven by the relative amounts of both parent IgG4 molecules (64). To prevent FAE in this antibody while making use of the other features of the IgG4 framework, Ser228 was substituted by Pro (Fig. 3 D).
Table 4.
Fc Portion Alignment of Currently Published X-Ray Structures
| Fc Portion 1 | Chain | Residues | Fc Portion 2 | PDB Entry | Chain | Residues | RMSD/Åa | |
|---|---|---|---|---|---|---|---|---|
| Anti-NPRA | H | 237–443 | Pembrolizumab | 5DK3 | B | 219–443 | 9.573 | NPRA versus 5DK3 (isolated Fc portions) |
| Anti-NPRA | H | 237–443 | Pembrolizumab | G | 219–443 | 0.939 | ||
| Anti-NPRA | K | 237–443 | Pembrolizumab | B | 219–443 | 9.166 | ||
| Anti-NPRA | K | 237–443 | Pembrolizumab | G | 219–443 | 0.799 | ||
| Anti-NPRA | H | 237–443 | rhu IgG4-Fc | 4C54 | A | 236–443 | 0.851 | NPRA versus 4C54 |
| Anti-NPRA | H | 237–443 | rhu IgG4-Fc | B | 236–443 | 0.851 | ||
| Anti-NPRA | K | 237–443 | rhu IgG4-Fc | A | 236–443 | 0.509 | ||
| Anti-NPRA | K | 237–443 | rhu IgG4-Fc | B | 236–443 | 0.719 | ||
| Anti-NPRA | H+K | 237–443 | rhu IgG4-Fc | A+B | 236–443 | 0.829 | ||
| Anti-NPRA | H | 237–443 | sdhu IgG4-Fc | 4C55 | A | 237–443 | 0.988 | NPRA versus 4C55 |
| Anti-NPRA | H | 237–443 | sdhu IgG4-Fc | B | 237–443 | 0.515 | ||
| Anti-NPRA | K | 237–443 | sdhu IgG4-Fc | A | 237–443 | 0.668 | ||
| Anti-NPRA | K | 237–443 | sdhu IgG4-Fc | B | 237–443 | 0.644 | ||
| Anti-NPRA | H+K | 237–443 | sdhu IgG4-Fc | A+B | 237–443 | 0.754 |
Values are generated by superimposition of individual Fc portions as outlined using PyMOL (29).
Interestingly, Scapin et al. reported recently an unusual structure of a therapeutic IgG4 antibody (pembrolizumab) with a Cγ2 domain rotated by 120° compared to all other Fc structures reported so far (9). One of the resolved glycan moieties faces the solvent and is not buried between the two Cγ2 domains as has been found in all other crystal structures to date (9). Our structural investigations of anti-NPRA do not reveal the unusual Cγ2 domain orientation found for pembrolizumab (5DK3 (9)), showing instead the common canonical Fc conformation. However, the anti-NPRA IgG4 full-length antibody structure studied here differs substantially from all other crystal structures reported so far in the relative Fab-Fc orientation.
We showed by 2D classification of negative-stain electron microscopy images that conformers resembling the described crystal structure also occur in solution. This is further substantiated by solution SAXS measurements, in which the Kratky analysis showed a distinct change between the crystal and solution structures with the solution structure resembling the T-shaped IgG2 structure. The following rigid body modeling showed a high flexibility of each individual domain, with the most favored model resembling the T-shaped conformation (Fig. 3), though λ conformers were well represented (Fig. 4). Similarly, previous SAXS data from a monoclonal IgG4 also showed a flexible behavior for the antibody with the preferred conformer in the λ-shape (61). In addition, MD simulations and 3D-RISM calculations revealed a less favorable hydration free energy for the λ conformer than for the Y conformer. The corresponding 2D class averages represent only 10% of the data set, suggesting a lower solubility of the λ conformer in this experiment. Nevertheless, a population of 2% of all investigated images exhibits a Fab-Fc angle similar to the λ conformer described above, even though the preferred angle is ∼90°.
Although the anti-NPRA IgG4 molecule possesses high flexibility, its biological function to its natural antigen remains unaffected. Its therapeutic antigen, the NPRA, can either bind to both Fab arms in the Y conformation or only to a single Fab-arm not involved in Fab-Fc interaction (e.g., FabKM in Fig. 3, A and C) in the λ conformation. Antigen binding might be affected when anti-NPRA adopts the λ conformation and the Fab-arm is oriented toward the Fc portion (e.g., FabHL in Fig. 3 A). This hardly depends on which conformation is preferred in solution. However, anti-NPRA has been shown to specifically recognize the NPRA extracellular domain bound to its natural natriuretic cyclic peptide ligands ANP and BNP in vivo. Anti-NPRA enhances ANP- and BNP-dependent cyclic guanosine monophosphate production in human embryonic kidney-presenting NPRA cells at physiologically relevant levels of ANP and BNP, resulting in a reduction of observed EC50 values when compared to its control (Fig. S1).
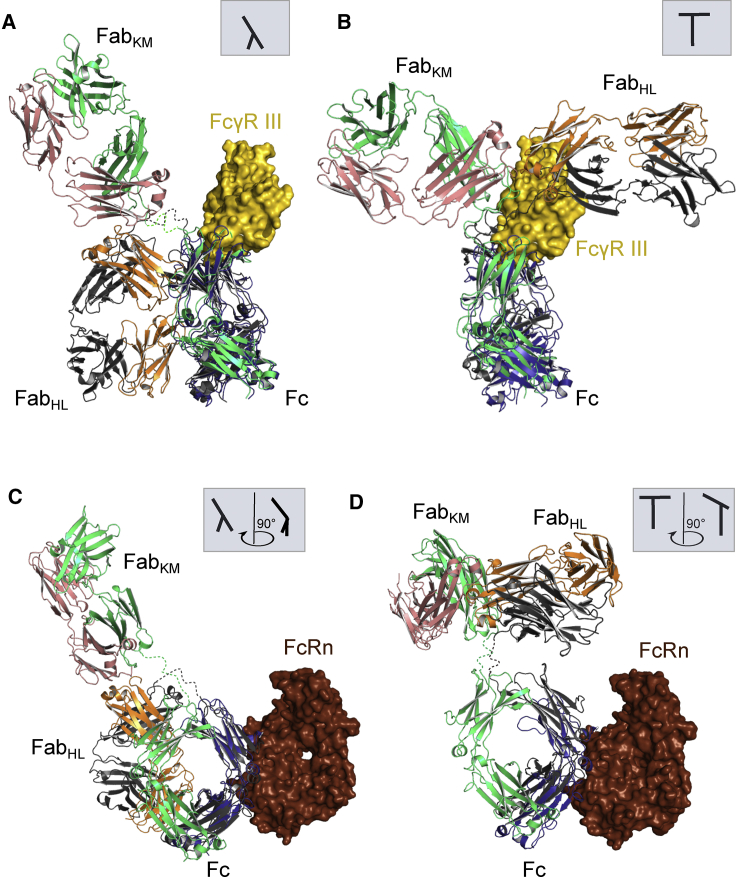
One of the key features of IgG antibodies is that they interact with the immune system via Fc domain binding to specific Fcγ receptors on immune cells, thereby provoking antibody-dependent cell-mediated cytotoxicity, complement activation, cytokine release, and antibody-dependent cell-mediated phagocytosis (13, 67, 68). Human immune cells express four activating Fcγ receptors: FcγRI (CD64), FcγRIIa (CD32a), FcγRIIc (CD32c), and FcγRIIIa (CD16a) (13, 67). The human FcγRs all have similar overall structures and bind IgGs in the same general way, although specific differences clearly exist between each receptor and each IgG isotype to permit differential binding and activation kinetics (68, 69). To assess anti-NPRA’s potential immune effector mechanisms, we focused on FcγRIII as a representative of its family and the FcRn, known to be responsible for the antibody clearance mechanism by FcRn-mediated recycling (half-life in human serum) (13, 67, 69). Superimposition of the co-crystal structure of IgG-Fc in complex with soluble recombinant FcγRIII with the T conformer of anti-NPRA IgG4 (Fig. 6 B) revealed a substantial overlap of both structures in the lower hinge and hinge-proximal Cγ2 domain with the anti-NPRA Fab constant domains. FcγRIII binding will not be affected by steric hindrance when the anti-NPRA λ-shaped conformation is considered (Fig. 6 A). In contrast, the superimposition of anti-NPRA with FcRn demonstrates that FcRn can bind without restriction between Cγ2 and Cγ3 to anti-NPRA independent of its structural conformation (Fig. 6, C and D). These findings underscore the importance of structural dynamics and conformational flexibility of antibodies in solution. Depending on the anti-NPRA‘s conformation and distribution in equilibrium, its interaction with FcγRIII might be impeded, or the anti-NPRA’s high structural flexibility might allow it to adapt its conformation and thus bind the receptor (see Supporting Materials and Methods, Aalberse et al. (64, 65)) or C1q. On the other hand, previous studies reported that the Fab conformations in IgG4 in solution might be the key driver for restricting access to the Fc portion and thereby impair functional activity (70, 71, 72). The complement Fc binding sites are hindered by the Fab regions, explaining the loss of activity. Consequently, the human IgG4 subclass does not activate complement (71). In summary, the crystal structure of human anti-NPRA IgG4 reported in this work is only the sixth atomic-resolution structure of a full-length mAb. The complete IgG4 antibody structure described illustrates a large degree of asymmetry (both interdomain and intradomain) and structural flexibility. It shows dynamics that allow the Fab and Fc domains to rotate, flex, and extend. All data and analyses point to a substantial conformational plasticity of the mAb in solution, which encompasses different conformational states, ranging from the unusual λ-shaped conformation observed in our x-ray crystal structure to more canonical T- and Y-shaped structures. The structure of anti-NPRA IgG4 provides snapshots of the wide range of conformations underlying the molecule′s IgG4-specific biological diversity. Ultimately, the high resolution of our crystal structure gives detailed insight into the structural biology of the mAb interacting with its biological targets, represented by the cyclic peptide ANP, the Fc-γ RIII receptor, and the FcRn.
Figure 6.
Proposed binding of anti-NPRA IgG4 to FcγRIII and FcRn receptors. Structures of the FcγRIII-IgG1 Fc complex (PDB: 1E4K) and the co-crystal structure of neonatal Fc receptor (PDB: 4NOU) are superimposed with anti-NPRA, with the A- and E-chains (both blue) used as reference, respectively. (A) Interactions of FcγRIII (gold) with the anti-NPRA Fc fragment (gray, chain H; green, chain K) are shown. Considering the λ-shaped conformation of anti-NPRA, the configuration illustrates that there is no steric hindrance for FcγRIII binding. (B) In contrast, considering the T-shaped conformation of anti-NPRA, FcγRIII binding is heavily affected. The figure illustrates a substantial structural overlap of FcγRIII with FabHL and the hinge region. (C) Shown is the proposed binding of the neonatal Fc receptor (dark red) with anti-NPRA Fc. The λ-shaped conformation of anti-NPRA allows for two binding possibilities. Depending on which chains are superimposed, either by superposing FcRn to heavy chain or to heavy chain K, a different relative orientation of FcRn to FabHL is obtained. The depicted configuration revealed no structural overlap of FcRn with FabHL, allowing it to exert effector function. Considering a FcRn position opposite from the one depicted in (C), FcRn could interfere sterically with FabHL and thus function may likely be affected. (D) For the T-shaped conformation, both Fabs are oriented such that sterical hindrance is unlikely. To see this figure in color, go online.
The structure has been deposited with the PDB (PDB: 6GFE).
Author Contributions
P.G., M.B., L.V.S., and S.H. prepared and designed the study. H.K. and Y.Z. set up the crystallization experiments. S.H. was responsible for the x-ray investigation. M.W. performed the TEM experiments, and S.K. performed the 2D image analysis. A.B.K., H.G., S.M.K, Y.A., and L.V.S. performed and analyzed the simulations. M.D.T. performed SAXS data analysis. The manuscript was written by M.B., S.H., and P.G., and all authors edited the manuscript.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany`s Excellence Strategy (EXC-2033) project number 390677874 to L.V.S. and S.M.K. and Emmy Noether grant SCHA 1574/3-1 to L.V.S.
Editor: Michael Sattler.
Footnotes
Michaela Blech and Stefan Hörer contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.03.036.
Contributor Information
Michaela Blech, Email: michaela.blech@boehringer-ingelheim.com.
Patrick Garidel, Email: patrick.garidel@boehringer-ingelheim.com.
Supporting Material
References
- 1.Kellner C., Derer S., Peipp M. Boosting ADCC and CDC activity by Fc engineering and evaluation of antibody effector functions. Methods. 2014;65:105–113. doi: 10.1016/j.ymeth.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Scallon B.J., Tam S.H., Raju T.S. Higher levels of sialylated Fc glycans in immunoglobulin G molecules can adversely impact functionality. Mol. Immunol. 2007;44:1524–1534. doi: 10.1016/j.molimm.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Raju T.S. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol. 2008;20:471–478. doi: 10.1016/j.coi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Lee C.C., Perchiacca J.M., Tessier P.M. Toward aggregation-resistant antibodies by design. Trends Biotechnol. 2013;31:612–620. doi: 10.1016/j.tibtech.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Harris L.J., Larson S.B., McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- 6.Saphire E.O., Parren P.W., Wilson I.A. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 7.Harris L.J., Skaletsky E., McPherson A. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 1998;275:861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- 8.Guddat L.W., Herron J.N., Edmundson A.B. Three-dimensional structure of a human immunoglobulin with a hinge deletion. Proc. Natl. Acad. Sci. USA. 1993;90:4271–4275. doi: 10.1073/pnas.90.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scapin G., Yang X., Strickland C. Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab. Nat. Struct. Mol. Biol. 2015;22:953–958. doi: 10.1038/nsmb.3129. [DOI] [PubMed] [Google Scholar]
- 10.Potter L.R., Abbey-Hosch S., Dickey D.M. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr. Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 11.Davies A.M., Rispens T., Sutton B.J. Structural determinants of unique properties of human IgG4-Fc. J. Mol. Biol. 2014;426:630–644. doi: 10.1016/j.jmb.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies A.M., Sutton B.J. Human IgG4: a structural perspective. Immunol. Rev. 2015;268:139–159. doi: 10.1111/imr.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dübel S., Reichert J.M. John Wiley & Sons; Hoboken, NJ: 2014. Handbook of Therapeutic Antibodies. [Google Scholar]
- 14.Saphire E.O., Stanfield R.L., Wilson I.A. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson I.C., Fowler S.B., Webster C.I. Monovalent IgG4 molecules: immunoglobulin Fc mutations that result in a monomeric structure. MAbs. 2013;5:406–417. doi: 10.4161/mabs.23941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothe C., Urlinger S., Urban M. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J. Mol. Biol. 2008;376:1182–1200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 17.Bergemann K., Eckermann C., Pisch-Heberle S. Handbook of Therapeutic Antibodies. Wiley-VCH; 2007. Production and downstream processing; pp. 199–237. [Google Scholar]
- 18.Jacobi A., Eckermann C., Subramanian G. Bioseparation and Bioprocessing. Wiley-VCH; 2007. Developing an antibody purification process; pp. 431–457. [Google Scholar]
- 19.Walker J.M. Humana Press; New York: 2002. Protein Protocols. [Google Scholar]
- 20.Garidel P., Kliche W., Thierolf M. Characterization of proteins and related analytical techniques. In: Mahler H.C., Borchard G., Lueßen H., editors. Protein Pharmaceuticals- Formulation, Analytics & Delivery. Editio Cantor Verlag; 2010. pp. 44–89. [Google Scholar]
- 21.Leslie A.G.W., Powell H.R. Evolving Methods for Macromolecular Crystallography. Springer; 2007. Processing diffraction data with MOSFLM; pp. 41–51. [Google Scholar]
- 22.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 24.Vonrhein C., Flensburg C., Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy A.J., Grosse-Kunstleve R.W., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smart O.S., Womack T.O., Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen V.B., Arendall W.B., III, Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrödinger LLC . 2008. The PyMOL Molecular Graphics System.https://pymol.org/2/ [Google Scholar]
- 30.Stanfield R.L., Zemla A., Rupp B. Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheres S.H. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 38.Hornak V., Abel R., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindorff-Larsen K., Piana S., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirschner K.N., Yongye A.B., Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 43.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 44.Padlan E.A. Anatomy of the antibody molecule. Mol. Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 45.Schlitter J. Estimation of absolute and relative entropies of macromolecules using the covariance matrix. Chem. Phys. Lett. 1993;215:617–621. [Google Scholar]
- 46.Beglov D., Roux B. Solvation of complex molecules in a polar liquid: an integral equation theory. J. Chem. Phys. 1996;104:8678–8689. [Google Scholar]
- 47.Kovalenko A., Hirata F. Three-dimensional density profiles of water in contact with a solute of arbitrary shape: a RISM approach. Chem. Phys. Lett. 1998;290:237–244. [Google Scholar]
- 48.Heil J., Kast S.M. 3D RISM theory with fast reciprocal-space electrostatics. J. Chem. Phys. 2015;142:114107. doi: 10.1063/1.4914321. [DOI] [PubMed] [Google Scholar]
- 49.Imai T., Harano Y., Hirata F. A theoretical analysis on hydration thermodynamics of proteins. J. Chem. Phys. 2006;125:24911. doi: 10.1063/1.2213980. [DOI] [PubMed] [Google Scholar]
- 50.Kovalenko A., Hirata F. Self-consistent description of a metal–water interface by the Kohn–Sham density functional theory and the three-dimensional reference interaction site model. J. Chem. Phys. 1999;110:10095–10112. [Google Scholar]
- 51.Luchko T., Gusarov S., Kovalenko A. Three-dimensional molecular theory of solvation coupled with molecular dynamics in Amber. J. Chem. Theory Comput. 2010;6:607–624. doi: 10.1021/ct900460m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beglov D., Roux B. An integral equation to describe the solvation of polar molecules in liquid water. J. Phys. Chem. B. 1997;101:7821–7826. [Google Scholar]
- 53.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Lefranc M.P., Giudicelli V., Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lefranc M.P., Pommié C., Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 56.Gelfand I.M., Kister A.E. Analysis of the relation between the sequence and secondary and three-dimensional structures of immunoglobulin molecules. Proc. Natl. Acad. Sci. USA. 1995;92:10884–10888. doi: 10.1073/pnas.92.24.10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies A.M., Rispens T., Sutton B.J. Crystal structure of the human IgG4 C(H)3 dimer reveals the role of Arg409 in the mechanism of Fab-arm exchange. Mol. Immunol. 2013;54:1–7. doi: 10.1016/j.molimm.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 58.Walenta E. Small angle x-ray scattering. Von O. GLATTER und O. KRATKY. London: Academic Press Inc. Ltd. 1982. ISBN 0–12–286280–5. X, 515 Seiten, geb. £ 43,60; US $ 81.00. Acta Polym. 1985;36 296–296. [Google Scholar]
- 59.König N., Paulus M., Tolan M. Antibodies under pressure: a small-angle X-ray scattering study of immunoglobulin G under high hydrostatic pressure. Biophys. Chem. 2017;231:45–49. doi: 10.1016/j.bpc.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 60.Lu Y., Harding S.E., García de la Torre J. Solution conformation of wild-type and mutant IgG3 and IgG4 immunoglobulins using crystallohydrodynamics: possible implications for complement activation. Biophys. J. 2007;93:3733–3744. doi: 10.1529/biophysj.107.108993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian X., Vestergaard B., Langkilde A.E. In-depth analysis of subclass-specific conformational preferences of IgG antibodies. IUCrJ. 2015;2:9–18. doi: 10.1107/S205225251402209X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corper A.L., Sohi M.K., Sutton B.J. Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody-antigen interaction. Nat. Struct. Biol. 1997;4:374–381. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]
- 63.Brady R.L., Edwards D.J., King D.J. Crystal structure of a chimeric Fab′ fragment of an antibody binding tumour cells. J. Mol. Biol. 1992;227:253–264. doi: 10.1016/0022-2836(92)90695-g. [DOI] [PubMed] [Google Scholar]
- 64.Aalberse R.C., Schuurman J. IgG4 breaking the rules. Immunology. 2002;105:9–19. doi: 10.1046/j.0019-2805.2001.01341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aalberse R.C., Stapel S.O., Rispens T. Immunoglobulin G4: an odd antibody. Clin. Exp. Allergy. 2009;39:469–477. doi: 10.1111/j.1365-2222.2009.03207.x. [DOI] [PubMed] [Google Scholar]
- 66.Salfeld J.G. Isotype selection in antibody engineering. Nat. Biotechnol. 2007;25:1369–1372. doi: 10.1038/nbt1207-1369. [DOI] [PubMed] [Google Scholar]
- 67.Strohl W.R., Strohl L.M. Elsevier; Amsterdam, the Netherlands: 2012. Therapeutic Antibody Engineering: Current and Future Advances Driving the Strongest Growth Area in the Pharmaceutical Industry. [Google Scholar]
- 68.Radaev S., Sun P. Recognition of immunoglobulins by Fcgamma receptors. Mol. Immunol. 2002;38:1073–1083. doi: 10.1016/s0161-5890(02)00036-6. [DOI] [PubMed] [Google Scholar]
- 69.Sondermann P., Kaiser J., Jacob U. Molecular basis for immune complex recognition: a comparison of Fc-receptor structures. J. Mol. Biol. 2001;309:737–749. doi: 10.1006/jmbi.2001.4670. [DOI] [PubMed] [Google Scholar]
- 70.Abe Y., Gor J., Dalby P.A. Masking of the Fc region in human IgG4 by constrained X-ray scattering modelling: implications for antibody function and therapy. Biochem. J. 2010;432:101–111. doi: 10.1042/BJ20100641. [DOI] [PubMed] [Google Scholar]
- 71.Rayner L.E., Hui G.K., Perkins S.J. The Fab conformations in the solution structure of human immunoglobulin G4 (IgG4) restrict access to its Fc region: implications for functional activity. J. Biol. Chem. 2014;289:20740–20756. doi: 10.1074/jbc.M114.572404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ugurlar D., Howes S.C., Gros P. Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science. 2018;359:794–797. doi: 10.1126/science.aao4988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.