Abstract
Replicating the human genome efficiently and accurately is a daunting challenge involving the duplication of upward of three billion base pairs. At the core of the complex machinery that achieves this task are three members of the B family of DNA polymerases: DNA polymerases α, δ, and ε. Collectively these multimeric polymerases ensure DNA replication proceeds at optimal rates approaching 2 × 103 nucleotides/min with an error rate of less than one per million nucleotides polymerized. The majority of DNA replication of undamaged DNA is conducted by DNA polymerases δ and ε. The DNA polymerase α-primase complex performs limited synthesis to initiate the replication process, along with Okazaki-fragment synthesis on the discontinuous lagging strand. An increasing number of human disorders caused by defects in different components of the DNA-replication apparatus have been described to date. These are clinically diverse and involve a wide range of features, including variable combinations of growth delay, immunodeficiency, endocrine insufficiencies, lipodystrophy, and cancer predisposition. Here, by using various complementary approaches, including classical linkage analysis, targeted next-generation sequencing, and whole-exome sequencing, we describe distinct missense and splice-impacting mutations in POLA1 in five unrelated families presenting with an X-linked syndrome involving intellectual disability, proportionate short stature, microcephaly, and hypogonadism. POLA1 encodes the p180 catalytic subunit of DNA polymerase α-primase. A range of replicative impairments could be demonstrated in lymphoblastoid cell lines derived from affected individuals. Our findings describe the presentation of pathogenic mutations in a catalytic component of a B family DNA polymerase member, DNA polymerase α.
Keywords: POLA1, polymerase alpha, intellectual disability, X-linked, microcephaly, growth retardation
Main Text
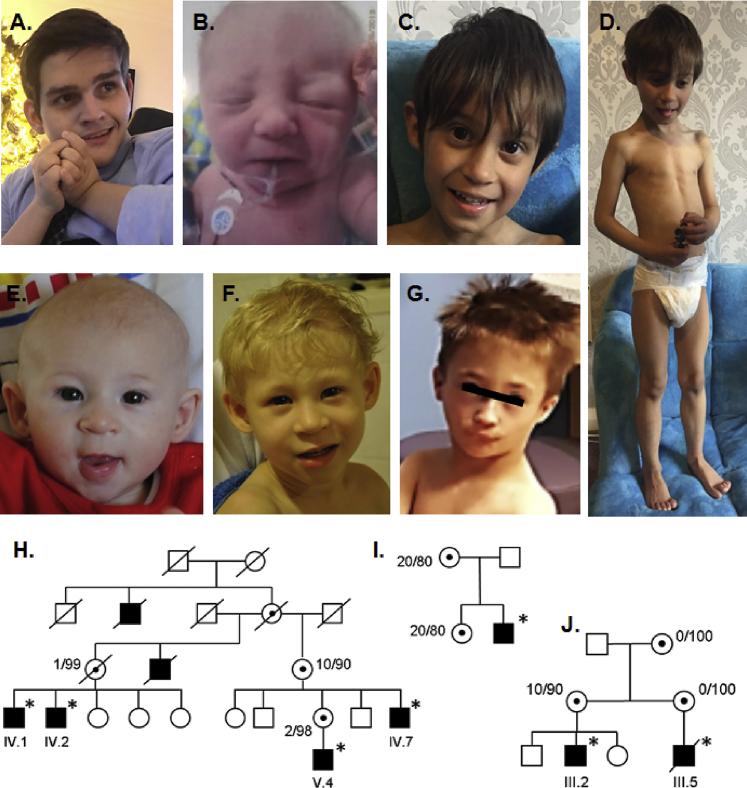
X-linked intellectual disability (XLID) is a heterogeneous disorder that can be classified as either non-syndromic, when cognitive impairment is the only feature, or as syndromic. In the latter, the cognitive impairment is associated with dysmorphic, metabolic, and/or neurological features. Until now, over 140 XLID-associated genes have been identified,1 mainly through the implementation of comparative genome hybridization and next-generation sequencing technologies.2, 3 Many of these genes converge into a few common functional networks because ID proteins often participate in interconnected cellular and molecular processes, including neurogenesis, neuronal migration, and synapse formation and function.4, 5 Here, we report that hypomorphic defects in the replicative DNA polymerase α cause a human XLID syndrome. In five families, we identified mutations in POLA1 (Xp22.1–p21.3, MIM: 312040), which encodes the p180 catalytic subunit of the heterotetrameric DNA polymerase α-primase (POLα). All affected individuals present with different degrees of intellectual disability and moderate to severe short stature, microcephaly, hypogonadism, and variable congenital malformations (Figure 1). Written informed consent was obtained from all parents on behalf of the affected individuals according to local ethical protocols and the principles of the Declaration of Helsinki. An overview of the clinical features of the affected individuals is presented in Table 1. More detailed clinical descriptions and pedigrees are provided in the Supplemental Note and Figure 1. The core clinical features consist of intellectual disability and developmental delay (ranging from mild to severe), pronounced proportionate short stature (ranging from −2 SD to −7.7 SD), and microcephaly (ranging from –3.1 SD to −7.8 SD), pointing toward a clear growth-deficiency syndrome of prenatal origin. Hypogonadism is also frequently evident. The index individual of family B also developed seizures and secondary neurological and orthopedic manifestations; these traits were not seen in the other individuals. In two affected individuals (in family C), a congenital heart malformation was present at birth. Although we cannot define a recognizable facial gestalt, mild upslant of the palpebral fissures is present in four affected individuals (Figures 1A–G).
Figure 1.
Clinical Pictures and Pedigrees
In clockwise order from top left: The index individual of family B at age 19 years (A); individual III-5 (family C) at newborn age (B); proband III-2 (family C) at age 5 years, demonstrating lack of subcutaneous fat, microcephaly, and proportionate short stature (C and D).
Lower panel: Pictures of the index individual of family D at age 6 months (E) and then at age 3 years (F); and a picture of the index individual of family E at the age of 4 years (G). All individuals displayed proportionate short stature and microcephaly, as well as a pronounced nasal bridge and a mild upslant of the palpebral fissures.
Below: Pedigrees of family A (H), family B (I), and family C (J). Carrier females showed skewing of X-inactivation (next to symbol). Asterisks indicate the affected individuals that were tested and carry the respective mutation.
Table 1.
Overview of the Clinical Features of Affected Individuals
|
Family A |
Family B |
Family C |
Family D |
Family E |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Individual V-4 | Individual IV-7 | Individual IV-1 | Individual IV-2 | Individual III-2 | Individual III-5 | ||||
| Gender | m | m | m | m | m | m | m | m | m |
| Country | Belgium | Belgium | UK | Australia | USA | ||||
| Gene | POLA1 | POLA1 | POLA1 | POLA1 | POLA1 | ||||
| Chromosome change (Hg19), GenBank: NM_016937.3 | c.236T>G | c.4142C>T | c.507+1G>A | c.445_507del | c.328G>A | ||||
| Protein change | p.Ile79Ser | p.Pro1381Leu | p.Lys149_Glu169del,Thr170_Ser1462 delins15∗ |
p.Lys149_Glu169del | p.Gly110Arg | ||||
| Mutation type | missense | missense | splice site | in frame deletion exon 6 | splice site | ||||
| Birth Parameters | |||||||||
| Birth (weeks) | 40 weeks | NA | NA | NA | 39 weeks | 38 weeks | 38 weeks | 38 weeks | 29 weeks |
| Birth weight (g) | 1,500 g | NA | NA | NA | 2,700 g | 1,786 g | 1,729 g | 1,688 g | 840 g |
| Birth length (cm) | 45 cm | NA | NA | NA | 46.5 cm | NA | NA | 44.5 cm | 41 cm |
| Birth OFC (cm) | NA | NA | NA | NA | 33 cm | 30 cm | NA | 28.5 cm | 31.5 cm |
| Growth | |||||||||
| Age | 6 years | 28 years | 46 years | 44 years | 16 years | 5 years | 14 months | 6 years, 11 months | 4 years, 5 months |
| Weight (kg) | 10.5 kg (−7,9 SD) | NA | 72 kg (+0.5 SD) | 62 kg (−0.5 SD) | 36.2 kg (−3.6 SD) | 9.4 kg (−7 SD) | 5.7 kg (−5 SD) | 14.6 kg (−4.5 SD) | 13.3kg (−1.9 SD) |
| Height (cm) | 98 cm (−4 SD) | 150 cm (−4.1 SD) | 158 cm (−2.9 SD) | 160 cm (−2.6 SD) | 137 cm (−5 SD) | 95.2 cm (−3.5 SD) | 59 cm (−7.7 SD) | 110.4 cm (−2.6 SD) | 95.8cm (−2 SD) |
| OFC (cm) | 42.9 cm (−5.7 SD) | 47.7 cm (−4.9 SD) | 51.2 cm (−2.9 SD) | 49.7 cm (−3.7 SD) | 49.4 cm (−3.7 SD) | 41 cm (−7.8 SD) | 38 cm (−6.6 SD) | 43 cm (−5.8 SD) | 46 cm (−3.1 SD) |
| Neurological | |||||||||
| Degree of DD/ID | mild (TIQ 71) | moderate (TIQ 57) | mild (TIQ 68) | moderate (TIQ 53) | severe | moderate | developmental delay | mild | mild (mainly speech delay) |
| Behavioral problems | ADHD | NP | NP | NP | hand stereotypies, autistic behavior | difficult behavior in association with frustration | NP | impulsive behavior, short attention span | shyness, weak eye contact, short attention |
| Hypotonia | childhood hypotonia | childhood hypotonia | NA | NA | childhood hypotonia | NP | yes | NP | yes |
| Epilepsy | NP | NP | NP | NP | epilepsy from age 3 months, therapy resistant | NP | NP | NP | NP |
| Brain abnormalities and/or MRI | normal MRI imaging | normal MRI imaging | NA | NA | cerebellar atrophy | slight thickening of pituitary stalk | cerebral atrophy, postoperative left thalamic bleed | normal MRI imaging | normal MRI imaging |
| Other | – | – | – | – | spasticity | – | vocal cord palsy | – | – |
| Facial Features | |||||||||
| Dysmorphic features | retrognathia | NP | NP | NP | large mouth, downturned corners of the mouth | short palpebral fissures; upslanting, prominent ears; brachymesophalangy | retrognathia, flat nasal bridge, small nose | upslanting palpebral fissures, long face | small ears, shallow orbits, upslanting palpebral fissures, fifth finger clinodactyly |
| Gastro-Intestinal | |||||||||
| NP | NP | NP | NP | gastrostomy | esophageal atresia with tracheosophageal fistula (repaired postnatal day 3) | NP | NP | tube feeding | |
| Heart | |||||||||
| normal | NA | NA | NA | normal | small ASD | pulmonary artery stenosis, VSD, pulmonary atresia | normal | normal | |
| Urogenital and Endocrine | |||||||||
| Hypogonadotropic hypogonadism | NA | yes | yes | yes | yes, small testes (10 ml) | NA | yes | NA | left testicular atrophy, small right testicle |
| Senses | |||||||||
| Vision | normal | normal | normal | normal | normal | normal | normal | normal | intermittent esotropia, astigmatism |
| Other | – | – | – | NIDDM | secondary scoliosis | sacral dimples | sacral dimples, bifid uvula | – | recurrent infections |
Abbreviations are as follows: NA = not assessed; NP = not present; OFC = occipital frontal circumference; ASD = atrial septum defect; NIDDM = non-insulin-dependent diabetes; ADHD = attention deficit hyperactivity disorder; VSD = ventral septal defect; and TIQ = total intelligence quotient.
Of the three mammalian replicative DNA polymerases (POLα, POLδ, and POLε), POLα-primase is the only polymerase that can initiate de novo DNA synthesis from licensed replication origins. It also mediates Okazaki-fragment synthesis during lagging-strand DNA replication.6, 7, 8 POLα is also involved in other cellular processes such as DNA-damage-response signaling from stalled replication forks, telomere maintenance, and epigenetic regulation.9, 10, 11, 12, 13, 14 Interestingly, a recurrent, deep-intronic mutation in POLA1 was recently found to cause X-linked reticulate pigmentary disorder (XLPDR; MIM: 301220), a primary immunodeficiency with autoinflammatory features, as well as skin hyperpigmentation and a prototypical facial gestalt.15, 16 This intronic mutation creates a novel exon 13a in the POLA1 transcript, reducing the total amount of p180-POLα protein. XLPDR-affected individuals do not exhibit intellectual disability, altered body growth, or smaller head circumference, and this might indicate tissue-specific differences in abnormal splicing. Conversely, our affected individuals do not show any of the XLPDR-related symptoms, except proband E, who suffered from recurrent infections.
We identified POLA1 mutations either by using classical linkage analysis and then Sanger sequencing of all 17 genes present in the 6 cM interval (LOD score 2.6) (family A),17 by a custom-designed microcephaly/microcephalic dwarfism Sure Select capture panel consisting of 63 genes (family C), by single whole-exome sequencing (WES) (family B), or by trio WES (families D and E). In family A, a missense mutation in exon 3 of POLA1 was identified, and this mutation, c.236T>G (p.Ile79Ser), segregates with the disease in all four affected individuals and obligate carrier mothers (Figure 1H). The sequence variant results in the replacement of an isoleucine, a non-polar amino acid, by a serine, a polar amino acid. This residue and its surrounding sequence are highly conserved, and p.IIe79Ser was predicted to be deleterious by various in silico methods (Figures S1 and S2). In family B, exome analysis identified a missense mutation, c.4142C>T, leading to a p.Pro1381Leu mutation. This mutation affects a conserved residue and is also present in the unaffected mother, maternal grandmother, and sister (Figures 1I, S1, and S2). In family C, a splice-site variant, c.507+1G>A, located in the donor splice site of intron 6 of POLA1 was identified. The variant was identified in the affected proband and his affected maternal cousin (Figure 1J). The c.507+1G>A splice-site variant was predicted by five different splicing prediction programs to completely abolish the donor splice site. We performed RNA studies that showed that c.507+1G>A prevents normal splicing and leads to the production of two abnormally-spliced transcripts (Figure S1). The larger c.507+1G>A transcript results from the activation of a cryptic splice donor site within intron 6, leading to an insertion of the first 60 nucleotides of intron 6. This is predicted to cause an insertion of 15 amino acids and the introduction of a premature termination codon, p.Thr170_Ser1462delins15∗, truncating the potential p180-POLα product upstream of the domains responsible for DNA binding and catalytic activity. The smaller c.507+1G>A transcript results from exon 6 skipping and leads to an in-frame deletion, i.e., p.Lys149_Glu169del, that is predicted to produce a protein product lacking 21 amino acids (Figure S1). In family D, a hemizygous deletion of exon 6 was identified via exome sequencing, and this deletion led to an in-frame deletion producing a protein lacking 21 amino acids, as seen for the smaller transcript in family C. This deletion arose de novo in the index individual (Figure S1). Exome sequencing in family E identified a de novo variant, c.328G>A, affecting the last nucleotide of exon 4 and leading to a p.Gly110Arg mutation. Bioinformatic analysis predicts a high probability of intron 4 missplicing upon c.328G>A replacement, and subsequent qRT-PCR analysis displayed a dramatic reduction of POLA1 mRNA in affected cells compared to cells derived from unaffected males; the reduction was even more profound than that observed in XLPDR-derived fibroblasts15 (Figure S1). None of the above identified variants are present in the dbSNP, 1000 Genomes, ExAC, or gnomAD databases, and we have submitted them to ClinVar (see Accession Numbers). All missense mutations affect conserved amino acids and are predicted to be deleterious by various in silico methods (Figure S2). In addition, in the three familial cases (families A–C), all obligate female carriers show significant to complete skewing of X inactivation (Figures 1H–J).
DNA polymerases are highly expressed during development, when rapid DNA replication and cell division is required.18, 19 To further investigate POLα/POLA1 in mammalian brain development, we assessed Pola1 expression by in situ hybridization in the embryonic and adult mouse brain. In the mouse forebrain, Pola1 is expressed in those zones containing proliferating cells in the developing embryonic neocortex (Figure 2A), as well as in the lateral and medial ganglionic eminences (not shown). After birth, the gene is transcribed in cells that remain proliferating in the ventricular and subventricular zone of the striatum (Figure 2B). These data suggest that Pola1 has a role in neurogenesis throughout life. Additionally, pola1 expression by in situ hybridization in developing zebrafish embryos shows early and intense staining in the developing brain,20, 21 whereas Polα activity appears highest in isolated neurons from developing rat-brain cerebral cortex when the mitotic activity is at its peak.22 Conversely, an insertional mutation in zebrafish pola1 (pola1hi1146Tg) led to central nervous system (CNS) necrosis, a small head and eyes, an inflated hindbrain ventricle, a thin and often curved body, and a rounder yolk with no extension at day 2.23 At days 3–5, the necrosis spread throughout the body, resulting in overt body wasting and a small head and eyes.23
Figure 2.
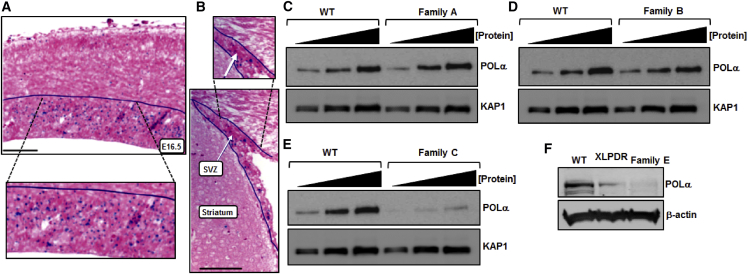
Pola1 Is Expressed in Proliferating Progenitors During Embryonic and Postnatal Neurogenesis in the Mouse Brain, and POLA1-Mutated Cells Exhibit Variable POLα Expression
(A) The Pola1 transcript (dark blue signal) is found in the proliferative zone of the embryonic neocortex. The scale bar represents 200 μm. The dashed lines indicate an expanded area of the image.
(B) Three weeks after birth, Pola1 (dark blue signal) is expressed in the subventricular zone (SVZ), where postnatal neurogenesis occurs. The scale bar represents 200 μm. The dashed lines indicate an expanded area of the image.
(C) Increasing amounts of whole-cell extract (WCE) from LCLs derived from a clinically unaffected, unrelated, normal wild-type (WT) male individual and a POLA1-mutant-affected individual from family A were assessed for POLα expression levels. No difference in expression was observed.
(D) Increasing amounts of WCE from LCLs derived from the WT and a POLA1-mutant-affected individual from family B were assessed for POLα expression levels. POLα expression was comparable.
(E) Increasing amounts of WCE from LCLs derived from the WT and a POLA1-mutant-affected individual from family C were assessed for POLα expression levels. Here, POLα was markedly reduced in affected cells compared to in WT LCLs.
(F) POLα levels were assessed via WCE derived from dermal fibroblasts from the WT, an XLPDR-affected individual, and the POLA1-mutant-affected individual from family E. POLα was reduced in both instances of POLA1 mutation.
We examined POLα protein levels in proband-derived cell lines from families A, B, C, and E (Figures 2C–F). Cell lines from the family D proband were unavailable. By using whole-cell extracts (WCEs) from lymphoblastoid cell lines (LCLs) derived from affected individuals from families A and B, we found POLα protein levels comparable to those of wild-type (WT) LCLs (Figures 2C and 2D). In contrast, WCEs derived from the family C proband’s LCLs or from dermal, primary fibroblasts from the family E proband both showed marked reduction in POLα levels (Figures 2E and 2F). These findings are consistent with the RT-PCR analysis from each of these two families (Figure S1). We next assessed POLα enrichment on chromatin by using LCLs derived from affected individuals. Although chromatin extracts from families A and B tended toward slightly reduced POLα levels compared to those of WT LCLs, the reduction did not reach statistical significance (Figures S3A and S3B). In contrast, chromatin extracts from family C’s LCLs showed an approximately 60% reduction in POLα levels compared to those of WT LCLs (Figure S3C). Furthermore, chromatin recruitment of the additional POLα-primase subunit proteins p68-POLA2 and p48-PRIM1 appeared unaffected in proband LCL extracts from families A, B, and C, suggesting the stoichiometry of the POLα-primase component subunits is largely preserved (Figure S3D).
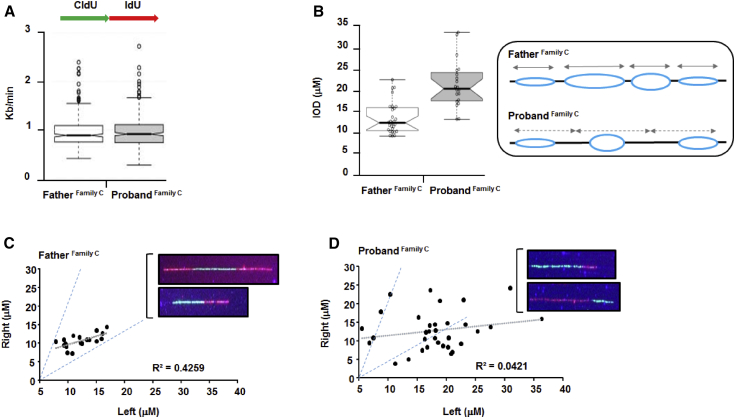
Reduced cellular proliferation represents a logical pathomechanism underlying growth retardation and microcephaly in human disorders such as Seckel syndrome (MIM: 210600), which presents with prototypical microcephalic primordial dwarfism, or Meier-Gorlin syndrome (MIM: 224690), in which the dwarfism is caused by mutations in multiple components of the DNA replication-licensing machinery.24, 25, 26 The C.elegans div-1 (division delayed) allele encoding the B subunit of DNA polymerase α-primase delays cell division and lethally disrupts cell polarity in embryos,27 whereas POL1 mutants of S. cerevisiae and a Pola1 mutant (p.Ser1180Phe) of the mouse mammary carcinoma line FM3A are each associated with temperature-sensitive growth delay.11, 28 Nonetheless, we did not observe a marked delay in proliferation of proband LCLs from families A, B, and C compared to in the WT (Figure S4). Therefore, we carefully assessed different aspects of DNA-replication capacity in family C’s LCLs specifically, as these showed a pronounced reduction in POLα-primase expression and chromatin localization (Figures 2E and S3C). POLα is characterized by limited processivity, and it also lacks 3′ exonucleolytic proofreading capacity. Therefore, in contrast to POLδ and POLε, POLα is unsuited to efficiently and accurately duplicate long DNA templates.29, 30 By using DNA-fiber-combing analysis of ongoing, unperturbed DNA replication in LCLs obtained from the proband of family C and his unaffected father, we observed similar rates of replication-fork progression (Figure 3A). This was perhaps not entirely unanticipated because POLα-primase doesn’t replicate the bulk of the genomic DNA, and POLA1 encodes a core product of a fundamentally essential cellular process, hence any viable defects in this gene would have to be hypomorphic. POLα is essential for viability;23, 27, 31 indeed, POLA1 has a negative residual-variation intolerance score of −0.795, indicating it is under substantial purifying selection.15, 32 This is further illustrated by the absence of microdeletions involving POLA1 in males, both in the control and diseases copy number variation (CNV) databases, as well as by the identification of a female with X-autosome translocation-disrupting POLA1. In this female, in contrast to what normally happens, the wild-type X chromosome remained active in all her cells, probably as a result of selection against cells that contained the non-functional POLA1.33 DNA-fiber-combing analysis did reveal a reduction in new initiation events in the family C proband’s LCLs of 5.9% (n = 170 fibers), compared to 9.6% (n = 178 fibers) observed in the paternal LCLs, a difference indicative of impaired “productive” replication initiation.34, 35 Consistent with this, we also observed increased inter-origin distance (IOD) in the family C proband’s LCLs compared to those of the father (Figure 3B). This would also be consistent with possible impairments in dormant origin firing.34, 35 Furthermore, we found an increase in asymmetric forks and an accumulation of longer replication tracts in the family C proband’s LCLs compared to those of the father (Figures 3C and 3D). Collectively, analyses of multiple replication-fork parameters in these POLA1-deficient LCLs demonstrated several phenotypes consistent with spontaneously diminished productive-replication initiation under unperturbed exponential growth conditions. These replication phenotypes are reminiscent of those recently reported for Polε impairment.36
Figure 3.
POLA1-Mutant LCLs Spontaneously Exhibit a Range of DNA-Replication Defects During Unperturbed, Exponential Growth
(A) Dual CldU (5-Chloro-2′-deoxyuridine)- and IdU (5-Iodo-2′-deoxyuridine)-labeled (for 20 min each, as indicated) DNA-fiber-combing analyses of unperturbed, exponentially growing LCLs from the unaffected father and family C proband (individual III-2) demonstrated similar replication-fork speeds between the fathers’ LCLs (mean: 0.95kb/min, median: 0.89, from n = 615 fibers) and the family C proband’s (mean: 1.01kb/min, median: 1.01kb/min, from n = 926 fibers). CldU fibers are shown in green and IdU fibers are shown in red.
(B) Origins from the family C proband’s (individual III-2) LCLs exhibited significantly elevated inter-origin distance (IOD) compared to that in those of the father during unperturbed, asynchronous growth conditions. The father’s IOD median was 12.62 μM (n = 28), compared to the family C proband’s IOD median of 20.75 μM (n = 20) (p < 0.05; Student’s t test). The schematic idealizes the most likely contrasting situation with regard to the bidirectional movement of fired origins (blue) in the father’s LCLs compared to that in those of the proband. In the proband, fewer new initiation events are seen (i.e., fewer new origins and reduced dormant origin-firing capacity), and those that have fired are thus compelled to traverse greater distances.
(C) In the XY scatterplot, the data show the length of fibers on the right- and left-hand-sides of fired origins and/or ongoing forks, visualized after the father’s LCLs were subjected to DNA fiber combing. Normally, functional replicating forks exhibit left-right symmetry reflective of coordinated bidirectional movement. The dotted gray line indicates linear regression (R2 = 0.4259 from n = 18) showing a strong clustering, which is consistent with symmetrical movement, of the forks. The dotted blue lines are guide lines drawn to encapsulate the lengths of all of the forks assessed. Representative symmetrical fibers from the paternal LCLs are shown inset.
(D) This XY scatterplot shows the length of fibers on the right- and left-hand-sides of fired origins and/or ongoing forks derived from the family C proband’s LCLs. The wide dispersal of the data points with regard to the blue guide lines (copied from the paternal XY scatterplot shown in [C]) indicates marked asymmetric movement of active replication forks. The dotted gray line denotes linear regression (R2 = 0.0421 from n = 33). Note also the preponderance of longer fork lengths (i.e., >15 μM) observed in these fibers compared to the lengths of those of the father’s fibers (shown in [C]). Representative fibers that are derived from the family C proband’s LCLs and that demonstrate asymmetry are shown inset.
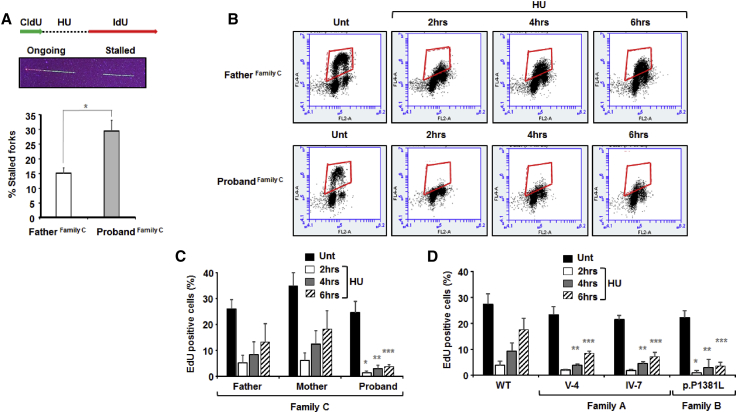
We next reasoned that additional impairments of DNA replication in POLA1 LCLs could be context dependent, and DNA replication under conditions of replication stress may represent that physiological context.37 Disrupting the temporally coordinated balance between stem-cell proliferation and differentiation programs profoundly impacts upon brain and body growth.38, 39 Rapidly proliferating murine embryonic stem cells exhibit constitutive replication stress and are highly dependent on replication-coupled pathways to preserve genome integrity and execute DNA replication efficiently and effectively.40 Therefore, we reasoned that DNA replication in POLA1-mutated cells may be hypersensitive to replication-stress conditions, particularly if dormant origin capacity was restricted due to a genetic defect of this nature.34, 35 We examined DNA fibers under conditions of replication stress by limiting deoxyribonucleotide availability via treatment with the ribonucleotide reductase inhibitor hydroxyurea (HU), and we observed an approximately 2-fold increase in stalled replication forks in combed fibers from the family C proband’s LCLs compared to paternal LCLs (Figure 4A). We next assessed DNA replication via pulse-labeled EdU incorporation within LCL populations in a kinetic fashion after HU treatment (Figures 4B–D). Figure 4B shows representative EdU flow-cytometry profiles from family C paternal and proband LCLs, either untreated (Unt) or at different times after HU treatment. The family C proband’s LCLs incorporated significantly less EdU upon HU-treatment compared to control LCLs (Figures 4B and 4C). This is a POLA1-dependent cellular phenotype, as demonstrated by siRNA of POLA1 in U2OS cells (Figure S5). Similarly, after treatment with HU, proband LCLs from family A and family B exhibited significantly reduced EdU incorporation, which is indicative of impaired DNA replication (Figure 4D). Collectively, these results show that LCLs from affected individuals with distinct POLA1 mutations exhibit reduced DNA replication under conditions of replication stress. A similar cellular response has been demonstrated for ORC1-mutated (MIM: 224690) and MCM5-mutated Meier-Gorlin syndrome (MIM: 617564) LCLs.41, 42 Indeed, pathogenic mutations in MCM4 (MIM: 609981) and in the GINS1 (MIM: 617827) component of the heterotetrametic Go-Ichi-Ni-San (GINS) complex, both encoding key components of the DNA replication apparatus, are each associated with cellular-proliferation impairments, growth delay, and natural killer (NK) cell deficiency.43, 44, 45
Figure 4.
POLA1-Mutant Proband LCLs Exhibit DNA-Replication Deficits After Experiencing Replication Stress
(A) The level of replication-fork stalling was investigated via a dual CldU- and IdU-labeling approach that incorporated a hydroxyurea (HU) treatment. LCLs were first labeled with CldU (for 20 min) and then treated with HU (2 mM, for 120 mins) before a second labeling with IdU (for 60 mins) to monitor fork recovery, as indicated. The middle panel shows a representative image of a labeled fiber demonstrating ongoing replication and a stalled replication fork. Under these conditions, an approximately 2-fold increase in the levels of stalled replication forks was observed in LCLs from the family C proband, relative to the levels in the paternal LCLs (∗p < 0.05, Student’s t test; error bars = standard deviation).
(B) The impact of mildly stressing conditions (125 μM HU) upon DNA replication was assessed in LCLs from family C via EdU-pulse incorporation (for 30 min) and flow cytometry. Representative flow cytometry panels are shown; the area boxed in red denotes EdU-positive cells. LCLs derived from the unaffected father and the proband were either untreated (Unt) or treated with HU, and EdU incorporation was measured at the times indicated post-treatment. Consistent with the DNA-fiber fork-rate analysis shown in Figure 3A, EdU incorporation in untreated LCLs was grossly comparable between the father and the proband. This was in contrast to the HU-treated LCLs, where the proband showed markedly fewer EdU-positive cells at each time point compared to the father.
(C) The bar chart shows EdU incorporation in untreated (Unt) and HU-treated LCLs from the father, mother, and proband of family C from 4× independent experiments (asterisks indicate p < 0.05 [Student’s t test], compared to the equivalent parental time points; error bars = standard deviation). The proband LCLs demonstrate significantly reduced EdU incorporation at each time point after HU treatment, compared to the parental LCLs under these conditions.
(D) The bar chart shows EdU incorporation in untreated (Unt) and HU-treated LCLs from a clinically normal, unrelated, wild-type (WT) male individual and affected individuals from family A and family B from 4× independent experiments (asterisks indicate p < 0.05 [Student’s t test], compared to the equivalent WT time point; error bars = standard deviation). LCLs from all of the probands demonstrate significantly reduced EdU incorporation compared to WT LCLs after HU treatment. This was most evident at 4 and 6 hr post-HU treatment.
In summary, we describe nine affected individuals, from five families, who present with a syndrome involving a spectrum of developmental delay/intellectual disability, growth failure, microcephaly, hypogonadism, and additional, isolated abnormalities; this syndrome is associated with five different mutations in POLA1, which encodes the catalytic subunit of the DNA polymerase α-primase. The growth impairments were evident prenatally, suggesting an early origin in utero. LCLs from the proband of one affected family spontaneously displayed altered replication-fork parameters, including reduced new-initiation events, increased IOD and fork asymmetry, and elongated replication tracts. All the POLA1-mutant LCLs we examined were additionally found to exhibit impaired DNA-replication capacity under conditions of replication stress. These data strongly suggest that cellular DNA-replication deficits during development may underlie many of the clinical features observed in these families.
Interestingly, a recurrent intronic variant in POLA1 has been shown to underlie XLPDR, a primary immunodeficiency associated with type I-interferon-derived autoinflammatory features.15 The elevated type I-interferon-signaling response underlying XLPDR has been shown to derive from a reduction in POLα-dependent synthesis of cytosolic RNA:DNA hybrid species.15 Importantly, XLPDR cells with this specific intronic POLA1 variant do not exhibit a proliferative impairment and, except for the recurrent infections observed in proband E, we observed no other phenotypic overlap with XLPDR.15
The remarkable fidelity of human DNA replication is a consequence of the combined and coordinated action of highly processive DNA polymerases, their intrinsic exonucleolytic proofreading activity, and post-replicative DNA mismatch repair (MMR). Although POLα-primase initiates DNA replication and Okazaki-fragment synthesis, it is not highly processive and does not possess an intrinsic proofreading activity. Processivity and proofreading are carried out by POLδ and POLε.29 Germline mutations in components of the MMR pathway result in dramatically elevated spontaneous mutation frequencies and are associated with hereditary, non-polyposis colorectal carcinoma (HNPCC) or Lynch syndrome (MIM: 120435).46, 47 Germline mutations within the exonucleolytic, domain-encoding regions of POLD1 (MIM: 174761) and POLE (MIM: 174762), each encoding the catalytic subunit of the replicative polymerases POLδ and POLε, respectively, have been identified as causing ultra-mutated colorectal (“polymerase proofreading-associated polyposis”) and endometrial cancers (MIM: 612591 and 615083).48, 49, 50, 51 Fascinatingly, differing mutations in POLE underlie a clinical spectrum that includes FILS syndrome (facial dysmorphism, immunodeficiency, livedo reticularis, and short stature; MIM: 615083)52 and IMAGe syndrome (intrauterine growth restriction, metaphyseal dysplasia, adrenal hypoplasia, and congenital and genitourinary anomalies in males; MIM: 614732), and both diseases are associated with variable immunodeficiency.36, 53 Additionally, a POLE2 (MIM: 602670) mutation has been identified in an individual with combined immunodeficiency, facial dysmorphism, and autoimmunity associated with compromised lymphocyte proliferation.54 Germline mutations in POLD1 have been described to underlie a range of congenital disorders, including MDP syndrome (mandibular hypoplasia, deafness, and progeroid; MIM: 615381), lipodystrophy, and atypical Werner’s syndrome with short stature (MIM: 277700).55, 56, 57, 58, 59, 60 Therefore, it appears that germline mutations in the core DNA-replication polymerases can present as a wide range of phenotypes and variably incorporate cancer predisposition, developmental and/or progeroid syndromes with or without growth failure, endocrine insufficiency, and variable immunodeficiency. Our findings make an important additional contribution to this expanding knowledge base: namely, that hitherto-undescribed hypomorphic POLA1 mutations affecting the catalytic subunit of DNA POLα-primase are associated with multifaceted cellular DNA-replicative deficits, and they underlie an X-linked syndrome of intellectual disability, microcephaly, growth failure, and hypogonadism.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors thank the families for their cooperation and acknowledge Andrew Jackson for helpful comments and assistance, Mark Greenslade for his work at Bristol Genetics Laboratory with family C, and Claudia Kerzendorfer for her contributions to the functional characterization of cells from family A. Thanks also to Adam Lopez, Luis Situentes-Dominguez, and Ann Ray. H.V.E. and K.D. are clinical investigators of the Fund for Scientific Research Flanders (FWO-Vlaanderen), Belgium. M.O’D. acknowledges program funding from Cancer Research UK (United Kingdom). Work with family E was supported by funds to E.B from the Pollock Family Center for Research in Inflammatory Bowel Disease.
Published: April 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.006.
Contributor Information
Hilde Van Esch, Email: hilde.vanesch@med.kuleuven.be.
Mark O’Driscoll, Email: m.o-driscoll@sussex.ac.uk.
Accession Numbers
ClinVar accession numbers for the variants reported in this paper were not available from ClinVar as of the date this article was finalized for press; please contact the corresponding authors for the numbers.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Align-GVGD, http://agvgd.hci.utah.edu/
Genic Intolerance, http://genic-intolerance.org/index.jsp
MutationTaster, http://mutationtaster.org/
Online Mendelian Inheritance in Man, http://www.omim.org
Provean (Protein Variation Effect Analyzer), http://provean.jcvi.org/index.php
PolyPhen, http://genetics.bwh.harvard.edu/pph/
Supplemental Data
References
- 1.Neri G., Schwartz C.E., Lubs H.A., Stevenson R.E. X-linked intellectual disability update 2017. Am. J. Med. Genet. A. 2018;176:1375–1388. doi: 10.1002/ajmg.a.38710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aerts S., Lambrechts D., Maity S., Van Loo P., Coessens B., De Smet F., Tranchevent L.-C., De Moor B., Marynen P., Hassan B. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 3.Tarpey P.S., Smith R., Pleasance E., Whibley A., Edkins S., Hardy C., O’Meara S., Latimer C., Dicks E., Menzies A. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chelly J., Khelfaoui M., Francis F., Chérif B., Bienvenu T. Genetics and pathophysiology of mental retardation. Eur. J. Hum. Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- 5.Gécz J., Shoubridge C., Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Burgers P.M.J., Kunkel T.A. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgers P.M.J. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stodola J.L., Burgers P.M. Mechanism of Lagging-Strand DNA Replication in Eukaryotes. In: Masai H., Foiani M., editors. DNA Replication: From Old Principles to New Discoveries. Springer Singapore; Singapore: 2017. pp. 117–133. [DOI] [PubMed] [Google Scholar]
- 9.Yan S., Michael W.M. TopBP1 and DNA polymerase-α directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michael W.M., Ott R., Fanning E., Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura M., Nabetani A., Mizuno T., Hanaoka F., Ishikawa F. Alterations of DNA and chromatin structures at telomeres and genetic instability in mouse cells defective in DNA polymerase α. Mol. Cell. Biol. 2005;25:11073–11088. doi: 10.1128/MCB.25.24.11073-11088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama Ji, Allshire R.C., Klar A.J.S., Grewal S.I.S. A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 2001;20:2857–2866. doi: 10.1093/emboj/20.11.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace J.A., Orr-Weaver T.L. Replication of heterochromatin: Insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114:389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert D.M. Replication timing and transcriptional control: Beyond cause and effect. Curr. Opin. Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 15.Starokadomskyy P., Gemelli T., Rios J.J., Xing C., Wang R.C., Li H., Pokatayev V., Dozmorov I., Khan S., Miyata N. DNA polymerase-α regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat. Immunol. 2016;17:495–504. doi: 10.1038/ni.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starokadomskyy P., Sifuentes-Dominguez L., Gemelli T., Zinn A.R., Dossi M.T., Mellado C., Bertrand P., Borzutzky A., Burstein E. Evolution of the skin manifestations of X-linked pigmentary reticulate disorder. Br. J. Dermatol. 2017;177:e200–e201. doi: 10.1111/bjd.15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Esch H., Zanni G., Holvoet M., Borghgraef M., Chelly J., Fryns J.-P., Devriendt K. X-linked mental retardation, short stature, microcephaly and hypogonadism maps to Xp22.1-p21.3 in a Belgian family. Eur. J. Med. Genet. 2005;48:145–152. doi: 10.1016/j.ejmg.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava V.K., Busbee D.L. Replicative enzymes, DNA polymerase alpha (pol alpha), and in vitro ageing. Exp. Gerontol. 2003;38:1285–1297. doi: 10.1016/j.exger.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Loeb L.A., Monnat R.J., Jr. DNA polymerases and human disease. Nat. Rev. Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 20.Thisse B., Thisse C. Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submission. 2004 http://zfin.org/ZDB-PUB-040907-1 [Google Scholar]
- 21.Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X.Q., Thisse C. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) ZFIN Direct Data Submission. 2001 http://zfin.org/ZDB-PUB-010810-1 [Google Scholar]
- 22.Raji N.S., Krishna T.H., Rao K.S. DNA-polymerase α, β, δ and ε activities in isolated neuronal and astroglial cell fractions from developing and aging rat cerebral cortex. Int. J. Dev. Neurosci. 2002;20:491–496. doi: 10.1016/s0736-5748(02)00079-5. [DOI] [PubMed] [Google Scholar]
- 23.Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S., Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcantara D., O’Driscoll M. Congenital microcephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:124–139. doi: 10.1002/ajmg.c.31397. [DOI] [PubMed] [Google Scholar]
- 25.O’Driscoll M. The pathological consequences of impaired genome integrity in humans; disorders of the DNA replication machinery. J. Pathol. 2017;241:192–207. doi: 10.1002/path.4828. [DOI] [PubMed] [Google Scholar]
- 26.Klingseisen A., Jackson A.P. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–2024. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Encalada S.E., Martin P.R., Phillips J.B., Lyczak R., Hamill D.R., Swan K.A., Bowerman B. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 2000;228:225–238. doi: 10.1006/dbio.2000.9965. [DOI] [PubMed] [Google Scholar]
- 28.Gutiérrez P.J.A., Wang T.S.-F. Genomic instability induced by mutations in Saccharomyces cerevisiae POL1. Genetics. 2003;165:65–81. doi: 10.1093/genetics/165.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T.A., Burgers P.M. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlov Y.I., Frahm C., Nick McElhinny S.A., Niimi A., Suzuki M., Kunkel T.A. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Eki T., Enomoto T., Murakami Y., Hanaoka F., Yamada M. Characterization of chromosome aberrations induced by incubation at a restrictive temperature in the mouse temperature-sensitive mutant tsFT20 strain containing heat-labile DNA polymerase α. Cancer Res. 1987;47:5162–5170. [PubMed] [Google Scholar]
- 32.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podolska A., Kobelt A., Fuchs S., Hackmann K., Rump A., Schröck E., Kutsche K., Di Donato N. Functional monosomy of 6q27-qter and functional disomy of Xpter-p22.11 due to X;6 translocation with an atypical X-inactivation pattern. Am. J. Med. Genet. A. 2017;173:1334–1341. doi: 10.1002/ajmg.a.38183. [DOI] [PubMed] [Google Scholar]
- 34.McIntosh D., Blow J.J. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb. Perspect. Biol. 2012;4:a012955. doi: 10.1101/cshperspect.a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alver R.C., Chadha G.S., Blow J.J. The contribution of dormant origins to genome stability: From cell biology to human genetics. DNA Repair (Amst.) 2014;19:182–189. doi: 10.1016/j.dnarep.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellelli R., Borel V., Logan C., Svendsen J., Cox D.E., Nye E., Metcalfe K., O’Connell S.M., Stamp G., Flynn H.R. Polε instability drives replication stress, abnormal development, and tumorigenesis. Mol. Cell. 2018;70:707–721.e7. doi: 10.1016/j.molcel.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotsantis P., Petermann E., Boulton S.J. Mechanisms of oncogene-induced replication stress: Jigsaw falling into place. Cancer Discov. 2018;8:537–555. doi: 10.1158/2159-8290.CD-17-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arai Y., Pulvers J.N., Haffner C., Schilling B., Nüsslein I., Calegari F., Huttner W.B. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 40.Ahuja A.K., Jodkowska K., Teloni F., Bizard A.H., Zellweger R., Herrador R., Ortega S., Hickson I.D., Altmeyer M., Mendez J., Lopes M. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 2016;7:10660. doi: 10.1038/ncomms10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerzendorfer C., Colnaghi R., Abramowicz I., Carpenter G., O’Driscoll M. Meier-Gorlin syndrome and Wolf-Hirschhorn syndrome: Two developmental disorders highlighting the importance of efficient DNA replication for normal development and neurogenesis. DNA Repair (Amst.) 2013;12:637–644. doi: 10.1016/j.dnarep.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Vetro A., Savasta S., Russo Raucci A., Cerqua C., Sartori G., Limongelli I., Forlino A., Maruelli S., Perucca P., Vergani D. MCM5: A new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur. J. Hum. Genet. 2017;25:646–650. doi: 10.1038/ejhg.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes C.R., Guasti L., Meimaridou E., Chuang C.-H., Schimenti J.C., King P.J., Costigan C., Clark A.J.L., Metherell L.A. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J. Clin. Invest. 2012;122:814–820. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gineau L., Cognet C., Kara N., Lach F.P., Dunne J., Veturi U., Picard C., Trouillet C., Eidenschenk C., Aoufouchi S. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J. Clin. Invest. 2012;122:821–832. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cottineau J., Kottemann M.C., Lach F.P., Kang Y.-H., Vély F., Deenick E.K., Lazarov T., Gineau L., Wang Y., Farina A. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J. Clin. Invest. 2017;127:1991–2006. doi: 10.1172/JCI90727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 47.Leach F.S., Nicolaides N.C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomäki P., Sistonen P., Aaltonen L.A., Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 48.Rayner E., van Gool I.C., Palles C., Kearsey S.E., Bosse T., Tomlinson I., Church D.N. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer. 2016;16:71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 49.Palles C., Cazier J.-B., Howarth K.M., Domingo E., Jones A.M., Broderick P., Kemp Z., Spain S.L., Guarino E., Salguero I., CORGI Consortium. WGS500 Consortium Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Church D.N., Briggs S.E.W., Palles C., Domingo E., Kearsey S.J., Grimes J.M., Gorman M., Martin L., Howarth K.M., Hodgson S.V., NSECG Collaborators DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Briggs S., Tomlinson I. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J. Pathol. 2013;230:148–153. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pachlopnik Schmid J., Lemoine R., Nehme N., Cormier-Daire V., Revy P., Debeurme F., Debré M., Nitschke P., Bole-Feysot C., Legeai-Mallet L. Polymerase ε1 mutation in a human syndrome with facial dysmorphism, immunodeficiency, livedo, and short stature (“FILS syndrome”) J. Exp. Med. 2012;209:2323–2330. doi: 10.1084/jem.20121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logan C.V., Murray J.E., Parry D.A., Robertson A., Bellelli R., Tarnauskaitė Ž., Challis R., Cleal L., Borel V., Fluteau A., SGP Consortium DNA polymerase epsilon deficiency causes IMAGe syndrome with variable immunodeficiency. Am. J. Hum. Genet. 2018;103:1038–1044. doi: 10.1016/j.ajhg.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frugoni F., Dobbs K., Felgentreff K., Aldhekri H., Al Saud B.K., Arnaout R., Ali A.A., Abhyankar A., Alroqi F., Giliani S. A novel mutation in the POLE2 gene causing combined immunodeficiency. J. Allergy Clin. Immunol. 2016;137:635–638.e1. doi: 10.1016/j.jaci.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weedon M.N., Ellard S., Prindle M.J., Caswell R., Lango Allen H., Oram R., Godbole K., Yajnik C.S., Sbraccia P., Novelli G. An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat. Genet. 2013;45:947–950. doi: 10.1038/ng.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shastry S., Simha V., Godbole K., Sbraccia P., Melancon S., Yajnik C.S., Novelli G., Kroiss M., Garg A. A novel syndrome of mandibular hypoplasia, deafness, and progeroid features associated with lipodystrophy, undescended testes, and male hypogonadism. J. Clin. Endocrinol. Metab. 2010;95:E192–E197. doi: 10.1210/jc.2010-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reinier F., Zoledziewska M., Hanna D., Smith J.D., Valentini M., Zara I., Berutti R., Sanna S., Oppo M., Cusano R. Mandibular hypoplasia, deafness, progeroid features and lipodystrophy (MDPL) syndrome in the context of inherited lipodystrophies. Metabolism. 2015;64:1530–1540. doi: 10.1016/j.metabol.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 58.Pelosini C., Martinelli S., Ceccarini G., Magno S., Barone I., Basolo A., Fierabracci P., Vitti P., Maffei M., Santini F. Identification of a novel mutation in the polymerase delta 1 (POLD1) gene in a lipodystrophic patient affected by mandibular hypoplasia, deafness, progeroid features (MDPL) syndrome. Metabolism. 2014;63:1385–1389. doi: 10.1016/j.metabol.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 59.Lessel D., Hisama F.M., Szakszon K., Saha B., Sanjuanelo A.B., Salbert B.A., Steele P.D., Baldwin J., Brown W.T., Piussan C. POLD1 germline mutations in patients initially diagnosed with Werner syndrome. Hum. Mutat. 2015;36:1070–1079. doi: 10.1002/humu.22833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicolas E., Golemis E.A., Arora S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene. 2016;590:128–141. doi: 10.1016/j.gene.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.