Abstract
NAC transcription factors (TFs) are important regulators of expressional reprogramming during plant development, stress responses, and leaf senescence. NAC TFs also play important roles in fruit ripening. In tomato (Solanum lycopersicum), one of the best characterized NACs involved in fruit ripening is NON-RIPENING (NOR), and the non-ripening (nor) mutation has been widely used to extend fruit shelf life in elite varieties. Here, we show that NOR additionally controls leaf senescence. Expression of NOR increases with leaf age, and developmental as well as dark-induced senescence are delayed in the nor mutant, while overexpression of NOR promotes leaf senescence. Genes associated with chlorophyll degradation as well as senescence-associated genes (SAGs) show reduced and elevated expression, respectively, in nor mutants and NOR overexpressors. Overexpression of NOR also stimulates leaf senescence in Arabidopsis thaliana. In tomato, NOR supports senescence by directly and positively regulating the expression of several senescence-associated genes including, besides others, SlSAG15 and SlSAG113, SlSGR1, and SlYLS4. Finally, we find that another senescence control NAC TF, namely SlNAP2, acts upstream of NOR to regulate its expression. Our data support a model whereby NAC TFs have often been recruited by higher plants for both the control of leaf senescence and fruit ripening.
Keywords: Aging, leaf, NAC, non-ripening, NOR, senescence, tomato, transcription factor
The fruit ripening transcription factor NOR positively regulates leaf senescence in tomato by directly controlling the expression of key senescence-associated genes.
Introduction
Transcription factors (TFs) of the NAC (for NAM, ATAF1/2, and CUC2) family play important roles for development and the response of plants to abiotic and biotic stresses (Puranik et al., 2012; Shao et al., 2015). A prominent process controlled by NAC TFs is leaf senescence, which is a complex physiological process of nutrient recovery to support the development and growth of newly forming organs, including new leaves, flowers, and seeds (Hendelman et al., 2013; Zhong et al., 2016). NAC TFs in diverse dicot and monocot plant species have been shown to control the onset and execution of senescence, for example in Arabidopsis thaliana (Guo and Gan, 2006; Kim et al., 2009; Balazadeh et al., 2010, 2014; Wu et al., 2012; Garapati et al., 2015; Kamranfar et al., 2018), rice (Oryza sativa; Zhou et al., 2013; Mao et al., 2017), wheat (Triticum aestivum; Uauy et al., 2006; Zhao et al., 2015), cotton (Gossypium hirsutum; Fan et al., 2015), and tomato (Solanum lycopersicum; Lira et al., 2017; Ma et al., 2018).
A master positive regulator of leaf senescence in Arabidopsis is ORE1 (ORESARA1; ANAC092; Kim et al., 2009; Balazadeh et al., 2010). Expression of ORE1 increases with leaf age, a process regulated at the transcriptional level by the ORE1 promoter, and post-transcriptionally by the miRNA miR164 (Kim et al., 2009). ORE1 controls the expression of a number of senescence-associated genes (SAGs) by directly binding to their promoters (Balazadeh et al., 2010) and, accordingly, overexpression or knocking out of ORE1 promotes or inhibits senescence, respectively (Kim et al., 2009; Balazadeh et al., 2010). Recently, the closest putative orthologs of ORE1 in tomato (i.e. SlORE1S02, SlORE1S03, and SlORE1S06) were also shown to positively control leaf senescence (Lira et al., 2017). In addition, inhibiting SlORE1S02 by RNAi not only delayed leaf senescence but also triggered an altered source–sink sugar partitioning, resulting in an increased number of fruits per plant with elevated sugar levels (Lira et al., 2017). Similarly, we recently showed that inhibiting expression of the SlNAP2 TF gene in transgenic tomato plants delays leaf senescence, which was accompanied by an increased yield of fruits (with elevated sugar content) probably due to extended photosynthesis in aging plants (Ma et al., 2018). SlNAP2 belongs to the NAP clade of NAC TF genes of which AtNAP from Arabidopsis was first studied with respect to leaf senescence (Guo and Gan, 2006) and was later shown also to control silique senescence (Kou et al., 2012). In rice, inhibiting OsNAP1 delayed leaf senescence but increased seed yield (Liang et al., 2014).
In addition, NAC TFs have been reported, or suggested, to be involved in ripening of fleshy fruits in several species, with a particular emphasis on tomato, an important fleshy fruit-bearing crop that is extensively used as a model vegetable for studies on fruit physiology and development; its nuclear genome has been sequenced (Tomato Genome Consortium, 2012). One of the best characterized examples in tomato is NON-RIPENING (NOR), which also affects fruit shelf life, an important economic trait. Mutations in the NOR gene (locus Solyc10g006880) lead to the formation of a truncated TF protein (nor mutant) or a NAC TF with a single amino acid substitution (alcobaca mutant, alc) (Giovannoni et al., 2004; Casals et al., 2012). Recently, a further mutation of the NOR gene, leading to an early stop codon, was identified in the tomato variety Penjar-1 grown in the Mediterranean area (Kumar et al., 2018). NOR acts upstream of ethylene synthesis and thereby controls fruit ripening (Barry and Giovannoni, 2007). ChIP assays demonstrated that NOR is a direct downstream target of RIN (Ripening Inhibitor), a MADS-box TF controlling fruit ripening (Martel et al., 2011; Fujisawa et al., 2013; Ito et al., 2017). Similarly, in melon (Cucumis melo), a NOR TF (CmNAC-NOR) was found to be involved in fruit ripening (Rios et al., 2017). In addition, NOR homologs control senescence in non-fleshy fruits such as the siliques of Arabidopsis where NARS1/NAC2 and NARS2/NAM redundantly and positively regulate silique senescence while leaf senescence is unaltered compared with the wild type (WT), indicating organ-specific functions of the two NAC TFs (Kunieda et al., 2008).
Besides NOR, other TFs of the NAC family in tomato have been reported to control fruit ripening, including SlNAC4 which positively regulates ripening, possibly through physical interaction with NOR and RIN (shown by yeast two-hybrid studies); furthermore, SlNAC4 was suggested to act as an upstream regulator of RIN (Zhu et al., 2014). Evidence for a positive role in regulating fruit ripening was also obtained for SlNAC48 and SlNAC19 (which is identical to SlNAP2) using a virus-induced gene silencing (VIGS) approach. The data suggest that both TFs, SlNAC19 and SlNAC48, act by affecting ethylene biosynthesis and signaling (Kou et al., 2016). SlNAC3 (recently named NOR-like1) shows high expression in fruits and is involved in seed development and fruit ripening (Han et al., 2012, 2014; Gao et al., 2018).
Evidence for an involvement of NAC TFs in fleshy fruit ripening was also obtained from studies performed on developing and ripening fruits of other species, including the octoploid strawberry cultivar Fragaria × ananassa (Moyano et al., 2018), the Chilean endemic strawberry Fragaria chiloensis (Carrasco-Orellana et al., 2018), and bilberry (Vaccinium myrtillus; Nguyen et al., 2018).
Taken together, many NAC TFs have been reported to control leaf senescence in different plant species, and some NACs have been firmly proven—or suggested—to control the ripening of fleshy or dry fruits. Considering this, we were interested to investigate whether the so-far best studied fruit ripening control NAC TF in tomato, namely NOR, additionally controls leaf senescence in this plant. Our data show that NOR acts as a positive transcriptional regulator of leaf senescence by directly and positively controlling the expression of several chlorophyll degradation- (CDGs) and senescence-associated genes (SAGs) in this species. The data suggest an evolutionary recruitment of NAC TFs from regulating leaf senescence towards the control of physiology during fruit ripening.
Materials and methods
General
Tomato orthologs of Arabidopsis genes were identified using the PLAZA 3.0 database (http://bioinformatics.psb.ugent.be/plaza/;Proost et al., 2015). Genes were annotated using the PLAZA 3.0 and Sol Genomics (https://solgenomics.net/) databases, and using information extracted from the literature. Oligonucleotide sequences are given in Table S1 at JXB online. Primers for quantitative real-time PCR (qRT-PCR) were designed using QuantPrime (www.quantprime.de;Arvidsson et al., 2008).
Plant material and growth conditions
Tomato (Solanum lycopersicum L., cultivar Moneymaker) was used as the WT. The nor mutant is in the Rutgers genetic background (Tomato Genetics Research Center, https://tgrc.ucdavis.edu; accession number LA3013). The mutant is due to a spontaneous mutation in the NOR gene. Seeds were germinated on full-strength Murashige and Skoog (MS) medium containing 2% (w/v) sucrose, and 3-week-old seedlings were transferred to soil containing a mixture of potting soil and quartz sand (2:1, v/v). Plants were grown in a growth chamber at 500 µmol photons m−2 s−1 (high-pressure sodium vapor lamps; Agrolux, https://www.agrolux.com) and 25 °C under a 14 h/10 h light/dark regime in individual pots (18 cm diameter). For experiments with Arabidopsis thaliana (L.) Heynh., accession Col-0 was used as the control. Seeds were germinated in soil (Einheitserde GS90; Gebrüder Patzer, Sinntal-Altengronau, Germany) in a climate-controlled chamber with a 16 h day length provided by fluorescent light at ~100 μmol m−2 s−1, day/night temperature of 20 °C/16 °C, and relative humidity of 60%/75%. After 2 weeks, seedlings were transferred to a growth chamber with a 16 h day (80 μmol m−2 s−1 or 120 μmol m−2 s−1), day/night temperature of 22 °C/16 °C, and 60%/75% relative humidity.
DNA constructs
Primer sequences are listed in Supplementary Table S1. Amplified fragments generated by PCR were sequenced by Eurofins MWG Operon (Ebersberg, Germany). For 35S:NOR-GFP, the full-length NOR ORF was amplified without its stop codon. The PCR product was cloned into the pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning kit (Invitrogen). The sequence-verified entry clone was then transferred to the pK7FWG2 vector (Karimi et al., 2002) by LR recombination (Invitrogen). For NOR-IOE, the NOR coding sequence was cloned into the pER10 vector (Zuo et al., 2002) made GATEWAY compatible. Constructs were transformed into tomato cv. Moneymaker using Agrobacterium tumefaciens GV2260, or into Arabidopsis using A. tumefaciens GV3101 (pMP90).
The DNA-binding protein–CELD (cellulose D) fusion vector pTacLCELD6xHis was used to construct NOR-CELD (Xue, 2005). The NOR coding sequence (without the stop codon) was amplified by PCR with a sense primer (including an NheI restriction site) and an antisense primer (including a BamHI restriction site) (Supplementary Table S1). The amplified DNA fragment was first inserted into pCR2.1 (Thermo Fisher Scientific) and then inserted N-terminally of CELD using the NheI and BamHI cloning sites of pTacLCELD6xHis to create an in-frame fusion.
Amino acid sequence alignment
Protein sequences were extracted from PLAZA 3.0 (http://bioinformatics.psb.ugent.be/plaza/). The protein alignment was done using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/); the output set-up was Pearson/FASTA. To highlight conserved regions within the alignments, the Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/color_align_cons.html) was employed.
Treatments
For estradiol (EST) induction, 3-week-old NOR-IOE seedlings were incubated in sterile water containing 15 µM EST [control treatment: 0.15% (v/v) ethanol]. The seedlings were kept on a rotary shaker for 6 h and then immediately frozen in liquid nitrogen. For abscisic acid (ABA) treatment, 3-week-old WT seedlings and detached young leaves from 10-week-old WT and NOR transgenic plants were placed in sterile water containing 40 µM ABA with constant light. The seedlings were kept on a rotary shaker for 2, 4, 8, or 16 h and harvested in liquid nitrogen. For dark-induced leaf senescence experiments, detached young leaves from 10-week-old WT and NOR transgenic plants were placed on moist filter papers in Petri dishes with the adaxial side facing upwards. The plates were kept in darkness at 22 °C for 2 weeks. Filter papers were changed every 5 d. Gene expression levels were determined by qRT-PCR.
Gene expression analysis
Total RNA was extracted using Trizol reagent (Life Technologies). Synthesis of cDNA and qRT-PCR using SYBR Green were performed as described (Balazadeh et al., 2008). PCR was performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Solyc04g009030) served as the reference gene for data analysis. Statistical significance was determined using Student’s t-test.
DNA-binding site selection
In vitro binding site selection was performed using the CELD fusion method with the pTacNOR-LCELD6xHis construct, employing biotin-labeled double-stranded oligonucleotides (Xue, 2005). The DNA binding activity of NOR–CELD was measured using methylumbelliferyl β-d-cellobioside as substrate (Xue, 2002). DNA binding assays with a biotin-labeled single-stranded oligonucleotide or a biotin-labeled double-stranded oligonucleotide without a target binding site were used as controls.
Chromatin immunoprecipitation
ChIP-qPCR was performed using leaves of mature 35S:NOR-GFP plants, and the WT served as control. ChIP was performed as described (Kaufmann et al., 2010) using anti-green fluorescent protein (GFP) antibody to immunoprecipitate protein–DNA complexes. qPCR primers were designed to flank the NOR-binding sites within the promoter regions of potential target genes. Primers annealing to a promoter region of Solyc04g009030 lacking a NOR-binding site were used as a negative control. Primers used for qPCR are listed in Supplementary Table S1.
Chlorophyll measurements
Chlorophyll content was determined using a SPAD analyser (N-tester; Hydro Agri). Alternatively (Fig. 4D), frozen leaf powder was suspended in 5 ml of 80% (v/v) acetone in water and homogenized for 1 min. Chlorophyll content was determined with a spectrophotometer at 663 nm and 646 nm as described by Arnon (1949).
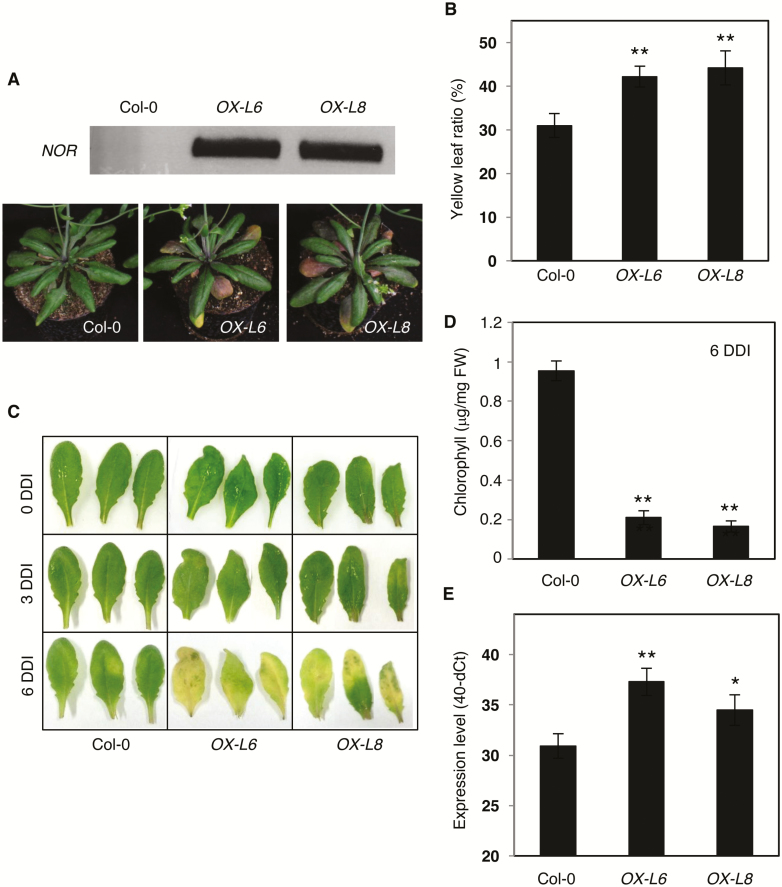
Fig. 4.
Overexpression of NOR in Arabidopsis promotes leaf senescence. (A) Phenotype of Arabidopsis Col-0 wild-type and NOR overexpression plants. The upper panel shows NOR transcript abundance in OX-L6 and OX-L8 plants, determined by end-point PCR; as expected, no NOR transcript is observed in the Arabidopsis WT. The lower panel shows the phenotype of 5-week-old plants (Col-0 and NOR overexpressors). (B) Yellow leaf ratio of 5-week-old Col-0, OX-L6, and OX-L8 plants. Yellow leaves showing >50% yellowing were counted and compared with the total leaf number. Data are means ±SD (n=5). (C) Dark-induced senescence. DDI, days after dark incubation. Note the more pronounced senescence in the two NOR overexpressors compared with Col-0 at 6 DDI. Leaves 5–7 detached from the various plants (separated by black vertical lines) were used in the experiment. (D) Chlorophyll content of (C), at 6 DDI of Col-0, OX-L6, and OX-L8 genotypes (n=5). (E) Expression of AtSAG12 in detached leaves 5–7 of Col-0, OX-L6, and OX-L8 plants at 6 DDI. The y-axis indicates the expression level (40-dCt). Data are means ±SD of three biological replicates. Asterisks in (B), (D), and (E) indicate a significant difference from the Col-0 wild type (Student’s t-test; *P≤0.05; **P≤0.01).
Ion leakage measurements
Ion leakage in leaves was determined as reported in Thirumalaikumar et al. (2018). Briefly, ion leakage during senescence was measured in control and dark-treated leaves of WT and NOR transgenic plants. Detached leaves were shaken in distilled water overnight at 4 °C and the initial conductivity of the samples was determined using an ion conductivity meter (SI Analytics, Mainz, Germany). Subsequently, the samples were boiled at 100 °C for 30 min and then kept at room temperature to cool down to 25 °C, after which the total conductivity was measured. Data are shown as the percentage of total conductivity.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NOR (NM_001247723.2); SlNAC3 (NM_001279348.2); SlNAP2 (XM_004236996.2); SlSAG15 (XM_010320381.2); SlSAG113 (XP_004239911.1); SlSGR1 (NP_001234723.1); SlPPH (XM_004229633.3); SlPAO (NP_0012 34535.2); SlYLS4 (XM_004245218); SlERT1B (NM_001361347); SlKFB20 (XM_010320257); SlABCG40 (XP_004247842.1); and SlGA2ox5 (NP_001234757.1).
Results
NOR is up-regulated during leaf senescence
NOR encodes a tomato NAC TF that harbours a conserved DNA-binding NAM domain at its N-terminus (Fig. 1A; Supplementary Fig. S1). At the protein level, NOR is closely related to SlNAC3/NOR-like1 from tomato, and to NARS1 and NARS2 from Arabidopsis (Fig. 1B; Supplementary Fig. S1). As shown below, NOR binds to the promoters of several direct target genes, demonstrating it to be a nuclear-localized protein.
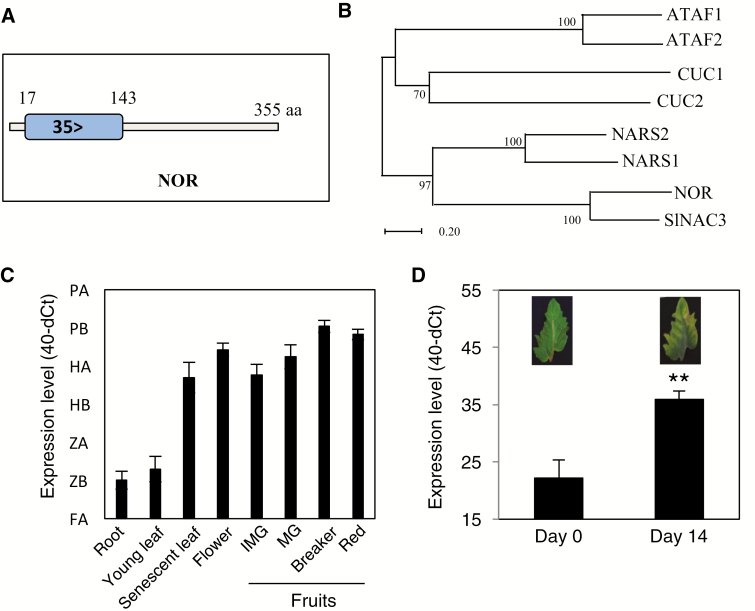
Fig. 1.
Relationship of NOR to other NAC factors and NOR expression during senescence. (A) Schematic presentation of the NAM domain of NOR. Numbers indicate amino acid positions. (B) Phylogenetic analysis of selected NAC proteins. The phylogenetic tree was constructed by MEGA 5.05 software using the Neighbor–Joining method with the following parameters: bootstrap analysis of 1000 replicates, Poisson model, and pairwise deletion. NOR and SlNAC3 are two tomato TFs and the others are from Arabidopsis. Gene codes of the Arabidopsis TFs are: ATAF1, At1g01720; ATAF2, At5g08790; NARS1, At3g15510; NARS2, At1g52880; CUC1, At3g15170; and CUC2, At5g53950. (C) NOR transcript abundance in different tissues of wild-type tomato plants (cv. Moneymaker). The y-axis indicates the expression level (40-dCt). Data are means ±SD of three biological replicates. (D) Expression of NOR in young detached leaves of 8-week-old WT plants before (day 0) and after 14 d of dark treatment. Leaves were excised from the top part of the stem. Data are means ±SD (n=3). Asterisks denote a significant difference from Day 0 (Student’s t-test, **P≤0.01).
NOR is hardly expressed in young leaves, but its expression increases during developmental and dark-induced senescence (Fig. 1C, D), indicating a possible function of the tomato TF for regulating leaf senescence. Of note, NOR shows high expression not only in senescent leaves, but also in flowers and fruits of different developmental stages (Fig. 1C).
Overexpression of NOR promotes leaf senescence
To test whether NOR indeed regulates leaf senescence, we first generated transgenic tomato (S. lycopersicum cv. ‘Moneymaker’) plants constitutively expressing NOR under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. We selected two lines (hereafter, OX-L5 and OX-L19; Supplementary Fig. S2A) for further analysis. Notably, NOR overexpression lines showed early leaf senescence (Fig. 2A). The ratio of yellow leaves (defined as leaves with >50% yellowing) to all leaves of 12-week-old OX plants was significantly higher in OX-L5 and OX-L19 plants than in the WT (Fig. 2B). Furthermore, the chlorophyll content of leaves from the same position (leaf no. 3) dropped faster during development in OX than in the WT (Fig. 2C). The precocious senescence of NOR overexpressors was not accompanied by an early flowering (days after sowing; Supplementary Fig. S3A), although the start of flowering was observed at a slightly reduced leaf number compared with the WT (Supplementary Fig. S3B). We did not observe a difference in flowering time between nor mutant and WT plants (Supplementary Fig. S3).
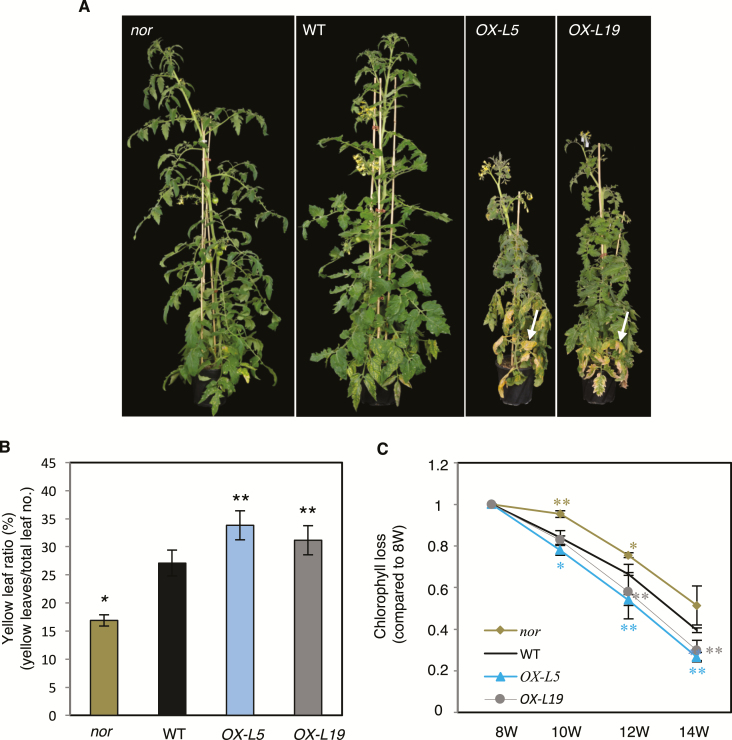
Fig. 2.
NOR promotes leaf senescence in tomato. (A) Phenotype of 12-week-old nor, WT, OX-L5, and OX-L19 plants. Note the early leaf senescence in NOR overexpressors (white arrows). (B) Yellow leaf ratio of 12-week-old WT, OX-L5, OX-L19, and nor plants. Yellow leaves showing >50% yellowing were counted and divided by the total number of leaves. Data are means ±SD (n=5). (C) Chlorophyll loss of the third true leaf (counted from the bottom of the stem) of 8- (8W), 10- (10W), 12- (12W), and 14-week-old (14W) WT, OX-L5, OX-L19, and nor plants. Chlorophyll content was measured by a SPAD meter and at each time point compared with 8W for each genotype (set to 1). Data are means ±SD of three biological replicates. Asterisks in (B) and (C) indicate significant differences from the WT (Student’s t-test, *P≤0.05; **P≤0.01).
We also observed a generally reduced shoot height of NOR overexpressors compared with the WT, while the nor mutant appeared slightly taller under our growth conditions (Fig. 2A). The reduced growth of NOR overexpressors (see also Supplementary Fig. S4A–C) may be due to increased expression of SlGA2ox5 (Supplementary Fig. S4D), a gibberellic acid (GA) degradation gene whose induction may result in reduced GA levels and thereby a reduced shoot elongation (Davière and Achard; 2013). SlGA2ox5 expression was also enhanced in plants expressing NOR from an EST-inducible promoter (hereafter, NOR-IOE), shortly (6 h) after EST treatment, suggesting SlGA2ox5 as a direct target of NOR, in accordance with the presence of a NOR-binding site in its promoter (Supplementary Table S3). Of note, expression of other GA metabolism genes was not significantly affected by NOR (Supplementary Table S4).
The tomato nor mutant exhibits retarded leaf senescence
Dark treatment is an efficient way to induce senescence in plants, as shown in many reports (Biswal and Mohanty, 1976; Chen and Kao 1991; Weaver et al., 2001). Here, we tested the effect of darkness on detached leaves which represent an excellent model to study senescence-related signaling events (e.g. Sakuraba et al., 2014); we examined the phenotypes of nor, WT, and OX-L19 after 14 d of dark treatment. Detached leaves from the overexpression line showed earlier de-greening in extended darkness than those of the WT. In contrast, leaves of the nor mutant remained green longer in darkness and their chlorophyll content remained high after treatment compared with the WT and OX-L19 (Fig. 3A, B). Moreover, ion leakage, an indicator of senescence often associated with membrane damage (Hou et al., 2013; Y.S. Kim et al., 2013; Bresson et al., 2018), was significantly elevated in OX-L19 compared with the WT, while it was reduced in nor (Fig. 3C). In accordance with this, expression of various SAGs and CDGs was up-regulated in OX-L19 plants compared with the WT, but down-regulated in nor (Fig. 3D; Supplementary Table S2). To substantiate the conclusion that NOR affects senescence-related genes, we also tested their expression during age-dependent and dark-induced senescence in WT tomato plants. As shown in Supplementary Fig. S5, all genes are significantly up-regulated in both senescence processes.
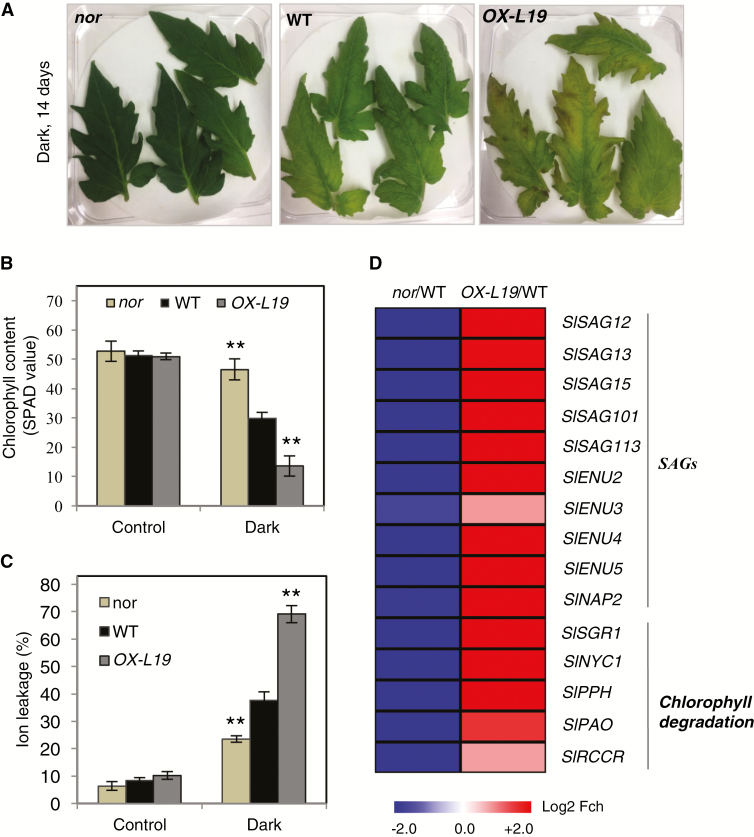
Fig. 3.
Dark-induced leaf senescence in NOR-modified plants. (A) Detached leaves of 8-week-old nor, WT, and OX-L19 plants after dark treatment. Young leaves from the top of the stem were detached and subjected to darkness for 14 d (Dark). (B) Chlorophyll content of leaves before darkness (control) and of dark-treated leaves. Chlorophyll content was measured using a SPAD meter. (C) Ion leakage of leaves before (control) and after dark treatment. (D) Heat map showing the fold change (log2) of the expression of SAGs and chlorophyll degradation genes in detached leaves of 8-week-old nor and OX-L19 plants, after dark treatment, compared with the WT. The full data are given in Suppelementary Table S2. In (B) and (C), asterisks indicate significant differences from the WT (Student’s t-test; **P≤0.01).
To further examine the function of NOR in regulating senescence, we generated NOR knock down lines by artificial miRNA (ami-NOR) in tomato cultivar Moneymaker. The ami-NOR construct targets 21 nucleotides (TGTACCATAGTTTGAAGGCTG) ~200 bp close to the 3' end of the NOR coding sequence. This region encodes the transactivation domain of the TF. We selected two lines (ami-L2 and ami-L35) with a reduced NOR transcript abundance as determined by end-point PCR (Supplementary Fig. S6A). The ami-NOR lines exhibited delayed senescence during dark treatment, similar to the nor mutant (Supplementary Fig. S6B, C).
NOR overexpression promotes leaf senescence in Arabidopsis
To test whether NOR also induced early leaf senescence in a heterologous species, we overexpressed it in transgenic A. thaliana plants. We selected two Arabidopsis lines expressing NOR for further analysis (hereafter, OX-L6 and OX-L8; Fig. 4A). As in tomato, overexpression of NOR promoted early leaf senescence in Arabidopsis (Fig. 4A), indicating functional conservation across species. OX plants had a higher ratio of yellow to all leaves than the WT at the same age (5 weeks) (Fig. 4B).
To test whether NOR overexpression promotes senescence in darkness, we detached leaves from the Arabidopsis OX-L6 and OX-L8 lines, and, after 6 d of dark incubation, observed much stronger senescence than in leaves of the WT control (Fig. 4C). Chlorophyll content after dark treatment was more strongly reduced in these lines than in the WT (Fig. 4D). Expression of the senescence-associated marker gene AtSAG12 (Noh and Amasino, 1999) was significantly up-regulated in these lines in comparison with the WT (Fig. 4E). From these results, we conclude that NOR positively regulates leaf senescence in both tomato and Arabidopsis.
A recent report by Kim et al. (2018) shows that expression of the NOR-related Arabidopsis genes NARS1 (also called ANAC056) and NARS2 (ANAC018) is elevated in aging leaves of Arabidopsis (see Supplementary fig. 1 of Kim et al.). Additionally, both genes show a relatively high expression in seeds (eFP browser). However, while silique senescence was delayed in the nars1/nars2 double mutant, no delay was observed for leaf senescence by Kunieda et al. (2008), indicating that NARS1 and NARS2 act differently from NOR.
Identification of the consensus DNA-binding sequence of NOR
Knowledge about the DNA-binding motif(s) of a TF under analysis strongly assists in unraveling the wider gene regulatory network it controls. We therefore performed an in vitro binding site selection assay using the earlier reported CELD fusion method (Xue, 2005) to identify NOR-binding sites. We first analyzed the binding activity of NOR towards 16 randomly selected TaNAC69 motifs, S1–S16, bound by the TaNAC69 TF from wheat (T. aestivum) (Supplementary Fig. S7A), considering the following rationale. Both NOR and TaNAC69 belong to the NAP subfamily of NAC TFs; the two proteins share ~53% overall amino acid similarity. Within their NAC domains, NOR and TaNAC69 share 79.4% similarity and 66.7% identify, suggesting that the two NACs might have a similar binding site. Previously, it was shown that S1 is a high-affinity binding sequence of TaNAC69 (Xue et al., 2006). In our results, NOR showed strong binding affinity for S1, with affinity decreasing progressively with substitutions. Overall, NOR bound to TaNAC69-selected motifs containing the YACG (or CGTR) core sequence (Supplementary Fig. S7A). Further analysis of the specificity of binding through base substitution, insertion, or deletion revealed that the mutation of nucleotides in the core motifs (e.g. S1m3 and S1m9) resulted in a strong reduction of NOR binding activity (Supplementary Fig. S7B). Taken together, our data suggest two high-affinity binding sites of NOR, CG(Y/C)(G/C)(5-7n)N(A/G)CGn(A/C/G)(A/C/T) and (C/T)ACGn(A/C)(A/T)(C/G/T)(C/T), as motif I and motif II, respectively.
Identification of NOR target genes
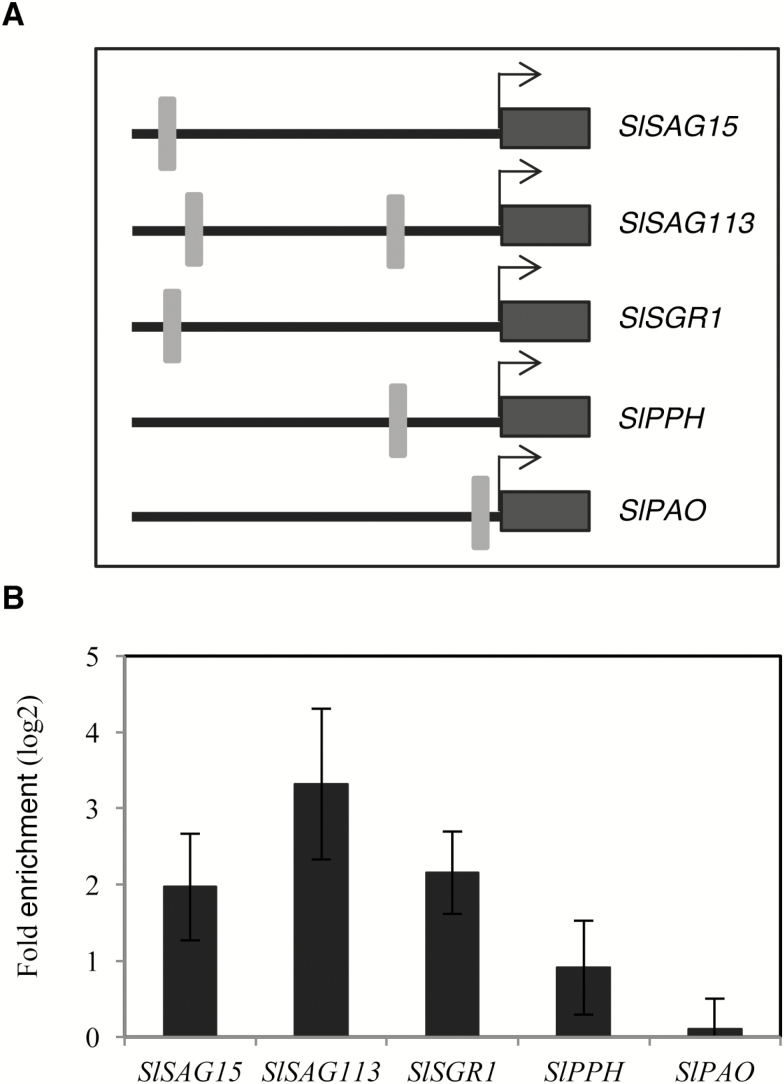
Although NOR is a TF well known for its function in fruit ripening, no direct target genes have, to our knowledge, been reported so far. Therefore, based on the results presented in Fig. 3D, we selected individual genes for further analysis to test whether they might be direct downstream targets of NOR. To this end, we chose several genes harboring the NOR-binding site within their 5' upstream regulatory regions, including SlSAG15, SlSAG113, SlSGR1, SlPPH, and SlPAO (Fig. 5A). ChIP-qPCR using tomato plants expressing NOR–GFP protein in the nucleus (Supplementary Fig. S2C) revealed direct binding of the NOR TF to the promoters of all genes except SlPAO (Fig. 5B). The absence of a detectable in planta binding of NOR to the SlPAO promoter might have several causes. One likely model is that the binding of NOR requires an open chromatin status which may not have been prevalent under the conditions in which we performed the experiment.
Fig. 5.
Direct regulation of SAGs by NOR. (A) Schematic diagram showing positions of NOR-binding sites in 1 kb promoters of selected genes. Arrows indicates the ATG translational start codon. Gray boxes indicate the NOR-binding sites and black boxes indicate the coding regions of the genes. Sequences of the gene promoters including the NOR-binding sites tested in the ChIP experiments are given in Supplementary Table S3. (B) ChIP-qPCR shows enrichment of SlSAG15, SlSAG113, SISGR1, and SlPPH promoter (1 kb) regions containing the NOR-binding site. Eight-week-old NOR-GFP plants (mature leaves ~3‒5) were harvested for the ChIP experiment. qPCR was performed to quantify the enrichment of the promoter regions. In the case of SlSAG113, which has two potential NOR-binding sites in its promoter (see A), we tested binding of NOR to the sequence proximal to the ATG start codon. Values were normalized to the values for Solyc04g009030 (promoter lacking a NOR-binding site). Data are the means ±SD of two independent biological replicates, each determined in three technical replicates.
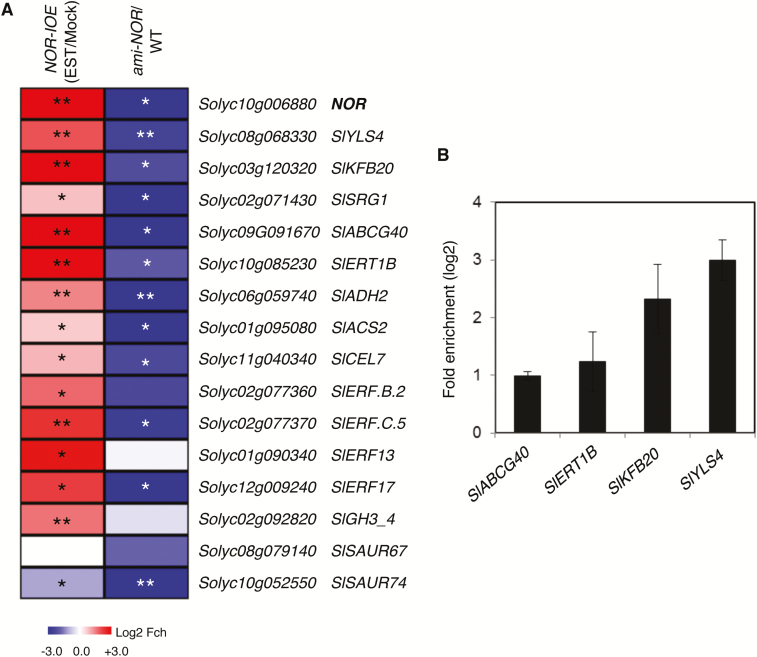
We next selected additional genes known to be regulated by natural or dark-induced senescence in tomato, or induced by abiotic stresses that trigger senescence (based on literature reports), and checked whether their promoters harbor a NOR-binding site. Considering that NOR regulates fruit ripening (Giovannoni et al., 2004; Casals et al., 2012; Kumar et al., 2018), we also included a few genes reported to control this process. We then tested whether expression of these genes is affected in tomato NOR-IOE plants. As shown in Supplementary Fig. S2B, expression of NOR was strongly enhanced in 3-week-old NOR-IOE seedlings 6 h after treatment with 15 µM EST, as expected. Similarly, all selected NOR-binding site-containing genes except two showed enhanced expression when NOR was induced (Fig. 6A; Supplementary Table S2).
Fig. 6.
Heat map of differentially expressed genes in NOR-IOE and ami-NOR plants. (A) Gene expression was analyzed by qRT-PCR in NOR-IOE seedlings treated with EST (15 µM) for 6 h and compared with expression in mock-treated [ethanol, 0.15% (v/v)] seedlings (left column), or in ami-NOR seedlings compared with wild-type (WT) seedlings. Seedlings were 3 weeks old. The color code indicates the log2 scale of the fold change; blue, down-regulated; red, up-regulated. Data represent means of three biological replicates. Data are means ±SD of three biological replicates. Asterisks indicate a significant difference from mock-treated samples (for NOR-IOE samples) or from the WT (for ami-NOR samples). Student’s t-test; *P≤0.05; **P≤0.01). The full data are given in Supplementary Table S2. (B) ChIP‒qPCR shows enrichment of SlABCG40, SlERT1B, SlKFB20, and SlYLS4 promoter regions containing the NOR-binding site within the 1 kb upstream promoter regions of the corresponding genes. Experimental conditions were as described in the legend to Fig. 5B. Sequences of the gene promoters including the NOR-binding sites tested in the ChIP experiments are given in Supplementary Table S3. Data are the means ±SD of two independent biological replicates, each determined in three technical replicates.
Among the genes up-regulated by NOR are the senescence-related genes SlYLS4, SlKFB20, and SlSRG1. SlYSL4 (Solyc08g068330), a homolog of Arabidopsis YLS4 (YELLOW LEAF SPECIFIC4), is expressed in a senescence-specific manner; the gene encodes an aspartate aminotransferase possibly involved in remobilizing leaf nitrogen during senescence (Yoshida et al., 2001). SlKFB20 (Solyc03g120320), a gene induced in tomato leaves during senescence, is a homolog of Arabidopsis AT1G80440, which encodes a kelch-repeat F-box protein targeting type-B ARR (Arabidopsis Response Regulator) proteins for degradation in the negative regulation of the cytokinin response (H.J. Kim et al., 2013a, b). Notably, cytokinins delay senescence (Hwang et al., 2012). SlSRG1 (Solyc02g071430) is closely related to SENESCENCE-RELATED GENE1 (SRG1) from Arabidopsis, which encodes a member of the Fe (II)/ascorbate oxidase gene family and is highly induced under low nitrogen conditions and during sucrose-induced senescence (Pourtau et al., 2006). SlABCG40 (Solyc09g091670), which encodes a protein belonging to the ATP-binding cassette (ABC) transporters, is one of the most up-regulated genes after EST treatment. It is induced by >120-fold after induction of NOR with EST in NOR-IOE lines. In Arabidopsis, ABCG40 encodes an ABC transporter protein involved in the cellular uptake of ABA (Kang et al., 2010), a phytohormone that triggers stomatal closure upon water shortage and stimulates leaf senescence in various species (Zhang et al., 2012; Zhao et al., 2017).
Three other genes analyzed, namely SlERT1B, SlADH2, and SlACS2, are involved in fruit ripening, and all are up-regulated after EST treatment in NOR-IOE plants. SlERT1B (Solyc10g085230) encodes a putative UDP-glycosyltransferase potentially involved in glycoalkaloid biosynthesis in tomato fruits (Itkin et al., 2013; Alseekh et al., 2015). SlADH2 (ALCOHOL DEHYDROGENASE2; Solyc06g059740) participates in the biosynthesis of volatiles and, accordingly, its transcript abundance increases during fruit ripening (Speirs et al., 1998); it is a direct target of RIN (Qin et al., 2012). SlACS2 (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE2; Solyc01g095080) encodes an ethylene biosynthesis gene highly expressed during fruit ripening. Down-regulation of SlACS2 lowers ethylene production and delays fruit ripening (Oeller et al., 1991).
We included further genes with likely functions in fruit ripening or leaf senescence in our analysis. One is SlCEL7 (Solyc11g040340), which encodes a putative endo-β-1,4-glucanase of the glycosyl hydrolase 9 (cellulase E) family (www.uniprot.org); SlCEL7 has been suggested to play a specific role for regulating the loosening of cell walls during fruit growth (Catalá et al., 2000). As seen in Fig. 6A (and Supplementary Table S2), expression of SlCEL7 was significantly elevated in NOR-IOE plants after EST induction, suggesting it to be a downstream target of NOR. In addition, the hormone-related genes ETHYLENE-RESPONSIVE TRANSCRIPTION FACTOR B.2 (SlERF.B.2, Solyc02g077360), SlERF.C.5 (Solyc02g077370), SlERF13 (Solyc01g090340), and SlERF17 (Solyc12g009240) were also significantly up-regulated after EST induction of the NOR TF (Fig. 6A), suggesting them to be downstream targets of NOR.
As the phytohormone auxin is involved in controlling leaf senescence and fruit ripening (Kim et al., 2011; Breitel et al., 2016), we also included three auxin-related genes in our analysis, namely SlGH3_4 (Solyc02g092820), which encodes a putative indole-3-acetic acid amido synthetase, an enzyme conjugating auxin to an inactive form thereby reducing cellular free auxin levels, and small auxin up-regulated RNA67 (SlSAUR67; Solyc08g079140). Expression of various GH3 genes has previously been shown to increase in leaves during developmental and dark-induced senescence, consistent with the decrease of free auxin levels in senescing leaves (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Kim et al., 2011). While SlGH3_4 was significantly up-regulated upon induction of NOR, SlSAUR67 was not affected. The expression of the third auxin-related gene SlSAUR74 (Solyc10g052550) was significantly reduced after NOR induction (Fig. 6A).
We next analyzed expression of the selected genes by qRT-PCR in ami-NOR lines. Almost all genes that were up-regulated in NOR-IOE plants after EST induction were down-regulated in ami-NOR, confirming the transcription activation role of NOR toward these genes (Fig. 6A).
Finally, we employed ChIP-qPCR to test binding of NOR to the promoters of selected downstream targets in vivo, including SlABCG40, SlERT1B, SlKFB20, and SlYLS4. As shown in Fig. 6B, NOR binds to all four promoters.
SlNAP2 affects NOR expression
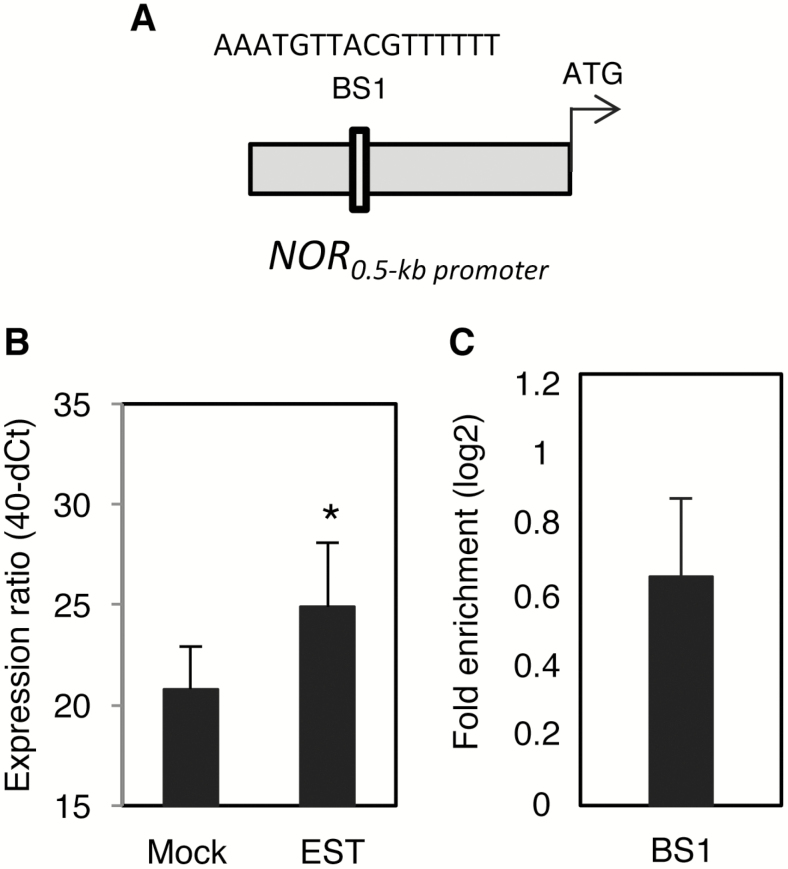
We previously reported that SlNAP2, a tomato NAC TF, functions as a positive regulator of leaf senescence by directly controlling the expression of various SAGs as direct targets. In addition, SlNAP2 controls the expression of several ABA-related genes (Ma et al., 2018). SlNAP2 has two related DNA-binding sites, called BS1 and BS2, which are present in the promoters of its direct gene targets (Ma et al., 2018). As previous work on Arabidopsis indicated regulatory connectivity between different NAC TFs to control senescence (e.g. Garapati et al., 2015; Kim et al., 2018), we here thought to investigate the possibility that NOR is a downstream affected gene target of SlNAP2. In accordance with this model, sequence analysis of the NOR promoter identified an SlNAP2 BS1-binding site 403 bp upstream of the ATG start codon (Fig. 7A). Furthermore, expression of NOR significantly increased in transgenic tomato plants expressing SlNAP2 from an EST-inducible promoter (SlNAP2-IOE; Ma et al., 2018) 6 h after EST treatment (Fig. 7B). Finally, SlNAP2 directly binds to the NOR promoter, as shown by ChIP-qPCR (Fig. 7C). Collectively, our data thus show that SlNAP2 functions as an upstream regulator of NOR.
Fig. 7.
SlNAP2 acts as an upstream regulator of NOR. (A) Schematic presentation of SlNAP2-binding site 1 (BS1) within the NOR promoter. The sequence of the binding site, which is located in the forward strand of the promoter, is indicated. (B) Expression of NOR in 3-week-old SlNAP2-IOE seedlings treated with estradiol (EST; 15 μM) for 6 h compared with ethanol [0.15% (v/v)]-treated seedlings (Mock). Gene expression was determined by qRT-PCR. Data represent means of three biological replicates. Asterisks indicate a significant difference from mock-treated plants (Student’s t-test; *P≤0.05). (C) ChIP-qPCR shows enrichment of the NOR promoter region containing the SINAP2-binding site 1 (BS1). Mature leaves (nos 3–5) harvested from 8-week-old SlNAP2-GPF plants were used for the ChIP experiment. Values were normalized to the values for Solyc04g009030 (promoter lacking a SlNAP2-binding site). Data are means ±SD of two independent biological replicates, each performed with three technical replicates.
NOR affects ABA-induced leaf senescence
The observation that SlABCG40, encoding a potential ABA transporter in tomato, is highly up-regulated upon NOR induction and the fact that SlNAP2 affects expression of NOR suggested an involvement of a SlNAP2–NOR signaling cascade in ABA-induced leaf senescence. To test this hypothesis, we analyzed the expression of SlNAP2 and NOR during dark-induced senescence, and after ABA treatment in non-senescent leaves. As shown in Supplementary Fig. S8A, expression of both genes increases during age-dependent senescence. In addition, expression of SlNAP2 increases rapidly (within 2–4 h) after ABA treatment, while expression of NOR shows a delayed response to ABA treatment, with a detectable increase only after 16 h of treatment (Supplementary Fig. S8B), in accordance with the observation that SlNAP2 acts upstream of NOR. We next tested the effect of ABA on NOR expression in tomato plants over- or underexpressing SlNAP2. As shown in Supplementary Fig. S8C, NOR was more highly expressed after ABA treatment in SlNAP2 overexpressors (SlNAP2-OX plants) than in non-ABA-treated SlNAP2-OX plants. In contrast, NOR expression was less affected by ABA treatment in WT plants and SlNAP2 knockdown plants (i.e. RNAi lines reported by Ma et al., 2018). Furthermore, ABA treatment accelerated leaf senescence more strongly in NOR overexpressors (OX-L19) than in WT and ami-NOR plants (Supplementary Fig. S8D, E). Taken together, our data (together with those of Ma et al., 2018) support the model that SlNAP2 and NOR act together to promote ABA-induced leaf senescence in tomato.
Discussion
NOR is a NAC TF well characterized for its role in fruit ripening in tomato (Barry and Giovannoni, 2007; Casals et al., 2012; Kumar et al., 2018). Also in melon (Cucumis melo), a NOR homolog has been shown to affect fruit ripening (Ríos et al., 2017). Recently, several other NAC TFs have been reported to control fruit ripening in tomato, including, for example, SlNAC4 (Zhu et al., 2014), SlNAC19 and SlNAC48 (Kou et al., 2016), and SlNAC3 (NOR-like1), the latter of which also functions in seed development (Han et al., 2012, 2014; Gao et al., 2018). With the data available so far, it appears that NAC TFs—in conjunction with TFs of other families—form interconnected regulatory networks to control fruit aging. For example, RIN, a long-known regulator of tomato fruit ripening of the MADS-box TF family, directly regulates NOR by binding to its promoter, as revealed by ChIP assay (Martel et al., 2011; Fujisawa et al., 2013). In addition, expression of NOR and RIN is reduced in SlNAC4 RNAi lines, which might indicate that it acts as an upstream regulator of NOR and RIN (Zhu et al., 2014). Furthermore, yeast two-hybrid assays revealed an interaction of SlNAC4 protein with NOR and RIN, although the functional relevance of this interaction in planta was not demonstrated (Zhu et al., 2014). Recently, experimental evidence showed that the basic leucine zipper (bZIP) TF SlAREB1 which, at the transcript level, is induced by ABA, may function as an upstream regulator of NOR, although direct in planta binding of SlAREB1 to the NOR promoter by, for example, ChIP was not demonstrated (Mou et al., 2018).
Although increasing evidence suggests an involvement of multiple NAC factors in tomato fruit development, a role for NACs in the regulation of leaf senescence in this vegetable crop has rarely been demonstrated despite the fact that NACs play diverse roles in the control of leaf senescence in other species (Podzimska-Sroka et al., 2015; Leng et al., 2017; Ma et al., 2018; Yang and Udvardi, 2018). Particularly detailed knowledge about the NAC-controlled senescence networks is available for Arabidopsis where multiple NAC TFs have been shown to positively or negatively regulate leaf senescence by binding to the promoters of diverse target genes to control different physiological processes underlying the complex syndrome of senescence (Guo and Gan, 2006; Kim et al., 2009; Wu et al., 2012; Garapati et al., 2015; Sakuraba et al., 2016; Oda-Yamamizo et al., 2016; Kamranfar et al., 2018; Kim et al., 2018; Li et al., 2018).
The situation is less clear in tomato, although aging in fruits and leaves may at least in part share identical TFs and gene regulatory networks. Recently, Lira et al. (2017) found that orthologs of ORE1, a central positive regulator of leaf senescence in Arabidopsis (Kim et al., 2009; Balazadeh et al., 2010), control leaf senescence in tomato leading to extended greenness upon down-regulation of SlORE1 gene expression. The increased fruit yield in such plants might be due to an extended photosynthetic lifetime of the leaves, providing carbon for fruit (sink) growth over a longer period than in WT plants, although another possibility is that SlORE1 genes directly control ripening processes in fruits. Similarly, down-regulation of the expression of the NAC TF gene SlNAP2 delays leaf senescence in tomato followed by an increased fruit yield (Ma et al., 2018). SlNAP2 binds to the promoters of several senescence-related genes, including SlSAG113 (Solanum lycopersicum SENESCENCE-ASSOCIATED GENE113) and the chlorophyll degradation genes SlSGR1 (S. lycopersicum senescence-inducible chloroplast stay-green protein 1) and SlPAO (S. lycopersicum pheide a oxygenase). SlNAP2 also directly controls the expression of several ABA-related genes including ABA transport, biosynthesis, and degradation genes, suggesting that it has an important function in controlling ABA homeostasis in senescing tomato leaves (Ma et al., 2018).
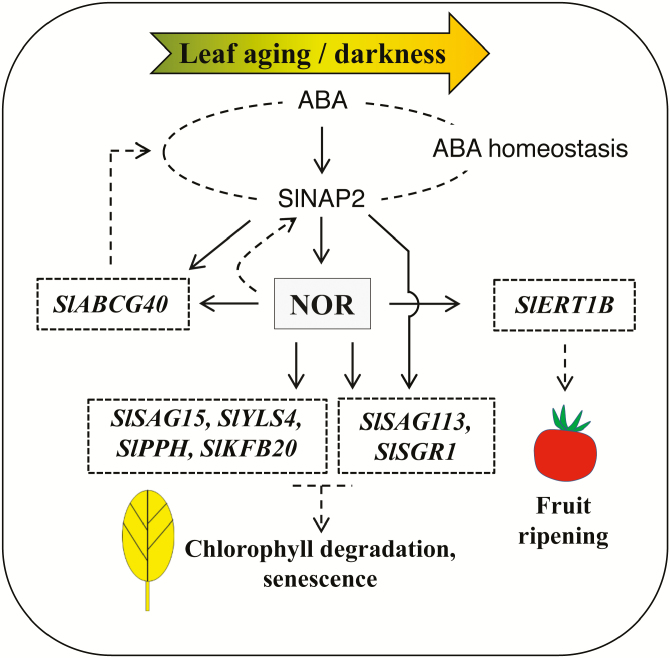
Here, we report that the long-known tomato fruit ripening factor NOR controls leaf senescence, thereby identifying a novel role for NOR for controlling development (see model in Fig. 8). Of note, overexpression of NOR in both transgenic tomato and Arabidopsis plants promotes developmental leaf senescence as well as dark-induced senescence. The role of NOR in regulating leaf senescence is related to changes in the expression of SAGs and CDGs. Expression of various senescence-responsive genes was enhanced in constitutive or EST-inducible NOR overexpressors, and we demonstrated binding of the NOR TF to their promoters by ChIP. As direct in vivo targets of NOR, we identified SlSAG15, SlSAG113, SlSGR1, and SlPPH, as well SlABCG40, SlERT1B, SlKFB20, and SlYLS4. Of note, SlSAG113, SlSGR1, and SlABCG40 were previously also identified as direct targets of SlNAP2 (Ma et al., 2018), strongly suggesting functional overlap of NOR and SlNAP2 in regulating leaf senescence-associated genes in tomato (Fig. 8). In accordance with this, both TFs belong to the same clade (the NAP clade) of NAC factors (Kou et al., 2014). This clade also includes the AtNAP gene, a well-known regulator of leaf senescence in A. thaliana (Guo and Gan, 2006). Interestingly, however, SlPAO did not appear to be a direct downstream target gene of NOR (this report; Fig. 5B), while we previously found it to be a direct target of SlNAP2 (Ma et al., 2018), indicating partial, but not complete, functional redundancy of both TFs with respect to the control of leaf senescence. Such a redundancy of NAC TFs for the control of senescence was recently highlighted for Arabidopsis by Li et al. (2018).
Fig. 8.
Model for the regulation of leaf senescence by NOR. NOR positively controls leaf senescence in tomato by directly regulating various senescence-associated genes including, besides others, SlSAG15, SlYLS4, SlPPH, SlKFB20, SlSAG113, and SlSGR1. It also directly regulates the expression of SlABCG40, an ABA transporter-encoding gene. The NAC transcription factor SlNAP2 enhances NOR expression by directly binding to its promoter and, together with NOR, it jointly regulates SlSAG113, SlSGR1, and SlABCG40. In addition, NOR enhances SlNAP2 expression, suggesting a positively acting feed-forward loop involving the two NAC factors. SlNAP2 has previously been reported to contribute to establishing ABA homeostasis during leaf senescence (Ma et al., 2018), to which the activation of SlABCG40 by NOR may contribute. NOR also directly and positively regulates the expression of the fruit ripening-related gene SlERT1B, consistent with its well-known role in this process. Solid lines indicate direct binding of the transcription factor (SlNAP2 or NOR) to target gene promoters, while dashed lines indicate indirect physiological connections.
Another important finding of our study is that SlNAP2 itself is affected, at the expression level, by NOR; more specifically, as shown in Fig. 3D, expression of SlNAP2 is significantly reduced in leaves of the nor mutant, while it is elevated in the NOR overexpressor line OX-19, suggesting that NOR acts upstream of SlNAP2. On the other hand, we found that expression of NOR is enhanced in SlNAP2-IOE plants shortly (6 h) after EST treatment (Fig. 7B), consistent with a model that places SlNAP2 upstream of NOR. In accordance with this, we found that ABA treatment triggered an increased expression of NOR in SlNAP2 overexpressors, and it supported a stronger leaf senescence in NOR overexpressors than in WT and ami-NOR plants (Supplementary Fig. S8). We previously found evidence that SlNAP2 plays an important role in establishing ABA homeostasis during the process of leaf senescence (Ma et al., 2018); the new data reported here about the involvement of NOR add further insights into this regulatory system (Fig. 8).
Collectively, the available experimental data therefore suggest that NOR and SlNAP2 together form a positively acting regulatory loop whereby the expressional activity of each NAC gene is enhanced by the other NAC TF. However, we note that unraveling the details of this regulatory interaction requires further detailed investigation in the future.
Together, the available data strongly suggest that NAC TFs controlling leaf senescence also affect age-dependent senescence (or ripening) of fleshy and non-fleshy fruits, across species. This observation raises a number of interesting questions, including the following. (i) How do NAC TFs exert their specific aging-related functions in photosynthetic leaves compared with those in fruits; that is, how are the target genes prevalent or specific for leaf senescence selected compared with target genes involved in fruit ripening? (ii) Related to this: do NAC TFs interact with other different TFs in leaves versus fruits to exert their molecular functions? (iii) To what extent do epigenetic marks affect which genes are primary targets of the senescence-related NACs in leaves versus fruits? (iv) In which way has evolution shaped the gene regulatory landscape of age-related NAC TFs in leaves compared with fruits? These questions lead to an even wider perspective which addresses the diversification of NAC functions at the organ, tissue, and cellular levels, an aspect not well understood at present. Future research clearly has to address this aspect in more detail.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Amino acid sequence alignment of selected NAC transcription factors.
Fig. S2. Selection of NOR transgenic lines.
Fig. S3. Flowering time of tomato wild-type and NOR-modified plants.
Fig. S4. Phenotype of NOR overexpressors at seedling stage.
Fig. S5. Heat map of the expression of senescence-related tomato genes during age-dependent senescence and dark-induced senescence.
Fig. S6. Dark-induced senescence in ami-NOR plants.
Fig. S7. Identification of the binding sequences of NOR.
Fig. S8. NOR affects ABA-induced leaf senescence.
Table S1. Oligonucleotide sequences.
Table S2. Data for results shown in heat maps.
Table S3. Promoters of NOR target genes.
Table S4. Expression of GA metabolism genes.
Acknowledgements
We thank Dr Karin Koehl and her team (Max Planck Institute of Molecular Plant Physiology) for plant care, and Gang-Ping Xue (CSIRO Agriculture and Food, Australia) for performing the binding site selection assays. We thank the University of Potsdam and the Max Planck Institute of Molecular Plant Physiology for supporting our research. This work was supported by the Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany. X.M. received a fellowship from the China Scholarship Council (CSC; file no. 201306510001).
References
- Alseekh S, Tohge T, Wendenberg R, et al. 2015. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. The Plant Cell 27, 485–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. 2008. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. 2008. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology 10(Suppl 1), 63–75. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Schildhauer J, Araújo WL, Munné-Bosch S, Fernie AR, Proost S, Humbeck K, Mueller-Roeber B. 2014. Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: transcriptomic and metabolomic consequences. Journal of Experimental Botany 65, 3975–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. 2010. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. The Plant Journal 62, 250–264. [DOI] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. 2007. Ethylene and fruit ripening. Journal of Plant Growth Regulation 26, 143–159. [Google Scholar]
- Biswal UC, Mohanty P. 1976. Dark stress-induced senescence of detached barley leaves. II. Alteration in absorption characteristic and photochemical activity of chloroplasts isolated from senescing leaves. Plant Science Letters 7, 371–379. [Google Scholar]
- Breitel DA, Chappell-Maor L, Meir S, et al. 2016. AUXIN RESPONSE FACTOR 2 intersects hormonal signals in the regulation of tomato fruit ripening. PLoS Genetics 12, e1005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson J, Bieker S, Riester L, Doll J, Zentgraf U. 2018. A guideline for leaf senescence analyses: from quantification to physiological and molecular investigations. Journal of Experimental Botany 69, 769–786. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Carrasco-Orellana C, Stappung Y, Mendez-Yañez A, Allan AC, Espley RV, Plunkett BJ, Moya-Leon MA, Herrera R. 2018. Characterization of a ripening-related transcription factor FcNAC1 from Fragaria chiloensis fruit. Scientific Reports 8, 10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals J, Pascual L, Canizares J, Cebolla-Cornejo J, Casanas F, Nuez F. 2012. Genetic basis of long shelf life and variability into Penjar tomato. Genetic Resources and Crop Evolution 59, 219–229. [Google Scholar]
- Catalá C, Rose JK, Bennett AB. 2000. Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiology 122, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Kao CH. 1991. Senescence of rice leaves XXX. Levels of endogenous polyamines and dark-induced senescence of rice leaves. Plant & Cell Physiology 32, 935–941. [Google Scholar]
- Davière JM, Achard P. 2013. Gibberellin signaling in plants. Development 140, 1147–1151. [DOI] [PubMed] [Google Scholar]
- Fan K, Bibi N, Gan S, Li F, Yuan S, Ni M, Wang M, Shen H, Wang X. 2015. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. Journal of Experimental Botany 66, 4669–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y. 2013. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. The Plant Cell 25, 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wei W, Zhao X, et al. 2018. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Horticultural Research doi: 10.1038/s41438-018-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapati P, Xue GP, Munné-Bosch S, Balazadeh S. 2015. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiology 168, 1122–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ, Tanksley SD, Vrebalov J, Noensie E. 2004. NOR gene for use in manipulation of fruit quality and ethylene response. US Patent No. 5,234,834 issued 13 July 2004. [Google Scholar]
- Guo Y, Gan S. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612. [DOI] [PubMed] [Google Scholar]
- Han Q, Zhang J, Li H, Luo Z, Ziaf K, Ouyang B, Wang T, Ye Z. 2012. Identification and expression pattern of one stress-responsive NAC gene from Solanum lycopersicum. Molecular Biology Reports 39, 1713–1720. [DOI] [PubMed] [Google Scholar]
- Han QQ, Song YZ, Zhang JY, Liu LF. 2014. Studies on the role of the SlNAC3 gene in regulating seed development in tomato (Solanum lycopersicum). Journal of Horticultural Science and Biotechnology 89, 423–429. [Google Scholar]
- Hendelman A, Stav R, Zemach H, Arazi T. 2013. The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. Journal of Experimental Botany 64, 5497–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou K, Wu W, Gan SS. 2013. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiology 161, 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. 2012. Cytokinin signaling networks. Annual Review of Plant Biology 63, 353–380. [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, et al. 2013. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Shima Y, Nakamura N, Kotake-Nara E, Kawasaki S, Toki S. 2017. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nature Plants 3, 866–874. [DOI] [PubMed] [Google Scholar]
- Kamranfar I, Xue GP, Tohge T, Sedaghatmehr M, Fernie AR, Balazadeh S, Mueller-Roeber B. 2018. Transcription factor RD26 is a key regulator of metabolic reprogramming during dark-induced senescence. New Phytologist 218, 1543–1557. [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. 2010. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proceedings of the National Academy of Sciences, USA 107, 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC. 2010. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nature Protocols 5, 457–472. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Chiang YH, Kieber JJ, Schaller GE. 2013a SCFKMD controls cytokinin signaling by regulating the degradation of type-B response regulators. Proceedings of the National Academy of Sciences, USA 110, 10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kieber JJ, Schaller GE. 2013b The rice F-box protein KISS ME DEADLY2 functions as a negative regulator of cytokinin signalling. Plant Signaling & Behavior 8, e26434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Park JH, Kim J, et al. 2018. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proceedings of the National Academy of Sciences, USA 115, E4930–E4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, Bressan RA, Narasimhan ML. 2011. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany 62, 3981–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC. 2013. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant & Cell Physiology 54, 1660–1672. [DOI] [PubMed] [Google Scholar]
- Kou X, Liu C, Han L, Wang S, Xue Z. 2016. NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Molecular Genetics and Genomics 291, 1205–1217. [DOI] [PubMed] [Google Scholar]
- Kou X, Wang S, Wu M, Guo R, Xue Z, Meng N, Tao X, Chen M, Zhang Y. 2014. Molecular characterization and expression analysis of NAC family transcription factors in tomato. Plant Molecular Biology Reporter 32, 501–516. [Google Scholar]
- Kou X, Watkins CB, Gan SS. 2012. Arabidopsis AtNAP regulates fruit senescence. Journal of Experimental Botany 63, 6139–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tamboli V, Sharma R, Sreelakshmi Y. 2018. NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chemistry 259, 234–244. [DOI] [PubMed] [Google Scholar]
- Kunieda T, Mitsuda N, Ohme-Takagi M, Takeda S, Aida M, Tasaka M, Kondo M, Nishimura M, Hara-Nishimura I. 2008. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. The Plant Cell 20, 2631–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Ye G, Zeng D. 2017. Genetic dissection of leaf senescence in rice. International Journal of Molecular Sciences 18, 2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Woo HR, Guo H. 2018. Genetic redundancy of senescence-associated transcription factors in Arabidopsis. Journal of Experimental Botany 69, 811–823. [DOI] [PubMed] [Google Scholar]
- Liang CZ, Wang YQ, Zhu YN, Tang JY, Hu B, Liu LC, Ou SJ, Wu HK, Sun XH, Chu JF, Chu CC. 2014. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proceedings of the National Academy of Sciences, USA 111, 10013–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira BS, Gramegna G, Trench BA, et al. 2017. Manipulation of a senescence-associated gene improves fleshy fruit yield. Plant Physiology 175, 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Y, Turečková V, Xue GP, Fernie AR, Mueller-Roeber B, Balazadeh S. 2018. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiology 177, 1286–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F. 2017. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiology 174, 1747–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. 2011. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiology 157, 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou W, Li D, Luo Z, Li L, Mao l, Ying T. 2018. SlAREB1 transcriptional activation of NOR is involved in abscisic acid-modulated ethylene biosynthesis during tomato fruit ripening. Plant Science 276, 239–249. [DOI] [PubMed] [Google Scholar]
- Moyano E, Martínez-Rivas FJ, Blanco-Portales R, Molina-Hidalgo FJ, Ric-Varas P, Matas-Arroyo AJ, Caballero JL, Muñoz-Blanco J, Rodríguez-Franco A. 2018. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria×ananassa fruits. PLoS One 13, e0196953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N, Suokas M, Karppinen K, Vuosku J, Jaakola L, Häggman H. 2018. Recognition of candidate transcription factors related to bilberry fruit ripening by de novo transcriptome and qRT-PCR analyses. Scientific Reports 8, 9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. 1999. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Molecular Biology 41, 181–194. [DOI] [PubMed] [Google Scholar]
- Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A. 2016. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Scientific Reports 6, 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. 1991. Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439. [DOI] [PubMed] [Google Scholar]
- Podzimska-Sroka D, O’Shea C, Gregersen PL, Skriver K. 2015. NAC transcription factors in senescence: from molecular structure to function in crops. Plants 4, 412–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A. 2006. Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224, 556–568. [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Research 43, D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. 2012. NAC proteins: regulation and role in stress tolerance. Trends in Plant Science 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Qin G, Wang Y, Cao B, Wang W, Tian S. 2012. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. The Plant Journal 70, 243–255. [DOI] [PubMed] [Google Scholar]
- Ríos P, Argyris J, Vegas J, et al. 2017. ETHQV6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. The Plant Journal 91, 671–683. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Han SH, Lee SH, Hörtensteiner S, Paek NC. 2016. Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Reports 35, 155–166. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G. 2014. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nature Communications 5, 4636. [DOI] [PubMed] [Google Scholar]
- Shao H, Wang H, Tang X. 2015. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Frontiers in Plant Science 6, 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs J, Lee E, Holt K, Yong-Duk K, Steele Scott N, Loveys B, Schuch W. 1998. Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiology 117, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalaikumar VP, Devkar V, Mehterov N, Ali S, Ozgur R, Turkan I, Mueller-Roeber B, Balazadeh S. 2018. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnology Journal 16, 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. 2006. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. 2006. Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology 141, 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM. 2001. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiology 127, 876–886. [PMC free article] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, et al. 2012. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. The Plant Cell 24, 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP. 2002. Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Research 30, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP. 2005. A CELD-fusion method for rapid determination of the DNA-binding sequence specificity of novel plant DNA-binding proteins. The Plant Journal 41, 638–649. [DOI] [PubMed] [Google Scholar]
- Xue GP, Bower NI, McIntyre CL, Riding GA, Kazan K, Shorter R. 2006. TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Functional Plant Biology 33, 43–57. [DOI] [PubMed] [Google Scholar]
- Yang J, Udvardi M. 2018. Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. Journal of Experimental Botany 69, 855–865. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. 2001. Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant & Cell Physiology 42, 170–178. [DOI] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan SS. 2012. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. The Plant Journal 69, 667–678. [DOI] [PubMed] [Google Scholar]
- Zhao D, Derkx AP, Liu DC, Buchner P, Hawkesford MJ. 2015. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biology 17, 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gao J, Im Kim J, Chen K, Bressan RA, Zhu JK. 2017. Control of plant water use by ABA induction of senescence and dormancy: an overlooked lesson from evolution. Plant & Cell Physiology 58, 1319–1327. [DOI] [PubMed] [Google Scholar]
- Zhong J, Powell S, Preston JC. 2016. Organ boundary NAC-domain transcription factors are implicated in the evolution of petal fusion. Plant Biology 18, 893–902. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Huang WF, Liu L, Chen TY, Zhou F, Lin YJ. 2013. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen G, Zhou S, Tu Y, Wang Y, Dong T, Hu Z. 2014. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant & Cell Physiology 55, 119–135. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, Chua NH. 2002. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. The Plant Journal 30, 349–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.