Abstract
Neohesperidosides are disaccharides that are present in some flavonoids and impart a bitter taste, which can significantly affect the commercial value of citrus fruits. In this study, we identified three flavonoid-7-O-di-glucosyltransferase (dGlcT) genes closely related to 1,2-rhamnosyltransferase (1,2RhaT) in citrus genomes. However, only 1,2RhaT was directly linked to the accumulation of neohesperidoside, as demonstrated by association analysis of 50 accessions and co-segregation analysis of an F1 population derived from Citrus reticulata × Poncirus trifoliata. In transgenic tobacco BY2 cells, over-expression of CitdGlcTs resulted in flavonoid-7-O-glucosides being catalysed into bitterless flavonoid-7-O-di-glucosides, whereas over-expression of Cit1,2RhaT converted the same substrate into bitter-tasting flavonoid-7-O-neohesperidoside. Unlike 1,2RhaT, during citrus fruit development the dGlcTs showed an opposite expression pattern to CHS and CHI, two genes encoding rate-limiting enzymes of flavonoid biosynthesis. An uncoupled availability of dGlcTs and substrates might result in trace accumulation of flavonoid-7-O-di-glucosides in the fruit of C. maxima (pummelo). Past human selection of the deletion and functional mutation of 1,2RhaT has led step-by-step to the evolution of the flavor-related metabolic network in citrus. Our research provides the basis for potentially improving the taste in citrus fruit through manipulation of the network by knocking-out 1,2RhaT or by enhancing the expression of dGlcT using genetic transformation.
Keywords: Bitterness, citrus, flavonoid, flavonoid-7-O-di-glucosides, flavonoid-7-O-glucoside, neohesperidoside
Coupled availability of the Cit1,2RhaT enzyme and flavanone-7-O-glucosides promote the biosynthesis of bitter-tasting neohesperidosides during early fruit development in citrus, and selection against Cit1,2RhaT has occurred during domestication.
Introduction
Glycosylation, the attachment of sugars to flavonoids, causes major modifications and is crucial in determining their physiological properties and functions. It can improve solubility and stability, facilitate transport and accumulation, and reduce the chemical toxicity of flavonoids (Vogt and Jones, 2000; Breada, 2001; Bowles et al., 2005; Koes et al., 2005). Importantly, it also significantly contributes to the flavor of citrus fruits (Frydman et al., 2004). The majority of flavonoid glycosides are C-linked through a carbon–carbon bond or O-linked through the hydroxyl group of aglycones, whilst the others, such as flavonoid di-glycosides, are usually attached via additional sugar groups to an existing sugar moiety of the glycoside (Jones and Vogt, 2001; Masada et al., 2009; Nagatomo et al., 2014). In plants, the sugars employed for glycosylation are mostly glucose and rhamnose, but arabinose, xylose, and glucuronic acid are also sometimes used (Jay et al., 2006; Williams, 2006).
Glycosyltransferases (GTs) are enzymes that catalyse the glycosylation between a glycosyl residue and an acceptor molecule, and are encoded by a large gene family in plants. In particular, UDP glycosyltransferases (UGTs), which have highly conserved (60–80% identity) motifs for putative secondary plant glycosyltransferases (PSPGs), are responsible for the glycosylation of various metabolites, using UDP-glucose as their sugar supplier (Paquette et al., 2003; Yonekura-Sakakibara and Hanada, 2011; Bönisch et al., 2014). Citrus UGTs mainly utilize the UDP-sugar supplier for their glycosylation processes, such as glucosylation and rhamnosylation, to form the most abundant flavanone di-glycosides (Frydman et al., 2004; Owens and McIntosh, 2009; Frydman et al., 2013; Devaiah et al., 2016).
The Citrus genus includes several major cultivated species, such as C. sinensis (sweet orange), C. reticulata (tangerine and mandarin), C. maxima (pummelo), C. paradisi (grapefruit), and C. medica (citron) (Xu et al., 2013). The global production of citrus exceeded 124 million tons in 2016 (FAO, 2016), which ranks it at the top among all fruit crops. Bitterness is an important attribute of taste and significantly affects consumers’ acceptance of various citrus fruits. Reducing bitterness by adding naringinase has become an important procedure in citrus juicing processes (Ni et al., 2012). The primary cause of bitterness is the taste of neohesperidoside (Rousseff et al., 1987; McIntosh and Mansell, 1997), a flavanone di-glycoside. Although neohesperidoside has board-spectrum functions in providing resistance to insects and protection against high and low temperatures (Xu et al., 2007; Agut et al., 2014; Chen et al., 2015a), reducing bitterness is required to improve the taste of citrus cultivars and hence to increase their economic value.
The GTs that affect the formation of the bitter-tasting flavanone glycoside have become targets of biotechnological applications in citrus species (Lewinsohn et al., 1989; Bar-Peled et al., 1993). They include 1,2-rhamnosyltransferase (1,2RhaT) and 1,6-rhamnosyltransferase (1,6RhaT). In pummelo, 1,2RhaT converts flavanone-7-O-glucosides (F7Gs) to bitter-tasting flavanone-7-O-neohesperidosides (McIntosh and Mansell, 1990; Bar-Peled et al., 1993; Frydman et al., 2004), whilst in sweet orange, mandarin, and citron, 1,6RhaT converts F7Gs to bitterless flavanone-7-O-rutinosides (see Supplementary Fig. S1 at JXB online). Interestingly, introgression of the 1,6RhaT gene into grapefruit and sour orange (C. aurantium) significantly reduces the accumulation of neohesperidoside due to direct and strong substrate competition (Frydman et al., 2013). 1,6RhaT is not functional in pummelo because of frameshifts, suggesting that the rutinoside branch is absence in pummelo fruit. It is therefore important to clarify the molecular mechanisms of the specific accumulation of bitter-tasting neohesperidoside in diverse citrus species if advances are to be made in improving the fruit quality. Another potential approach in pummelo fruit is that new GTs that can compete with 1,2RhaT for the F7G substrate could be used indirectly to effectively reduce the level of neohesperidosides whilst also increasing the levels of bioactive glycosides, which are beneficial to human health.
cDNAs encoding various sugar–sugar GTs have been cloned from higher plants and ectopically expressed for functionally characterization. These include flavanone/flavone-7-O-glucoside-1,2-rhamnosyltransferase from C. maxima (Frydman et al., 2004), anthocyanidin-3-O-glucoside-1,2-glucosyltransferase from Ipomoea nil (Morita et al., 2005), anthocyanidin-3-O-glucoside-1,2-glucuronyltransferase from Bellis perennis (Sawada et al., 2005), favonol-3-O-glucoside-1,6-glucosyltransferase from Catharanthus roseus (Masada et al., 2009), and flavonoid-7-O-glucoside-1,6-rhamnosyltransferase from C. sinensis (Frydman et al., 2013). Although several flavonoid di-glycosides have been identified in various higher plants (Masada et al., 2009; Chen et al., 2015b), there have been few studies on the enzymes of flavor-related metabolic networks in citrus species. Understanding the glycosylation mechanisms would facilitate metabolic engineering to produce more bioactive glycosides that would benefit human health.
In this study, three genes with high levels of sequence homology to Cit1,2RhaT were identified from a citrus genome database. Functional analyses showed that they encoded flavonoid di-glycosyltransferases, and thus they were named as CitdGlcTs. The CitdGlcTs could transfer a glucose and Cit1,2RhaT could transfer a rhamnose to F7G to form F7GG and neohesperidoside, respectively. The direct competition for F7G between the CitdGlcTs and Cit1,2RhaT may be a key factor for the formation of either bitterless F7GG or bitter neohesperidoside, and it has the potential to be manipulated to improve the taste and level of bioactive flavonoids in pummelo fruit.
Materials and methods
Plant material and collection of samples
The citrus accessions used in this study were common cultivars and most were maintained by the National Citrus Breeding Center in Huazhong Agricultural University, Wuhan, China. The 50 accessions of Citrus, Poncirus, and Fortunella are listed in Supplementary Table S1. Hybrid populations of ‘Red tangerine’ × ‘Trifoliate orange’ and ‘Hirado butun’ pummelo × ‘Fairchild’ tangelo were established in 2003, and fruit from 74 and 20 F1 plants were collected from these two populations, respectively. In addition, fruit samples of C. maxima ‘Fenghuangyu’, C. maxima ‘Kao Pan’ pummelo, C. maxima ‘Huanonghongyu’, and C. maxima × C. paradisi ‘Hirado Butun’ pummelo were collected at five developmental stages, namely 60, 90, 120, 150, and 210 d post-anthesis (DPA). Three biological replicates per accession were analysed, with each replicate consisting of six fruit from three different plants, and the juice-sac tissues were collected for flavonoid detection and total RNA extraction. Leaf samples of all the accessions were collected at a young stage in the spring season and used for genomic DNA extraction. Both fruit and leaf samples were immediately frozen in liquid nitrogen and stored at –80 °C until use.
Tobacco Bright Yellow 2 (BY2) callus was kindly provided by Prof. Botao Song at the College of Horticulture and Forestry, Huazhong Agricultural University.
BLAST analysis, cluster analysis, and gene cloning
BLAST analysis was performed using Cm1,2RhaT (GenBank accession no. AY048882) as the query sequence against the C. sinensis Annotation Project database (http://citrus.hzau.edu.cn/cgi-bin/orange/blast) and the Phytozome 12 database (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=BLAST). Sequences with >80% homology were selected to develop a graphical representation of Cit1,2RhaT and Cit1,2RhaT-like genes (CitdGlcT-1, CitdGlcT-2, and CitdGlcT-3) from different citrus germplasms regarding location, copy number, and coding-region sequences.
The coding regions of Cit1,2RhaT and Cit1,2RhaT-like genes were then amplified from the genomic DNA of young leaves of Citrus, Poncirus, and Fortunella extracted using a DN-Plant DNA Mini Kit (Aidlab, China) using primers designed based on the C. maxima and C. sinensis genome sequences (Supplementary Table S2).
To perform the cluster analysis, the sequences of Cm1,2RhaT and Cm1,2RhaT-like together with another 19 plant flavonoid GTs identified from the GenBank (https://www.ncbi.nlm.nih.gov/) and genome databases were aligned using ClustalW and viewed with GENEDOC 3.2. Phylogenetic analysis was performed using the MEGA 5.0 software with the neighbor-joining method based on ClustalW multiple analysis.
Flavonoid determination
Flavonoid aglycones and glycosides were identified using a 1200 Series Rapid Resolution HPLC system coupled with QTOF 6520 mass spectrometer (Agilent). A Zorbax Eclipse Plus C18 (1.8 mm, 2.1×100 mm) and a reverse-phase analytical column (Agilent) was used for separation at 35 °C. The mobile phase consisted of 0.1% formic acid in deionized water (A) and 0.1% formic acid in acetonitrile (B). The program for separation and source conditions for electrospray ionization were adopted from the methods described by Liu et al (2016) and Chen et al (2015b), respectively. Mass spectrometry analysis was performed using MassHunter (Agilent). Qualitative analyses were performed by comparing the retention times, UV spectra, MS, and ESI-MS/MS spectra with commercially available standards.
For quantitative analysis using HPLC, a 1525 Binary HPLC pump coupled with a 2998 Photodiode Array Detector and a 717plus Autosampler were used (Waters). Samples were fractionated at room temperature using a C18 Hypersil GOLD column (4.6 ×250 mm, 5 µm, Thermo scientific) at an overall flow rate of 1.0 ml min–1. The mobile phase was the same as that for the LC/MS analysis. The gradient elution system was as previously described by Chen et al. (2015b). The UV-Vis spectra were recorded from 210–400 nm with a detection wavelength of 280 nm. Quantification of the flavonoid glycosides was carried out using a calibration curve (y=16064x–3531, R2=0.99996) for a naringin standard, as flavonoids have similar structural and spectral properties.
Production of transgenic Fortunella plants over-expressing Cm1,2RhaT
The epicotyls of seedlings of F. hindisti ‘Hong Kong kumquat’ were used as the explants for genetic transformation because the kumquat has a short juvenile phase. The full-length coding region of Cm1,2RhaT was amplified from the genomic DNA of C. maxima ‘Fenghuangyu’ via PCR using the primers 5´-GCTCTAGAATGGATAC CAAGCATCAAG-3´ (XbaI site in italics) and 5´-CGAGCTCTTATTCAGA TTTCTTGACA AGCTG-3´ (SacI site in italics). The amplified products were digested with XbaI and SacI, and inserted into the corresponding sites of the binary plant expression vector pBI121, resulting in final construct pBI-Cm1,2RhaT. The construct was transferred into epicotyls using an Agrobacterium-mediated transformation protocol as previously described (Cao et al., 2012). Using kanamycin selection medium, six putative transgenic shoots were generated from 173 epicotyl explants. Two of these plants (16-1 and 18-1) survived to maturity and produced fruit. They were tested by PCR analysis using a forward primer annealing to the 35S promoter and a reverse primer annealing to the Cm1,2RhaT gene to confirm the presence of the transgene, and tested by qRT-PCR analysis using the primers 5´-CCTGAGGTCCTTTTCCAACCA-3´ and 5´-GGATTCCGGCAGCTTCCATT-3´ to determine the expression of the transgene. The plants were confirmed to be transgenic and were used for flavonoid profile analysis.
Expression of Cit1,2RhaT, CitdGlcT-1, and CitdGlcT-2 in tobacco BY2 cells
The coding sequences of Cit1,2RhaT, CitdGlcT-1, and CitdGlcT-2 were cloned into the binary vector PH7WG2D under the control of the CAMV-35S promoter and translationally fused to the green fluorescent protein (GFP) reporter gene, via the BP and LR reaction of the Gateway system (Pak et al., 2009; Kuma et al., 2015; Rohani et al., 2016). The recombinant vector was then transferred into Agrobacterium tumefaciens GV3101 using a heat-shock method. A single bacterial colony containing the vector (selected on an LB plate with spectinomycin) was used for inoculation in liquid LB medium (~50 ml) and grown at 28 °C until the absorbance reached 0.6–0.8 at 600 nm. BY2 fresh callus was transferred from solid medium to liquid Murashige and Skoog medium for generation of suspension cell cultures, and grown for 4 d to reach the exponential growth phase (Supplementary Fig. S2A, B). The BY2 suspension cultures were then infected by adding 100 μl of the Agrobacterium cells to 4 ml of BY2 cells. The BY2 and Agrobacterium cells were then co-cultivated for 48 h to generate transgenic BY2 cells (Supplementary Fig. S2C). The co-cultivated BY2 cells were transferred to solid selection medium containing cephalosporins (400 μg ml–1) and hygromycin (50 μg ml–1) and cultured for ~3 weeks to produce friable transgenic calli (Supplementary Fig. S2D, E). These were then collected and transferred to a fresh selection medium and cultured for 4 weeks to produce large calli (Supplementary Fig. S2F, G). At this stage, the fluorescence of the transgenic calli were detected using an epifluorescence stereomicroscope with a GFP filter with excitation at 488 nm. Detection of a strong GFP signal was positive indication for the BY2 cells containing the transgene construct (Fig. S2H, I). These methods for establishing suspension cell cultures and for transformation were modified from Frydman et al. (2004, 2013) and optimized for our study.
Biotransformation assays in tobacco BY2 cells
To carry out biotransformation assays, fresh cultures (2 g each) of wild-type (WT) and transgenic (BY2-Cit1,2RhaT, BY2-CitdGlcT-1, and BY2-CitdGlcT-2) cells were grown on agar medium for 7 d and then 1 g fresh cells were added to 20 ml liquid media in a 50 ml triangular flask and cultured for another 4 d to obtain the scattered and highly active cells (Supplementary Fig. S2J). Then 50 μl of flavonoid substrate stock solution (10 mg ml–1) was added to the flask. The cells were further cultured for 48 h, then collected by centrifugation, followed by a treatment of freeze-drying at –54 °C using a Lyolab 3000 (Heto-Holten, Denmark) and extraction with 90% methanol (5 ml) to release the end-point products (Chen et al., 2015b). After extraction, the cell debris was centrifuged down and the supernatant was collected and dried to 1 ml using a vacuum Concentrator 5301 (Eppendorf), and then filtered through a 0.22-µm Micropore filter before HPLC and LC/MS analysis.
Quantitative analysis of reaction products from the WT and transgenic cells was performed with three biological replicates. The net amount of product was defined as the amount of product in the transgenic cells minus that in the WT control cells.
RNA extraction, RNA sequencing, and qRT-PCR
To analyse the expression profiles of flavonoid biosynthesis-related genes, total RNAs were isolated from juice-sac tissues of C. maxima ‘Fenghuangyu’, ‘Kao Pan’, ‘Huanonghongyu’, and ‘Hirado Butun’ at 60, 90, 120, 150, and 210 DPA using a modified Trizol extraction protocol (Liu et al., 2016). RNA sequencing (RNA-seq) was performed as previously described (Shen et al., 2017). Briefly, the mRNA used for RNA-seq library construction was purified from the ‘Fenghuangyu’ total RNA samples using oligo-dT attached to magnetic beads; its concentration was measured using a Qubit 2.0 fluorometer (Life Tech, China) and its integrity was confirmed using a 2100 Bioanalyzer (Agilent). The RNA-seq libraries were prepared using a NEBNext Ultra RNA Library Prep Kit (New England Biolabs) according to the manufacturer’s instructions, and sequenced using an Illumina HiSeq 2000. For gene expression analysis, the number of sequence reads was calculated and then normalized to FPKM (fragments per kilobase of exon per million fragments mapped) (OuYang et al., 2016).
To validate the differential expression identified by RNA-seq, quantitative real-time RT-PCR (qRT-PCR) was performed on a Prism 7900HT (Applied Biosystems) with the SYBR Green system. All gene expression analyses were performed with three independent biological replicates. The gene-specific primers used in the analysis are listed in Supplementary Table S2. The actin gene was used as the reference control.
Treatment of mature juice sacs of C. maxima with naringenin-7-O-glucoside
Fresh juice sacs of C. maxima ‘Wanbeiyu’, ‘Huanonghongyu’, and ‘Fenghuangyu’ were separated from the fruit peel at 180 DPA and 1-g samples of tissue were placed in individual Petri dishes (2.5 cm diameter) containing 3 ml distilled water. There were 18 dishes for each cultivar. Three dishes per cultivar were supplemented with 0, 100, 200, 300, 500, or 1000 μg of naringenin-7-O-glucoside. The dishes were incubated at 30 °C for 60 h before the tissues were tested for naringenin-7-O-di-glucoside production using HPLC and LC/MS.
Results
Identification of three CitdGlcT genes related to 1,2RhaT in citrus genomes
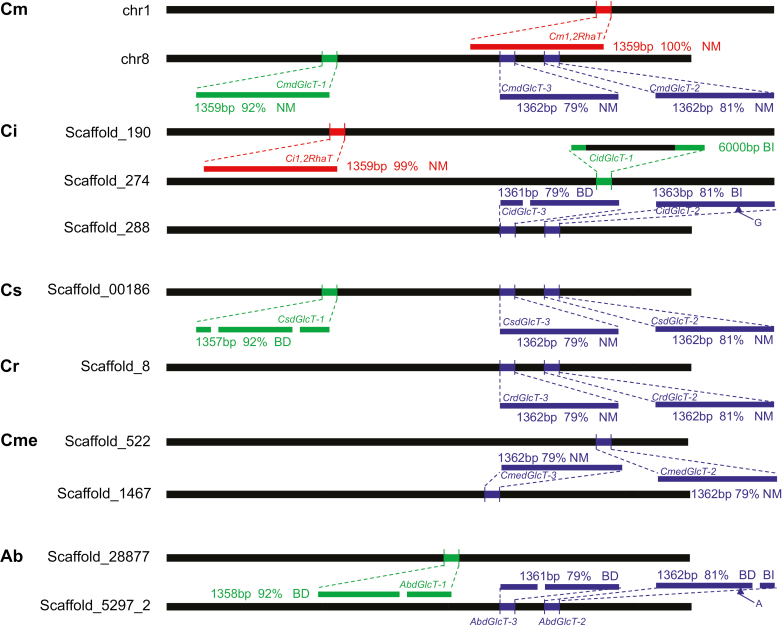
1,2RhaT and three dGlcT genes with high nucleotide sequence homology to Cm1,2RhaT were identified in the genomes of various citrus species (Fig. 1). The CitdGlcT genes could be classified into two categories according to their homology to Cm1,2RhaT: CitdGlcT-1 had high similarity with 92% homology, whilst CitdGlcT-2 and CitdGlcT-3 showed medium-high similarity (79–81% homology). In addition, CitdGlcT-2 and CitdGlcT-3 were arranged as tandem repeats in the species. The genome of C. maxima ‘Wanbeiyu’ contained CmdGlcT-1, CmdGlcT-2, and CmdGlcT-3 on chromosome 8, and Cm1,2RhaT on chromosome 1. All four genes were found in the C. ichangensis genome, but only Ci1,2RhaT was functional as insertions or deletions caused frameshifts in the three CidGlcT genes. In C. sinensis, CsdGlcT-1, CsdGlcT-2, and CsdGlcT-3 were found on the same Scaffold_00186, whereas CsdGlcT-1 was non-functional because of two single-base deletions at nucleotide positions 279 and 1289. The 1,2RhaT gene was not found in the C. sinensis genome assembly. The C. clementina and C. medica genomes contained dGlcT-2 and dGlcT-3, but 1,2RhaT and dGlcT-1 were not found. In Atalantia buxifolia, Ab1,2RhaT was not found and all three AbdGlcT genes were non-functional because of frameshifts caused by various insertions and deletions. Of these citrus species, only C. maxima and C. ichangensis can accumulate the bitter-tasting neohesperidoside, suggesting that 1,2RhaT and dGlcT-1 may be relevant to its accumulation.
Fig. 1.
Schematic view of chromosomal loci of Cit1,2RhaT, CitdGlcT-1, CitdGlcT-2, and CitdGlcT-3 in six citrus species. The red segments represent Cit1,2RhaT; green represents CitdGlcT-1; and blue represents CitdGlcT-2 and CitdGlcT-3. CitdGlcT-2 and CitdGlcT-3 share 95% nucleotide sequence homology in the same germplasm. The genes were mostly identified by searching genome databases, but identification of CitdGlcT-2 and CitdGlcT-3 in Cs and Cr was based on both genome databases and our own PCR-amplification-sequencing results. The percentage values indicate the nucleotide sequence homology to Cm1,2RhaT (GenBank accession no. AY048882, used as the reference sequence). NM, normal sequence; BD, base deletion; BI, base insertion; Cm, Citrus maxima; Ci, C. ichangensis; Cs, C. sinensis; Cr, C. reticulate; Cme, C. medica; Ab, Atalantia buxifolia.
Cit1,2RhaT determines neohesperidoside accumulation in the fruit of Citrus and Poncirus
To examine the correlation between the 1,2RhaT and dGlcT genotypes and the bitter-tasting neohesperidoside (i.e. naringin) accumulation phenotype, assorted paired primers were used to amplify 1,2RhaT and dGlcT-1 from the DNA of 50 accessions belonging to three citrus genera, Citrus, Poncirus, and Fortunella (Supplementary Table S1). Citrus and Poncirus accumulated naringin in their fruits if the 1,2RhaT gene contained no frameshifts, as verified by sequencing the PCR-amplified gene fragments. Other accessions did not accumulate naringin if the 1,2RhaT gene could not be amplified or contained various frameshifts. In contrast, dGlcT-1 was found to be functional only in C. maxima accessions ‘Wanbeiyu’ and ‘Liangpinyu’, and showed no obvious correlation with the accumulation of naringin. PCR amplification together with HPLC analysis of the 50 accessions thus revealed that 1,2RhaT played a crucial role in neohesperidoside accumulation in Citrus and Poncirus.
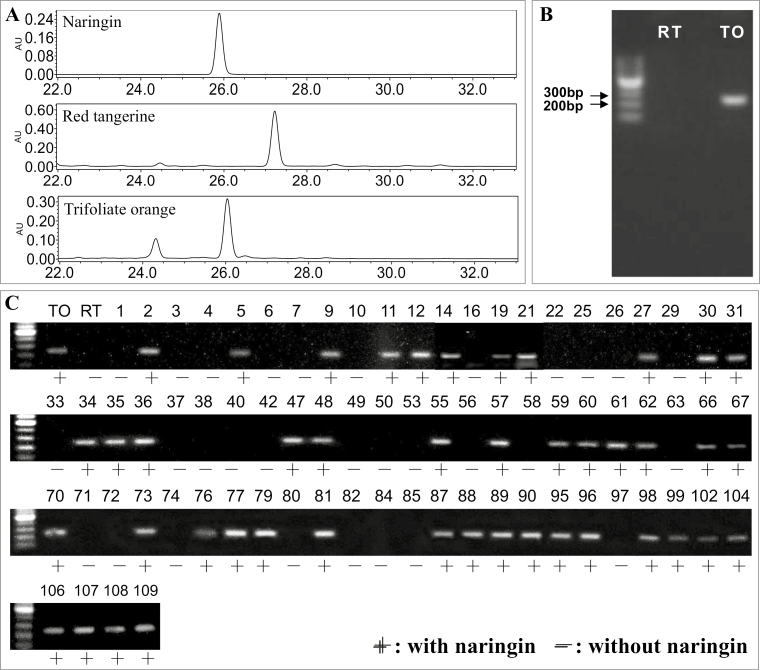
To confirm this role of 1,2RhaT, populations of C. reticulata ‘Red tangerine’, P. trifoliata ‘Trifoliate orange’, and their F1 hybrids were analysed (Fig. 2). ‘Trifoliata orange’ contained 1,2RhaT and accumulated normal levels of naringin, while ‘Red tangerine’ neither contained 1,2RhaT nor accumulated naringin (Fig. 2A; Supplementary Table S1). HPLC analysis showed that 44 of 74 F1 hybrids accumulated naringin in the fruit (Fig. 2C), fitting a 1:1 ratio (Chi-square test, P<0.05). This indicated that a dominant heterozygous gene controlled the accumulation of naringin. By using primers designed to amplifying 244 bp of 1,2RhaT, a specific PCR fragment was detected in all hybrids with naringin but not in any hybrid without naringin. These results suggested that the accumulation of naringin was co-segregated with 1,2RhaT in the hybrid population. In addition, we analysed 21 randomly selected hybrids of ‘Hirado butun’ pummelo (C. maxima × C. paradisi) and ‘Fairchild’ tangelo (C. reticulata × C. paradisi). ‘Hirado butun’ pummelo contained two alleles of 1,2RhaT and accumulated naringin, whereas ‘Fairchild’ tangelo did not contain 1,2RhaT and was without naringin (Supplementary Fig. S3A, B; Supplementary Table S1). All the hybrid offspring accumulated naringin in their fruit and showed the specific DNA fragment representing 1,2RhaT (Supplementary Fig. S3C). These results suggested that both alleles of 1,2RhaT could confer the accumulation of neohesperidoside.
Fig. 2.
Co-segregation between 1,2RhaT and naringin accumulation in the progeny of ‘Red tangerine’ × ‘Trifoliata orange’ (Citrus reticulata × Poncirus trifoliata). (A) Naringin is the most abundant and representative neohesperidoside in citrus fruit and was detected in mature fruits of ‘Trifoliata orange’ but not in those of ‘Red tangerine’. (B) A DNA fragment of 1,2RhaT could be amplified from ‘Trifoliata orange’ (TO) but not from ‘Red tangerine’ (RT) using PCR primers 5´-GTGGATTATTGCTCAGCGA-3´ and 5´-ATTGGTACCCCGAAAACCAT-3´. (C) Strong co-segregation was detected between the presence of 1,2RhaT and naringin in F1 progeny (individuals numbered 1–109). The DNA ladders are 100 bp.
Two Fortunella accessions were found to contain the 1,2RhaT gene with eight single-nucleotide polymorphisms (SNPs) compared to Cm1,2RhaT, and neither naringin nor any other neohesperidosides were detected in their fruit (Supplementary Table S1). To determine the possible reasons for the absence of neohesperidoside in Fortunella, the cDNA of Cm1,2RhaT under the control of the CaMV35S was transferred in transgenic plants of F. hindisti ‘Hong Kong kumquat’. Plants of two independent transgenic lines were grown to maturity and were confirmed to over-express Cm1,2RhaT. However, neohesperidosides were not detected in the fruit of these transgenic plants (Supplementary Fig. S4). These results suggested that the cause of the undetectable levels of flavanone glycosides in Fortunella was not the eight SNPs in the Fh1,2RhaT gene. Previous studies have shown that Fortunella fruit accumulate a high level of 3′,5′-di-glucopyranosylphloretin, but no F7G (Ito et al., 2017). Thus, it is possible that the absence of the F7G substrate is the reason for the lack of neohesperidosides in Fortunella fruit.
CitdGlcTs are flavonoid-7-O-di-glucosyltransferases
Phylogenetic analysis classified CitdGlcT-1, CitdGlcT-2, and CitdGlcT-3 into cluster E together with Cit1,2RhaT in an unrooted neighbor-joining tree (Supplementary Fig. S5). Enzymes grouped into different clusters are known to catalyse glycosylation at different positions of flavonoids using different types of sugars. For example, enzymes in clusters A–C are known to catalyse glucosylation, galactosylation, and rhamnosylation at position 3, 5, and 7 of flavonoids, respectively. Enzymes in cluster D catalyse rhamnosylation and xylosylation of flavonoid-O-glucosides to form flavonoid-O-di-glycosides, and enzymes in cluster E catalyse glucosylation, rhamnosylation, and glucuronosylation with substrate flavonoid-O-glucosides to form flavonoid-O-di-glucosides or flavonoid-O-neohesperidosides. Notably, clusters A–C were further grouped together to form Group I, and they all catalyse the formation of flavonoid mono-glycosides from flavonoid aglycones. Clusters D and E together formed Group II, and they catalyse the formation of flavonoid di-glycosides from flavonoid mono-glycosides. The classification of CitdGlcT-1, CitdGlcT-2, and CitdGlcT-3 into cluster E suggested that these CitdGlcTs are probably sugar–sugar GTs.
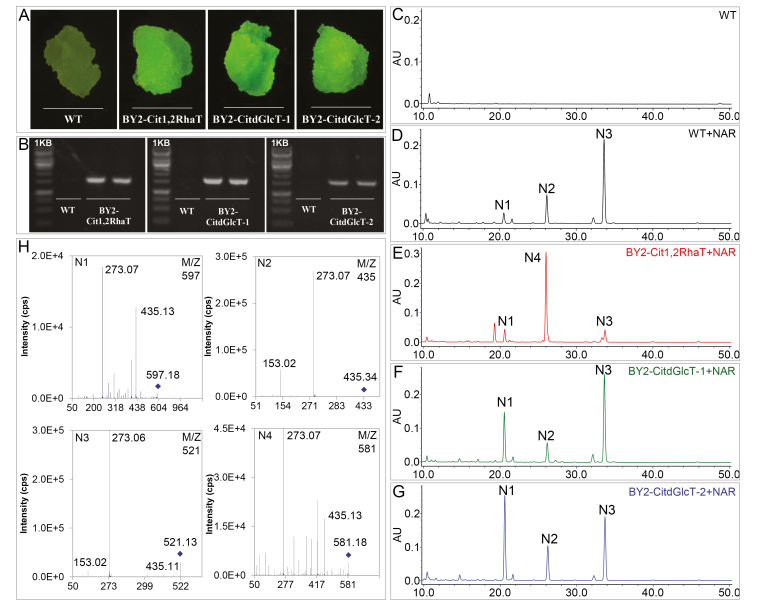
To verify the catalytic activity of the CitdGlcTs and to compare them with Cit1,2RhaT, tobacco BY2 cells were transformed using a binary vector over-expressing Cit1,2RhaT, CitdGlcT-1, or CitdGlcT-2 fused to the GFP reporter. The transformation was confirmed by viewing the GFP fluorescence and by PCR amplification of DNA of Cit1,2RhaT, CitdGlcT-1, and CitdGlcT-2 from the BY2 cells (Fig. 3A, B). The lack of any flavonoid peak from WT cells in HPLC analysis (Fig. 3C) indicated that this BY2 cell culture system could not produce flavonoids to a detectable level, making it a suitable system for subsequent biotransformation assays.
Fig. 3.
Testing the functions of Cit1,2RhaT, CitdGlcT-1, and CitdGlcT-2 using tobacco BY2 transgenic cell cultures. BY2 calli produced on medium containing hygromycin (50 μg ml–1) after Agrobacterium infection were confirmed to be transgenic by detection of GFP fluorescence (A) and amplification of DNA fragments using PCR primers specifically annealing to the transgene sequences (B). wild-type (WT) BY2 calli did not produced any flavonoids as determined by HPLC analysis (C), but produced three flavonoid compounds N1, N2, and N3, after application of naringenin (NAR) as a substrate (D). BY2-Cit1,2RhaT transgenic cells produced a new compound, N4 (E). BY2-CitdGlcT-1 and BY2-CitdGlcT-2 transgenic cells produced a higher level of N1 compound to the WT cells (F, G). LC/MS analysis (H) showed that N1 (m/z, 597→435→273) was naringenin-7-O-di-glucoside, N2 (m/z, 435→273) was naringenin-7-O-glucoside, N3 (m/z, 521→435→273) was naringenin-7-O-malonylglucoside, and N4 (m/z, 581→435→273) was naringin.
After applying naringenin (a flavanone aglycone) as a substrate, two weak (N1, N2) and one strong (N3) flavonoid peaks were detected in the WT BY2 cells (Fig. 3D) and these were identified as naringenin-7-O-di-glucoside, naringenin-7-O-glucoside, and naringenin-7-O-malonylglucoside, respectively, using LC-MS analysis (Fig. 3H). N2 was converted from naringenin by a flavonoid-7-O-glucosyltransferase (7GlcT). N1 and N3 were converted from N2 by a dGlcT and an acyltransferase, respectively. In BY2-Cit1,2RhaT transgenic cells, one strong peak, N4, was detected while the N1 and N3 peaks were weak, and N2 was not detected (Fig. 3E). N4 was identified as naringenin-7-O-neohesperidoside (Fig. 3H). The strong accumulation of N4 and reduced levels of N2 and N3 indicated that most N2 was converted to N4 by the transgenic Cit1,2RhaT and the remaining small amounts of N2 were converted to N3. In the BY2-CitdGlcT-1 and BY2-CitdGlcT-2 cell cultures, a significant peak of N1 was detected, although the N2 and N3 peaks were similar to those in the WT (Fig. 3F, G). N1 was identified as naringenin-7-O-di-glucoside (Fig. 3H). In addition, when flavone apigenin and flavonol quercetin were added as substrates, similar products were detected in the BY2-CitdGlcT-1 and BY2-CitdGlcT-2 transgenic cell cultures with HPLC and LC-MS (Supplementary Fig. S6). These results indicated that CitdGlcT-1 and CitdGlcT-2 could convert flavonoid-7-O-glucosides into flavonoid-7-O-di-glucosides but not into flavonoid-7-O-neohesperidoside.
F7Gs are the preferred substrate of CitdGlcTs and Cit1,2RhaT
To further determine the substrate specificity of Cit1,2RhaT, CitdGlcT-1, and CitdGlcT-2, 500 μg of 12 different flavonoid compounds that represent three different flavonoid subclasses (flavanone, flavone, and flavonol), were added to transgenic BY2 cells (Table 1). In the BY2-CitdGlcT-1 cells there was greater net accumulation of flavanone-7-O-di-glucosides (F7GGs) (103–137 μg) compared with flavone-7-O-di-glucosides (6–51 μg) and flavonol-7-O-di-glucosides (0.3–11 μg). A similar trend of accumulation levels was also seen in the BY2-CitdGlcT-2 cells. In the BY2-Cit1,2RhaT cells, net accumulation of products was 371–406 μg for flavanone and 229–327 μg for flavone, but flavonol was not detectable. These results showed that F7Gs are the preferred substrate for both the dGlcTs and 1,2RhaT.
Table 1.
Glucosyltransferase activity of 1,2RhaT and dGlcTs for different flavonoid substrates
| Subclass | Substrate applied | Structure | Substituent group | BY2-CitdGlcT-1 and BY2-CitdGlcT-2 | BY2-Cit1,2RhaT | |||
|---|---|---|---|---|---|---|---|---|
| Reaction product | Net amount (μg)1 | Reaction product | Net amount(μg)1 | |||||
| CitdGlcT-1 | CitdGlcT-2 | |||||||
| Flavanone |
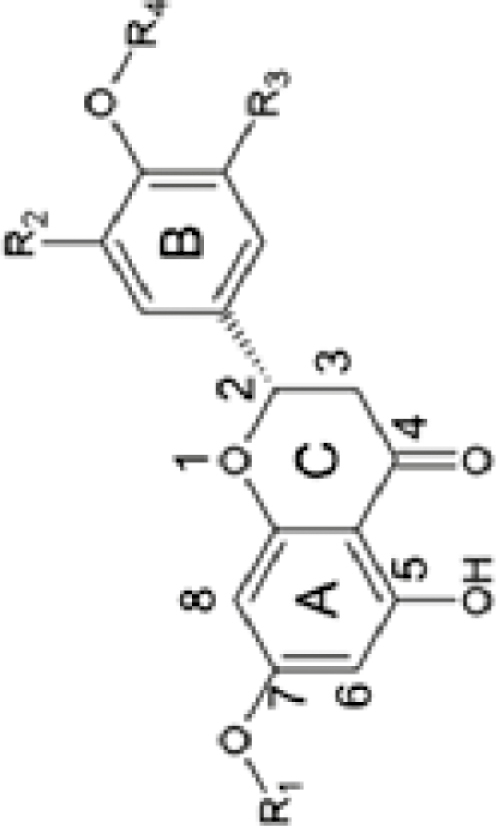
Naringenin Hesperetin Nar-7-O-Glu |
|
R1=H, R2=H, R3=H, R4=H R1=H, R2=H, R3=OH, R4=CH3 R1=Glu, R2=H, R3=H, R4=H |
Nar-7-O-di-Glu (m/z 597.18) Hes-7-O-di-Glu (m/z 627.19) Nar-7-O-di-Glu (m/z 597.18) |
137.09±5.80a 112.84±0.55b 103.48±7.14c |
201.88±49.23a 135.54±5.20b 141.31±18.04b |
Naringin (m/z 581.19) Hesperedin (m/z 611.20) Naringin (m/z 581.19) |
406.54±1.41b 415.49±11.31a 371.61±18.67c |
| Flavone |
Apigenin Luteolin Diosmetin |
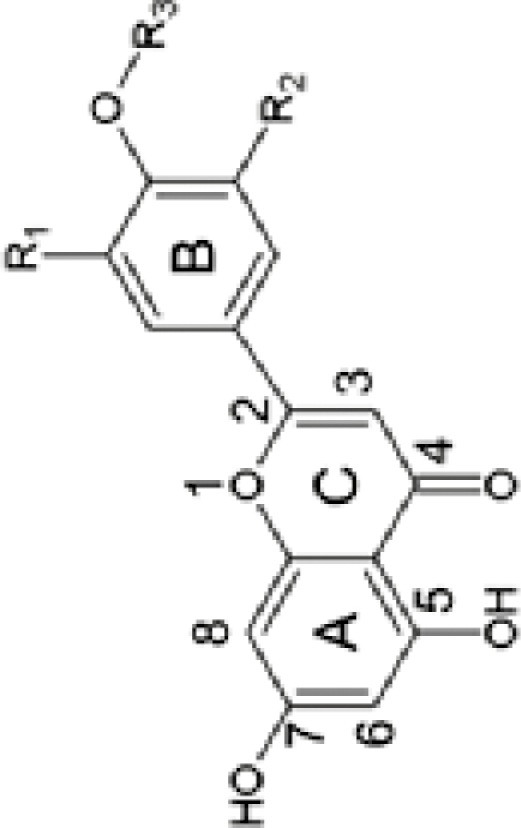
|
R1=H, R2=H, R3=H R1=OH, R2=H, R3=H R1=OH, R2=H, R3=CH3 |
Api-7-O-di-Glu (m/z 595.17) Lut-7-O-di-Glu (m/z 611.16) Dio-7-di-O-Glu (m/z 625.26) |
43.88±0.61d 30.99±0.64e 6.41±0.46gh |
51.59±3.58c 7.22±0.07f 21.69±0.92d |
Api-7-O-Neo (m/z 579.17) Lut-7-O-Neo (m/z 595.16) Dio-7-O-Neo (m/z 609.16) |
327.56±6.67d 229.41±6.88f 270.36±4.11e |
| Flavonol |
Kaempferol Quercetin Kael-3-O-Glu Kae-7-O-Glu Que-3-O-Glu Que-7-O-Glu |
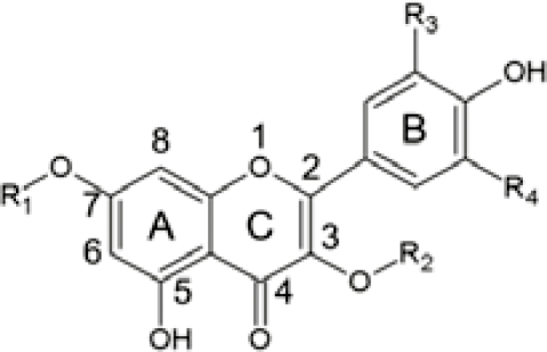
|
R1=H, R2=H, R3=H, R4=H R1=H, R2=H, R3=OH, R4=H R1=H, R2=Glu, R3=H, R4=H R1=Glu, R2=H, R3=H, R4=H R1=H, R2=Glu, R3=OH, R4=H R1=Glu, R2=H, R3=OH, R4=H |
Kae-7-O-di-Glu (m/z 611.16) Que-7-O-di-Glu (m/z 627.16) Kae-3-O-di-Glu (m/z 611.16) Kae-7-O-di-Glu (m/z 611.16) Que-3-O-di-Glu (m/z 627.16) Que-7-O-di-Glu (m/z 627.16) |
2.41±0.13i 8.68±0.10fg 1.46±0.38i 1.67±0.04i 1.75±0.15i 9.23±0.76f |
2.42±0.98g 10.58±4.52ef 0.31±0.26g 0.58±0.24g 1.56±0.05g 10.95±0.44e |
Kae-7-O-Neo (None detected) Que-7-O-Neo (None detected) Kae-7-O-Neo (None detected) Kae-7-O-Neo (None detected) Que-7-O-Neo (None detected) Que-7-O-Neo (None detected) |
0 0 0 0 0 0 |
1Net amount of reaction products catalysed by 1,2RhaT or dGlcTs with 0.5 mg substrate. Data are means (±SD) of three biological replicates. Different letters indicate significant differences as determined using Duncan’s test (P<0.05). Net amount refers to the reaction product accumulation in the transgenic cell culture minus that in the wild-type cell culture.
Uncoupled availability of CitdGlcT enzymes and F7G substrates leads to trace level accumulation of F7GGs in pummelo fruit
Using the tobacco BY2 system, we demonstrated that CitdGlcTs are flavonoid-7-O-di-glucosyltransferases and that they could convert F7Gs to F7GGs. However, although the products of Cit1,2RhaT were predominantly flavonoids, F7GGs were hardly detectable during fruit development of pummelo (Supplementary Fig. S7; Gattuso et al., 2007; Zhang et al., 2011; Chen et al., 2015b). Given that both F7Gs and CitdGlcTs are available in pummelo, it is intriguing why no F7GGs are detectable.
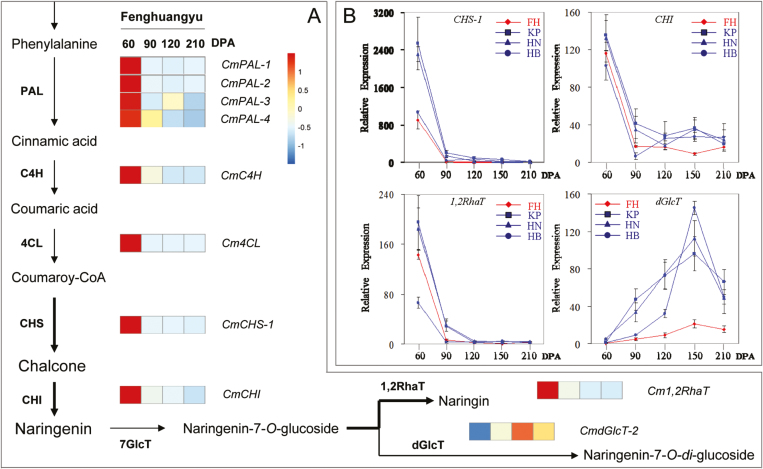
To answer this question, we first determined the expression patterns of flavonoid biosynthesis and glycosylation genes using RNA-seq analysis of tissues of C. maxima ‘Fenghuangyu’ fruit juice sacs. The expression levels of CmPAL, CmC4H, Cm4CL, CmCHS-1, and CmCHI were high at 60 DPA and had declined sharply at 90 DPA, after which they remained low through to fruit maturation (Fig. 4). As CHS and CHI are rate-limiting factors in flavonoid biosynthesis, the results suggested that the flavonoid substrates that could be used for glycosylation in pummelo fruit were largely produced at the young stage rather than at the mature stage. Cm1,2RhaT showed the same expression pattern as CmCHS-1 and CmCHI. In contrast, CmdGlcT-2 showed a different expression pattern with very low levels of transcripts at 60 DPA, although its transcript level was relatively high at 120 DPA (Fig. 4A). These RNA-seq results were confirmed by qRT-PCR analyses of CmCHS-1, CmCHI, Cm1,2RhaT, and CmdGlcT-2 expression in the juice sac tissues of four pummelo cultivars during fruit development (Fig. 4B). The expression levels of CmdGlcT-1 and CmdGlcT-3 were extremely low in the juice sacs at all developmental stages of the fruit (Supplementary Table S3), indicating they were not active. Collectively, the RNA-seq analysis and qRT-PCR results suggested that the presence of Cm1,2RhaT activity was coupled with the availability of F7Gs for the formation of neohesperidosides. However, the presence of CmdGlcT-2 was uncoupled with the availability of F7Gs. This may therefore explain why high levels of neohesperidosides but no F7GGs were detected in pummelo fruit.
Fig. 4.
Expression profiles of genes related to flavanone biosynthesis- in pummelo fruit undergoing ripening. (A) RNA-seq analysis of genes in the juice sacs of fruit of Citrus maxima ‘Fenghuangyu’ at 60–210 d post-anthesis (DPA). The RNA-seq FPKM vales of the genes are listed in Supplementary Table S3. (B) qRT-PCR analysis of selected genes in the juice sacs of fruits of four pummelo cultivars from 60–210 DPA: FH, ‘Fenghuangyu’; KP, ‘Kao Pan’; HN, ‘Huanonghongyu’; and HB, ‘Hirado Butun’. Data for ‘Fenghuangyu’ (the genotype used for RNA-seq) are highlighted in red.
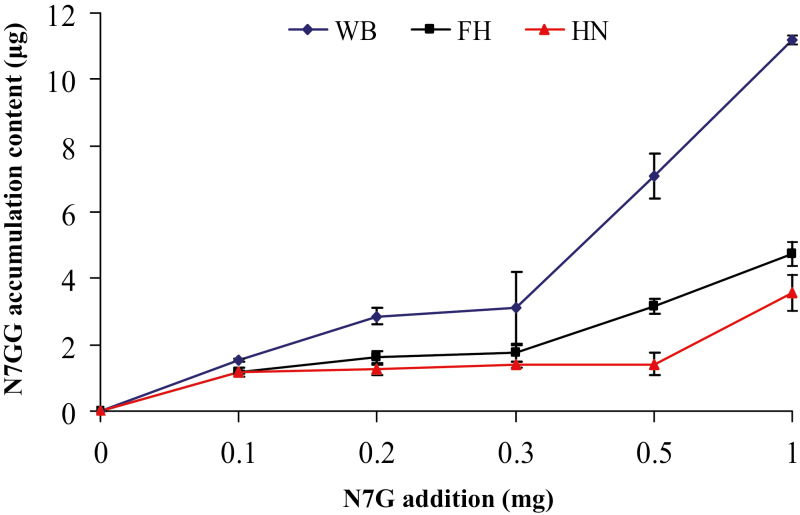
To confirm that CmdGlcT-2 in the fruit juice sacs was able to convert F7Gs to F7GGs, we added naringenin-7-O-glucoside to three C maxima cultivars, ‘Wanbeiyu’, ‘Huanonghongyu’, and ‘Fenghuangyu’ at 180 DPA. Several new metabolites were detected in these treated juice sacs including naringenin-7-O-di-glucoside, although naringenin acyl-glycosides were the major products (Supplementary Fig. S8). The content of naringenin-7-O-di-glucoside gradually increased as the amount of substrate was increased (Fig. 5), indicating that dGlcT-2 in mature fruit was capable of converting naringenin-7-O-glucoside to naringenin-7-O-di-glucoside. Based on all the observations, we concluded that citrus species do not produce F7GGs because of uncoupled availability of dGlcT enzymes and F7G substrates.
Fig. 5.
Levels of naringenin-7-O-di-glucoside (N7GG) in the juice sacs of fruit of different pummelo (Cirus maxima) species after application of naringenin-7-O-glucoside (N7G). Juice sacs were separated from the fruit at 180 d post-anthesis and incubated with naringenin-7-O-glucoside for 60 h. WB, ‘Wanbeiyu’; FH, ‘Fenghuangyu’; HN, ‘Huanonghongyu’.
Discussion
The genes encoding 1,2RhaT and dGlcTs might share a common evolutionary origin
We aimed to identify the enzymes that are either directly or indirectly involved in the accumulation of neohesperidosides in citrus fruits because they significantly contribute to imparting a bitter flavor (Rousseff et al., 1987; Frydman et al., 2004). We focused on Cit1,2RhaT because it is a known enzyme for neohesperidoside synthesis, and on CitdGlcTs because they have high sequence homology with Cit1,2RhaT. By analysing associations between neohesperidoside accumulation and different genotypes of Cit1,2RhaT and CitdGlcTs in various citrus accessions and in two breeding populations, we found that Cit1,2RhaT rather of CitdGlcTs was accountable for the accumulation in both Citrus and Poncirus. Cit1,2RhaT and CitdGlcTs share a common evolutionary origin, and exhibit a related but distinct functionality. They transfer two different sugars to the same flavonoid substrate, F7G, to form either F7GR or F7GG. CitdGlcTs may compete with Cit1,2RhaT for the same substrate to indirectly affect neohesperidoside levels if they are manipulated to be expressed in the same tissue at the same time.
In the genome of C. maxima ‘Wanbeiyu’ (Wang et al., 2017), Cm1,2RhaT and CmdGlcT-1, which share 92% homology in their nucleotide sequence, were found to be located on chromosomes 1 and 8, respectively (Fig. 1). The DNA sequences of the upstream, downstream, and coding regions of dGlcT-1 were more similar to those of Cm1,2RhaT than to CmdGlcT-2 and CmdGlcT-3, suggesting that it was most likely a duplication of an original 1,2RhaT. Furthermore, the expression profile of 1,2RhaT in the juice sacs was more similar to dGlcT-1 than dGlcT-2 during fruit development (Supplementary Fig. S9). In the C. reticulata genome, dGlcT-2 and dGlcT-3 were present but 1,2RhaT and dGlcT-1 were not (Fig. 1). Wu et al. (2018) found that modern mandarins are admixtures of wild C. reticulate and pummelo, which implies a plausible role of pummelo introgression in the selection of palatable mandarins. This might have led to the introgression of pummelo alleles of dGlcT-2 and dGlcT-3 but not of 1,2RhaT and dGlcT-1 into the mandarins, because mandarins harboring the latter might have been abandoned due to their bitter flavor. It remains possible that dGlcT-1 was a duplication of 1,2RhaT, while dGlcT-2 and dGlcT-3 were duplications of dGlcT-1, especially given that genome duplications have been identified between chromosomes 1 and 8 during citrus evolution (Xu et al., 2013). 1,2RhaT alleles might have played an important protective role in ancestral citrus germplasms (i.e. against insect pests) by promoting the accumulation of bitter-tasting neohesperidosides (Agut et al., 2014). However, the functional change from 1,2RhaT to dGlcTs might suggest that the flavor of citrus fruit has evolved from a more bitter to a less bitter taste. Interestingly, this would correspond to human preference for eating the fruit.
Domestication of citrus has involved artificial selection against 1,2RhaT
The molecular basis for the development of bitter and non-bitter-tasting citrus germplasms during domestication remains as an open question, although Frydman et al. (2004, 2013) showed have shown a strong correlation between fruit bitterness and levels of the 1,2RhaT enzyme, and a potential impact of artificial selection for non-bitter-tasting germplasms. Our study provided strong genetic evidence for artificial selection of 1,2RhaT mutations during citrus domestication, and leads us to propose a model to explain the regulatory mechanism that controls bitterness in sweet oranges (Fig. 6).
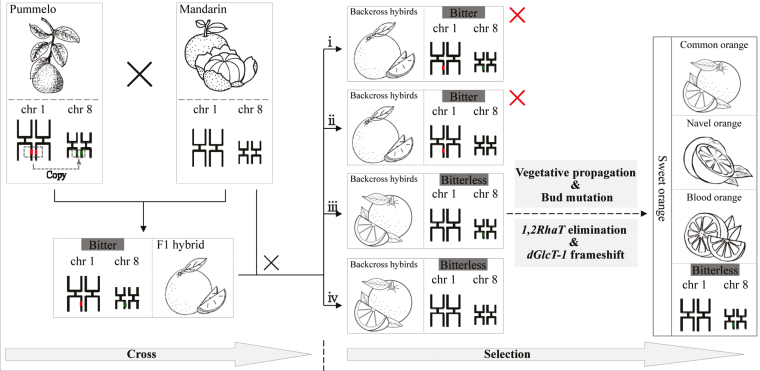
Fig. 6.
Proposed model of selection for bitterless sweet oranges. The pummelo genome contains both 1,2RhaT and dGlcT-1, but mandarin contains neither. Their F1 hybrid is heterozygous for both 1,2RhaT and dGlcT-1. Backcross progeny of the F1 hybrid and mandarin should include four genotypes with: (i) both 1,2RhaT and dGlcT-1; (ii) only 1,2RhaT; (iii) only dGlcT-1; or (iv) neither 1,2RhaT nor dGlcT-1. Types (iii) and (iv) may have been selected by animals and humans as a result of their less-bitter taste whilst the other two genotypes may have been eliminated due to their bitterness. Further bud mutations and subsequently selection and vegetative propagation might have resulted in the formation of navel and blood oranges, in addition the common orange, i.e. the three types of oranges present today. Red segments on chrosome 1 (chr 1) represent t 1,2RhaT; green segments on chr 8 represent dGlcT-1. Red crosses indicate the backcross-hybrids eliminated during selection.
Sweet oranges were derived from a cross between pummelo and mandarin, and therefore should have inherited a 1,2RhaT gene from pummelo. However, we found that modern sweet oranges do not have the 1,2RhaT gene or accumulate any flavanone neohesperidosides. Eight sweet oranges that we tested contained a dGlcT-1 gene with two single-base deletions but no 1,2RhaT gene (Supplementary Table S1). In addition, our results showed that CsdGlcT-1 in sweet orange had 99% homology with CmdGlcT-1 of pummelo. We therefore inferred that the initial F1 hybrids of pummelo and mandarin contained both 1,2RhaT and dGlcT-1 from the pummelo parent. Four genotypes could be produced from a backcross of the F1 and mandarin that contained: (i) both 1,2RhaT and dGlcT-1, (ii) only 1,2RhaT, (iii) only dGlcT-1, and (iv) neither of the two genes (Fig. 6). As a result of selection against bitter-tasting fruit, the and progenies with 1,2RhaT (i, ii) might have been eliminated, whereas the progenies without this gene (iii, iv) might have been maintained (Fig. 6). However, all the sweet orange accessions analysed so far in our studies have been type (iii) (Supplementary Table S1; Chen et al., 2015b). Further investigations are needed to determine the reason why no type (iv) have been found. Vegetative propagation and selection of bud mutations could have resulted in the formation of the three modern sweet orange types. It seems clear that a lot period of selection has led to the elimination of 1,2RhaT.
Uncoupled availability of enzymes and substrates is present in flavonoid biosynthesis in citrus
The RNA-seq data showed that the expression levels of CmPAL, CmC4H, Cm4CL, CmCHS, and CmCHI were highest at 60 DPA but this was followed by a dramatic decrease at 90 DPA, especially for CHS-1 and CHI (Fig. 4). This was consistent with previous reports on CHS and CHI expression patterns in citrus fruit (Moriguchi et al., 1999, 2001). Cm1,2RhaT, showed the same expression pattern as CmCHS-1 and CmCHI during fruit development. In contrast, the expression levels of CmdGlcT-2 were at their lowest at the fruitlet stage and increased during the later stages of fruit development. The RNA-seq profiles of CmCHS-1, CmCHI, Cm1,2RhaT, and CmdGlcT-2 were verified by qRT-PCR analysis of the juice sacs of four selected pummelo genotypes (Fig. 4B). As CHS and CHI are the maintenance and rate-limiting enzymes of flavonoid biosynthesis in citrus, their expression patterns suggested that large amounts of flavonoid aglycones were available before but not after 90 DPA. The coincident expression patterns between CHS, CHI, and 1,2RhaT explains the high level of accumulation of neohesperidosides in citrus. In contrast, opposite expression patterns between the rate-limiting genes and dGlcT-2 explain the trace levels of F7GGs. Although the expression level of dGlcT-2 peaked at 150 DPA (Fig. 4), the low expression levels of CHS-1 and CHI were unable to supply sufficient substrates to synthesize F7GGs and other flavanone glycosides during fruit maturation. However, when supplied with sufficient naringenin-7-O-glucoside, the mature juice sac tissues of pummelo could produce considerable amounts of naringenin glycosides (Supplementary Fig S8), suggesting that there were active GTs present in the mature citrus fruit.
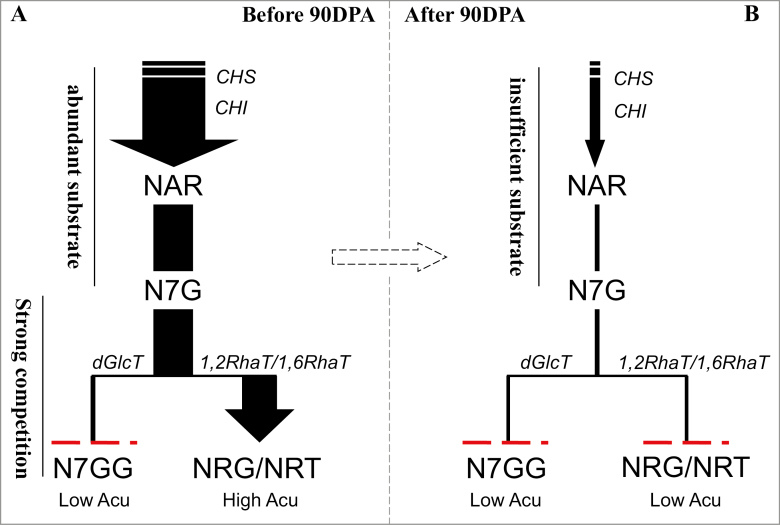
These results revealed that coupled availability of the Cit1,2RhaT enzyme and F7G promote the biosynthesis of neohesperidosides at the early development stages of citrus fruit (Fig. 7A). At later stages of fruit maturation, the decreased supply of flavonoid aglycone substrates might be the main cause of the reduction of biosynthesis of flavonoid glycosides (Fig. 7B). The peak stage of the flavonoid biosynthesis is probably before 90 DPA.
Fig. 7.
Proposed model of the conversion of naringenin-7-O-glucoside (N7G) to three different di-glucosides by glycosyltransferases in citrus. At the young fruit development stage (A), only a small amount of naringenin-7-O-di-glucoside (N7GG) is produced even though there is an abundant supply of the substrate N7G, because the latter is mostly converted to either naringin (NRG) or narirutin (NRT) by the highly expressed enzymes 1,2RhaT or 1,6RhaT, respectively, depending on the species. At the mature fruit development stages (B), small amounts of N7GG, NRG, or NRT are produced because there is only a small amount of N7G. The dashed red lines represent the much lower level of metabolite accumulation. There is a critical period after 90 d post-anthesis (DPA) when a sharp change in flavonoid biosynthesis occurs in the fruit of pummelo (Cirus maxima). Low Acu, low accumulation; High Acu, high accumulation.
In conclusion, guided by human selection, the deletion or functional mutation of 1,2RhaT has led to the step-by-step evolution of the flavor-related metabolic network in citrus. Our study provides a foundation for future breeding programmes for flavor improvement in citrus fruit. Based on the findings of this study, (1) genetic markers may be developed for early selection of bitterless citrus breeding lines before fruiting to accelerate the breeding programme, (2) transgenic approaches may be used to knock-out 1,2RhaT to eliminate the bitterness caused by neohesperidosides in pummelo fruit, and (3) manipulation of the competition for F7G between 1,2RhaT and dGlcT in pummelo fruit may be used to achieve a balance between taste and levels of bioactive flavonoids, which are beneficial for human health.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The biosynthesis pathway of flavanone glycosides in citrus.
Fig. S2. Illustration of the genetic transformation of flavonoid biosynthetic genes into tobacco BY2 suspension cells.
Fig. S3. Effect of 1,2RhaT on naringin accumulation in F1 hybrid fruit of ‘Hirado Butun’ pummelo × ‘Fairchild’ tangelo.
Fig. S4. Flavonoid profiles in transgenic ‘Hong Kong kumquat’ fruit over-expressing Cm1,2RhaT.
Fig. S5. Phylogenetic analysis of functionally characterized flavonoid glycosyltransferases.
Fig. S6. Testing the functions of CitdGlcT-1 and CitdGlcT-2 using tobacco BY2 transgenic cell cultures.
Fig. S7. Content of two major flavanone glycosides in the juice sacs of four C. maxima cultivars at five fruit developmental stages.
Fig. S8. Flavonoid profiles of maturation juice sacs of C. maxima ‘Wanbeiyu’ after application of naringenin-7-O-glucoside.
Fig. S9. Relative expression levels of five flavanone-biosynthesis-related genes in C. maxima ‘Wanbeiyu’ fruit juice sacs at five fruit developmental stages.
Table S1. Relationship of 1,2RhaT with accumulation of naringin in different citrus genotypes.
Table S2. Primers used for gene cloning and qRT-PCR analysis in this study.
Table S3. Normalized sequence reads from RNA-seq of juice sacs at four different development stages of pummelo ‘Fenghuangyu’ fruit.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFD1000204), the Special Fund for Agro-scientific Research in the Public Interest (grant no. 201303093), and the National Natural Science Foundation of China (NSFC, grant nos. 31672102 and 31521092).
Glossary
Abbreviations:
- 1,2RhaT
1,2-rhamnosyltransferase
- BY2
bright yellow 2
- DPA
days post-anthesis
- F7Gs
flavanone-7-O-glucosides
- F7GGs
flavanone-7-O-di-glucosides
- GTs
glycosyltransferases
References
- Agut B, Gamir J, Jacas JA, Hurtado M, Flors V. 2014. Different metabolic and genetic responses in citrus may explain relative susceptibility to Tetranychus urticae. Pest Management Science 70, 1728–1741. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Fluhr R, Gressel J. 1993. Juvenile-specific localization and accumulation of a rhamnosyltransferase and its bitter flavonoid in foliage, flowers, and young citrus fruits. Plant Physiology 103, 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D, Isayenkova J, Lim EK, Poppenberger B. 2005. Glycosyltransferases: managers of small molecules. Current Opinion in Plant Biology 8, 254–263. [DOI] [PubMed] [Google Scholar]
- Bönisch F, Frotscher J, Stanitzek S, Rühl E, Wüst M, Bitz O, Schwab W. 2014. Activity-based profiling of a physiologic aglycone library reveals sugar acceptor promiscuity of family 1 UDP-glucosyltransferases from grape. Plant Physiology 166, 23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breada WS. 2001. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Zhang J, Xu J, Ye J, Yun Z, Xu Q, Xu J, Deng X. 2012. Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. Journal of Experimental Botany 63, 4403–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li S, Xu J, Ding F, Wang Z, Cheng Y, Deng X. 2015a. Concentration and distribution of main bitter compounds in fruit tissues of ‘Oroblanco’ (Citrus grandis L.× Citrus paradisi Macf.). Scientia Horticulturae 193, 84–89. [Google Scholar]
- Chen J, Zhang H, Pang Y, Cheng Y, Deng X, Xu J. 2015b. Comparative study of flavonoid production in lycopene-accumulated and blonde-flesh sweet oranges (Citrus sinensis) during fruit development. Food Chemistry 184, 238–246. [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Owens DK, Sibhatu MB, et al. . 2016. Identification, recombinant expression, and biochemical analysis of putative secondary product glucosyltransferases from Citrus paradisi. Journal of Agricultural and Food Chemistry 64, 1957–1969. [DOI] [PubMed] [Google Scholar]
- FAO 2016. Citrus fruit – fresh and processed. Statistical bulletin 2016. Rome: FAO. [Google Scholar]
- Frydman A, Liberman R, Huhman DV, Carmeli-Weissberg M, Sapir-Mir M, Ophir R, W Sumner L, Eyal Y. 2013. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication. The Plant Journal 73, 166–178. [DOI] [PubMed] [Google Scholar]
- Frydman A, Weisshaus O, Bar-Peled M, Huhman DV, Sumner LW, Marin FR, Lewinsohn E, Fluhr R, Gressel J, Eyal Y. 2004. Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. The Plant Journal 40, 88–100. [DOI] [PubMed] [Google Scholar]
- Gattuso G, Barreca D, Gargiulli C, Leuzzi U, Caristi C. 2007. Flavonoid composition of Citrus juices. Molecules 12, 1641–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Fujimoto S, Suito F, Shimosaka M, Taguchi G. 2017. C-Glycosyltransferases catalyzing the formation of di-C-glucosyl flavonoids in citrus plants. The Plant Journal 91, 187–198. [DOI] [PubMed] [Google Scholar]
- Jay M, Viricel MR, Gonnet JF. 2006. C-glycosylflavonoids. In: Andersen OM, Markham KR, eds. Flavonoids, chemistry, biochemistry and applications. Boca Raton, FL: CRC Press/Taylor & Francis Group, 857–916. [Google Scholar]
- Jones P, Vogt T. 2001. Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213, 164–174. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Kuma KM, Lopes-Caitar VS, Romero CC, Silva SM, Kuwahara MK, Carvalho MC, Abdelnoor RV, Dias WP, Marcelino-Guimarães FC. 2015. A high efficient protocol for soybean root transformation by Agrobacterium rhizogenes and most stable reference genes for RT-qPCR analysis. Plant Cell Reports 34, 1987–2000. [DOI] [PubMed] [Google Scholar]
- Lewinsohn E, Britsch L, Mazur Y, Gressel J. 1989. flavanone glycoside biosynthesis in citrus: chalcone synthase, UDP-glucose:flavanone-7-O-glucosyl-transferase and -rhamnosyl-transferase activities in cell-free extracts. Plant Physiology 91, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Long J, Zhu K, Liu L, Yang W, Zhang H, Li L, Xu Q, Deng X. 2016. Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Scientific Reports 6, 25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masada S, Terasaka K, Oguchi Y, Okazaki S, Mizushima T, Mizukami H. 2009. Functional and structural characterization of a flavonoid glucoside 1,6-glucosyltransferase from Catharanthus roseus. Plant & Cell Physiology 50, 1401–1415. [DOI] [PubMed] [Google Scholar]
- McIntosh CA, Mansell RL. 1990. Biosynthesis of naringin in Citrus paradisi: UDP-glucosyl-transferase activity in grapefruit seedlings. Phytochemistry 29, 1533–1538. [Google Scholar]
- McIntosh CA, Mansell RL. 1997. Three-dimensional distribution of limonin, limonoate A-ring monolactone, and naringin in the fruit tissues of three varieties of Citrus paradisi. Journal of Agricultural and Food Chemistry 45, 2876–2883. [Google Scholar]
- Moriguchi T, Kita M, Tomono Y, EndoInagaki T, Omura M. 1999. One type of chalcone synthase gene expressed during embryogenesis regulates the flavonoid accumulation in citrus cell cultures. Plant & Cell Physiology 40, 651–655. [DOI] [PubMed] [Google Scholar]
- Moriguchi T, Kita M, Tomono Y, Endo-Inagaki T, Omura M. 2001. Gene expression in flavonoid biosynthesis: correlation with flavonoid accumulation in developing citrus fruit. Physiologia Plantarum 111, 66–74. [Google Scholar]
- Morita Y, Hoshino A, Kikuchi Y, et al. . 2005. Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2”-O-glucosyltransferase, due to 4-bp insertions in the gene. The Plant Journal 42, 353–363. [DOI] [PubMed] [Google Scholar]
- Nagatomo Y, Usui S, Ito T, Kato A, Shimosaka M, Taguchi G. 2014. Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (buckwheat) cotyledon. The Plant Journal 80, 437–448. [DOI] [PubMed] [Google Scholar]
- Ni H, Chen F, Cai H, Xiao A, You Q, Lu Y. 2012. Characterization and preparation of Aspergillus niger naringinase for debittering citrus juice. Journal of Food Science 77, C1–C7. [DOI] [PubMed] [Google Scholar]
- OuYang Q, Tao N, Jing G. 2016. Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genomics 17, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DK, McIntosh CA. 2009. Identification, recombinant expression, and biochemical characterization of a flavonol 3-O-glucosyltransferase clone from Citrus paradisi. Phytochemistry 70, 1382–1391. [DOI] [PubMed] [Google Scholar]
- Pak JH, Chung ES, Shin SH, Jeon EH, Kim MJ, Lee HY, Chung YS. 2009. Enhanced fungal resistance in Arabidopsis expressing wild rice PR-3 (OgChitIVa) encoding chitinase class IV. Plant Biotechnology Reports 3, 147–155. [Google Scholar]
- Paquette S, Møller BL, Bak S. 2003. On the origin of family 1 plant glycosyltransferases. Phytochemistry 62, 399–413. [DOI] [PubMed] [Google Scholar]
- Rohani FR, Chibag M, Kawaharada M, Asano T, Oshima Y, Mitsuda N, Yamazaki M. 2016. An MYB transcription factor regulating specialized metabolisms in Ophiorrhiza pumila. Plant Biotechnology 33, 1–9. [Google Scholar]
- Rousseff RL, Martin SF, Youtsey CO. 1987. Quantitative survey of narirutin, naringin, hesperidin and neohesperidin in citrus. Journal of Agricultural and Food Chemistry 35, 1027–1030. [Google Scholar]
- Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T. 2005. UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. The Journal of Biological Chemistry 280, 899–906. [DOI] [PubMed] [Google Scholar]
- Shen C, Guo H, Chen H, Shi Y, Meng Y, Lu J, Feng S, Wang H. 2017. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Scientific Reports 7, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Jones P. 2000. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends in Plant Science 5, 380–386. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu Y, Zhang S, et al. . 2017. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nature Genetics 49, 765–772. [DOI] [PubMed] [Google Scholar]
- Williams CA. 2006. Flavone and flavonol O-glycosides. In: Andersen OM, Markham KR, eds. Flavonoids, chemistry, biochemistry and applications. Boca Raton, FL: CRC Press/Taylor & Francis Group, 749–856. [Google Scholar]
- Wu GA, Terol J, Ibanez V, et al. . 2018. Genomics of the origin and evolution of Citrus. Nature 554, 311–316. [DOI] [PubMed] [Google Scholar]
- Xu G, Ye X, Chen J, Liu D. 2007. Effect of heat treatment on the phenolic compounds and antioxidant capacity of citrus peel extract. Journal of Agricultural and Food Chemistry 55, 330–335. [DOI] [PubMed] [Google Scholar]
- Xu Q, Chen LL, Ruan X, et al. . 2013. The draft genome of sweet orange (Citrus sinensis). Nature Genetics 45, 59–66. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. 2011. An evolutionary view of functional diversity in family 1 glycosyltransferases. The Plant Journal 66, 182–193. [DOI] [PubMed] [Google Scholar]
- Zhang M, Duan C, Zang Y, Huang Z, Liu G. 2011. The flavonoid composition of flavedo and juice from the pummelo cultivar (Citrus grandis (L.) Osbeck) and the grapefruit cultivar (Citrus paradisi) from China. Food Chemistry 129, 1530–1536. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.