Phylogenetic and structural findings reveal novel insights into the origin, evolution and structure of nuclear matrix constituent proteins, establishing their origin in Charophyta and demonstrating their functional conservation.
Keywords: Bioinformatics analysis, immunofluorescence microscopy, NMCP/CRWN proteins, phylogenetic analysis, plant lamina, protein structural analysis, Physcomitrella patens
Abstract
Nuclear matrix constituent proteins (NMCPs), the structural components of the plant lamina, are considered to be the analogues of lamins in plants based on numerous structural and functional similarities. Current phylogenetic knowledge suggests that, in contrast to lamins, which are widely distributed in eukaryotes, NMCPs are taxonomically restricted to Streptophyta. At present, most information about NMCPs comes from angiosperms, and virtually no data are available from more ancestral groups. In angiosperms, the NMCP family comprises two phylogenetic groups, NMCP1 and NMCP2, which evolved from the NMCP1 and NMCP2 progenitor genes. Based on sequence conservation and the presence of NMCP-specific domains, we determined the structure and number of NMCP genes present in different Streptophyta clades. We analysed 91 species of embryophytes and report additional NMCP sequences from mosses, liverworts, clubmosses, horsetail, ferns, gymnosperms, and Charophyta algae. Our results confirm an origin of NMCPs in Charophyta (the earliest diverging group of Streptophyta), resolve the number and structure of NMCPs in the different clades, and propose the emergence of additional NMCP homologues by whole-genome duplication events. Immunofluorescence microscopy demonstrated localization of a basal NMCP from the moss Physcomitrella patens at the nuclear envelope, suggesting a functional conservation for basal and more evolved NMCPs.
Introduction
The lamina is a conserved fundamental nuclear structure underlying the nuclear envelope in eukaryotes, which basically consists of a filamentous network of coiled-coil proteins (Ciska and Moreno Diaz de la Espina, 2014; Turgay et al., 2017). Lamins are the main structural components of the lamina in metazoa and directly promote the association of numerous lamin-binding proteins to the inner nuclear membrane, nuclear pore complexes, and chromatin (Goldberg et al., 2008; Gruenbaum and Medalia, 2015; Turgay et al., 2017). Lamins are present in all metazoa, as well as some other distant eukaryotes (Batsios et al., 2012; Kruger et al., 2012; Kollmar, 2015; Koreny and Field, 2016), and phylogenetic studies on lamins suggest an early origin in the last eukaryotic ancestor (Koreny and Field, 2016).
All lamins share a highly conserved domain architecture, with a central coiled-coil rod domain (CCD) containing four coils, a short N-terminal head with a CDK1 phosphorylation site, and a C-terminal tail. The tail contains several conserved domains: a nuclear localization signal, an IgG fold, and a terminal CAAX box. Lamins associate in vitro by parallel dimerization of the rod domain and head to tail association of dimers, forming protofilaments that associate laterally to form filaments (Davidson and Lammerding, 2014), although their molecular organization in the native lamina has been resolved only very recently (Turgay et al., 2017). Lamins are involved in diverse nuclear functions, such as the control of nuclear shape and architecture; connection of the nucleoskeleton to the cytoskeleton; chromatin organization, positioning, and expression; DNA replication, repair, and transcription; and cell proliferation and differentiation (Gruenbaum and Foisner, 2015).
Plants have a well-organized nuclear lamina (Moreno Diaz de la Espina et al., 1991; Minguez and Moreno Diaz de la Espina, 1993; Fiserova et al., 2009; Fiserova and Goldberg, 2010; Ciska and Moreno Diaz de la Espina, 2014) but they lack lamin genes. Organization of the plant lamina is based on plant specific coiled-coil proteins, without sequence homology to lamins but with a conserved structure, that act as functional homologues of lamins (Ciska and Moreno Diaz de la Espina, 2013, 2014; Koreny and Field, 2016; Poulet et al., 2017). Few proteins of the plant lamina have been characterized so far. Among these, only the nuclear matrix constituent proteins (NMCPs) behave as structural components (Masuda et al., 1997; Ciska et al., 2013; Sakamoto and Takagi, 2013), while the rest are NMCP-binding proteins, such as SUNs (Graumann, 2014), KAKU4 (Goto et al., 2014), ARP7 (Mochizuki et al., 2017), NEAPs (Pawar et al., 2016), MYB3, SINAT, and BIM1 (Mochizuki et al., 2017). NMCPs, also known as CROWDED NUCLEI (CRWN) in Arabidopsis (Wang et al., 2013), are currently considered to be the lamin analogues in plants based on numerous structural and functional similarities, such as their coiled-coil organization, domain layout, subnuclear distribution, low solubility, interaction with SUN proteins, and implication in the regulation of nuclear shape and size, nuclear envelope and heterochromatin organization, germination, and plant immunity (Dittmer et al., 2007; Kimura et al., 2010; Ciska et al., 2013; Ciska and Moreno Diaz de la Espina, 2013; Wang et al., 2013; Goto et al., 2014; Graumann, 2014; Kimura et al., 2014; Zhao et al., 2016; Guo et al., 2017).
Currently available phylogenetic information suggests that in contrast to lamins, which are widely distributed in eukaryotes, the structural proteins forming the plant lamina (i.e. the NMCPs) are taxonomically restricted to the clade Streptophyta (Koreny and Field, 2016).
Previous phylogenetic analysis of NMCP proteins, restricted to the moss Physcomitrella patens and to flowering plants (angiosperms), revealed that the NMCPs in angiosperms have evolved from two progenitor genes, NMCP1 and NMCP2. Dicots have two to three NMCP1-type proteins, whereas monocots have only one representative of this type. Both monocots and dicots contain a single NMCP2 protein. The moss P. patens has two proteins that evolved from the NMCP progenitor gene (Kimura et al., 2010; Ciska et al., 2013; Wang et al., 2013; Ciska and Moreno Diaz de la Espina, 2014). In a general evolutional analysis of plant nuclear envelope proteins, Poulet et al. (2017) reported two NMCP2-type homologues in the gymnosperm Picea abies, and established the origin of the NMCP family in mosses. Lastly, in a comparative genomic analysis of lamina proteins in eukaryotes, a few putative NMCP sequences were reported in charophyte algae (Koreny and Field, 2016).
Prediction of the secondary structure of NMCP proteins in angiosperms revealed a highly conserved tripartite structure similar to that of lamins, with a long central CCD formed by two segments with highly conserved ends separated by a linker, predicted to dimerize. Angiosperm NMCPs have also several conserved regions: a CDK1 phosphorylation site in the head domain, a nuclear localization signal and its adjacent region, an actin-binding domain, a segment of acidic amino acids, the C-terminus of the protein, and several phosphorylation sites in the tail domain (Ciska et al., 2013; Ciska and Moreno Diaz de la Espina, 2013, 2014; Ciska et al., 2018).
Here, we present new phylogenetic, sequence, and structural information about basal NMCPs. This evidence extends the presence of NMCPs to Charophyta algae, the earliest divergent group of streptophytes, in agreement with the results of Koreny and Field (2016) that identify some NMCP sequences in Charophyta, as well as to other Embryophyta other than angiosperms, such as Marchantiophyta, Briophyta, Pterydophyta, and gymnosperms. Our results provide important novel insights into basal NMCPs and the evolutionary history of these proteins. With the available tools, we establish the origin of NMCPs at least in Streptophyta after their branching from Chlorophyta, which lack NMCP proteins. We also demonstrate that the basal NMCP protein from P. patens, PpaNMCP1, which has a different coiled-coil domain organization from that of the most evolved NMCPs of angiosperms, is a component of the nuclear envelope, supporting the functional conservation of this structure from basal embryophytes (mosses) to angiosperms.
Materials and methods
Bioinformatics tools and analysis
Genome searches were performed using BLASTP against the Phytozome v12 (Goodstein et al., 2012), KEGG, congenie.org, and gymnoPLAZA (Proost et al., 2015) databases, Additional searches were performed using the NCBI database and the transcriptomic database 1KP (Matasci et al., 2014). In addition, a BLASTP search was performed using BLAST2GO against the translated transcriptome of Equisetum giganteum (Vanneste et al., 2015). Multiple alignments were carried out using Clustal Omega and MUSCLE, and the phylogenetic analysis was performed using MEGA7 (Kumar et al., 2016). A search for the best model within the Maximum Likelihood statistical method was performed, and the Maximum Likelihood method based on the JTT matrix model was chosen for the phylogenetic tree reconstruction as the best-suited model. All positions with less than 95% site coverage were eliminated, meaning that less than 5% alignment gaps, missing data, or ambiguous bases were allowed at any position. The reliability of the topology was inferred from bootstrap analyses of 1000 replicates.
The coiled coils and polymerization state were predicted using MARCOIL (https://toolkit.tuebingen.mpg.de/) (Delorenzi and Speed, 2002) and Multicoil2 (http://groups.csail.mit.edu/cb/multicoil2/cgi-bin/multicoil2.cgi), respectively. As a threshold, a 0.5 probability of coiled coil was used, and 28 amino acids (four repeats of a heptad pattern) as necessary to form a stable coiled coil.
Using the MEME 4.12.0 suite (Bailey et al., 2009) 20 conserved regions between 6 and 25 residues in length were found in a set of sequences representing the whole Streptophyta group as well as in each phylogenetic cluster. Multiple alignments were visualized and analysed in Jalview (Waterhouse et al., 2009).
Expression data for selected genes were found on the Electronic Fluorescent Pictograph (eFP) browser (http://bar.utoronto.ca/) (Winter et al., 2007).
Plant materials
Protonemata of Physcomitrella patens (Hedw.) Bruch & Schimp subsp. patens (Ashton and Cove, 1977) were grown in 9 cm Petri dishes on BCDATG medium solidified with 0.8% (w/v) agar (A-9799, Sigma) (Nishiyama et al., 2000). The medium was covered with a sheet of Cellophane to facilitate peeling off of protonemata from the medium. The dishes were kept at 25 °C under continuous dim light. The protonemata were maintained by subculture on to fresh medium at regular intervals.
Cloning of cDNA
RNA was extracted from the protonemata using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. First-strand cDNAs were primed with an oligo(dT) and synthesized using the Thermo Scientific Verso cDNA Synthesis Kit (Thermo Fisher Scientific, USA). Then, the 5′-terminal 629 nucleotides of the open reading frame of Pp1s76_81V6, named Pp1 (Ciska et al., 2013), were amplified by PCR using the primers 5′-ATGTACACACCGCAGGGGAGA and 5′-AACTCTCGAGCTTGAGCGAGCTGT. The cDNA fragment was ligated into the pTAC-1 vector (BioDynamics Laboratory, Japan) with the TA cloning technique. The vector was transformed into Escherichia coli strain DH5α.
Production and validation of the anti-Pp1NMCP antibody
The nucleotide sequence corresponding to the N-terminal 203 amino acids of Pp1 was amplified by PCR using the primers 5′- GCCTCTGTCGACTACACACCGCAG, which includes an extended sequence with the cleavage site for SalI, and the downstream primer 5′-TTATCACTCTCGAGCTTGAGCGAGCTG. The amplified fragments were digested with SalI and inserted into the multi-cloning site of the pEcoli-Nterm 6xHN vector (Clontech). The vector was transformed into E. coli BL21 (DE3). Expression of the recombinant protein was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside, and the cells were incubated for 4 h at 30 °C. The protein was extracted from an insoluble fraction of the cell homogenate with a buffer containing 6 M urea, and the 6xH-tagged protein was purified using a metal affinity chromatography resin (Profinity iIMac resin, BioRad). The protein was finally dissolved in PBS containing 0.02% sodium dodecyl sulfate and submitted to SIGMA Genosys, which performed immunization of rabbits, collection of the antiserum, and purification of polyclonal antibody. Controls of the anti-Pp1NMCP antibody were performed by western blot of total protonemata proteins and the 6xHN-tagged immunization polypeptide. The bacterially expressed 6xHN-tagged protein containing the N-terminal region of Pp1 was purified by affinity chromatography using Profinity iMac resin (BioRad). The major fraction was separated by using the Laemmli SDS-PAGE system with 12.5% polyacrylamide gel. The protonemata of P. patens were pulverized with liquid nitrogen in a mortar and pestle, and suspended in 20 mM 2-(N-morpholino)ethanesulfonic acid (MES)-KOH (pH 5.8) supplemented with 30 mM KCl, 20 mM NaCl, 3 mM MgCl2, 2 mM CaCl2,10% (w/v) glycerol, 0.25 M sucrose, 0.1% (w/v) Triton X-100, Complete (Roche), and 2 mM DTT. The homogenate was centrifuged at 40 000 × g for 10 min. The pellet was dissolved in an extraction buffer containing 8 M urea and centrifuged at 36 000 × g for 10 min. The supernatant was mixed with an equal volume of the 2× sample buffer for electrophoresis and separated by the Laemmli SDS-PAGE system using 7.5% polyacrylamide gels. Proteins resolved by electrophoresis were transferred to a polyvinylidene fluoride membrane. The membranes were incubated in a diluted solution (1:1000) of the Pp1-specific antibody and then with a peroxidase-conjugated second antibody. Peroxidase activity was detected with the SuperSignal Femto HRP chemifluorescent detection kit (Thermo), using a charge-coupled imaging device. The immunization peptide was used as a positive control for validation.
Immunofluorescence microscopy
Protonemata collected from the culture medium were treated with a cell-wall-degrading enzyme mixture containing 2% cellulase ‘Onozuka’ RS (Wako Chemicals), 1% hemicellulase (from mung beans, Sigma), 0.2% pectolyase Y-23 (Wako Chemicals), and a proteinase inhibitor mixture (Nacarai Tesque) at 25 °C for 20 min, and then fixed with 2.7% formaldehyde in MES-KOH (pH 5.8) containing 30% ethanol, 2 mM MgCl2, 3 mM CaCl2, and 100 mM KCl for 40 min at 0 °C, followed by washing a few times with PBS. They were then fixed on APS-coated slides and permeabilized with 0.2% Triton X-100. Immunofluorescence was performed using the anti-PpNMCP1 antibody (1:200) and Alexa Fluor 555-conjugated donkey anti-rabbit IgG plus IgM antibody (Agilent Technologies). Images were taken under a confocal laser scanning microscope (TCS SP5, Leica Microsystems) equipped with differential interference contrast optics.
Results and discussion
Genomic searches and number of NMCPs present in different species
We analysed the presence and number of genes encoding NMCPs in the genomes of 55 species across the Embryophyta and, additionally, the presence of NMCPs in transcriptomes from 36 species among gymnosperms, spikemosses, clubmosses, ferns, and a horsetail (see Supplementary Dataset S1 at JXB online), focusing on the clades scarcely described before.
We also investigated the presence of NMCP sequences in algae, since Koreny and Field (2016) described a few putative NMCP sequences in Charophyta that are thought to be the progenitors of land plants (Becker and Marin, 2009; Hori et al., 2014; Delwiche and Cooper, 2015). These sequences were used in standard BLASTP searches against the NCBI database and produced matches with Embryophyta NMCPs. Although the KEGG, Phytozome, and picoPLAZA databases contain sequenced algae genomes, none belongs to the Charophyta (most are part of the Chlorophyta). To our knowledge, the only full genome of a charophyte available is that of Klebsormidium nitens (formerly Klebsormidium flaccidum) (Hori et al., 2014). We were not able to find additional NMCP members among algae (using sequences of Charophyta and the liverwort Marchantia polymorpha in the searches). Taking into account the low level of sequence conservation in this group and the scarce genomic data available, it is extremely difficult to identify new NMCP homologues in this group. On the other hand, the closest matching sequences in chlorophyte and other algae are myosins or other coiled-coil sequences (golgins, cingulin), due to the typical heptad pattern of coiled coils (HPPHPPP, where H represents hydrophobic amino acids and P polar amino acids). It is unlikely that these matches are significant, due to the low scores and e-values, as well as ‘false positives’ frequently observed in coiled-coil protein analysis (Koreny and Field, 2016).
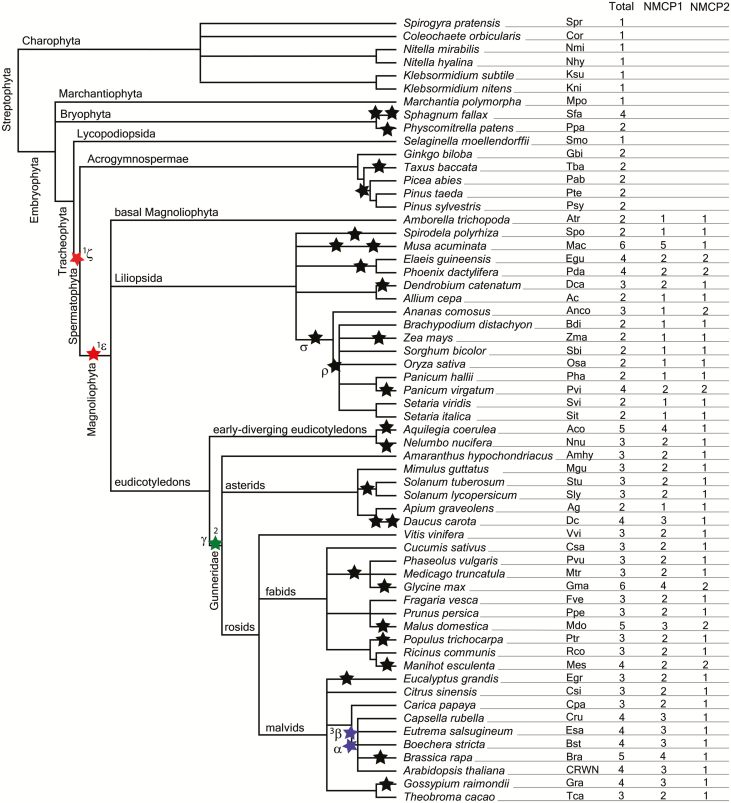
We collected sequences of a number of NMCP homologues in plant species, aligned all sequences using Clustal Omega, and investigated possible sequence duplications, errors, and missing regions. Charophyta algae and some basal embryophytes evidenced a single protein, while the rest rendered two, three, or more proteins (Fig. 1). We investigated the mechanisms of gene duplication possibly influencing the number of NMCPs in different species, the most common being whole-genome duplication (WGD) and tandem duplications (Wang et al., 2011b; Panchy et al., 2016). Extensive genome-scale synteny and phylogenomic analyses support a role for WGDs in the following lineages: two WGDs (α and β) in crucifers (Brassicaceae), one (γ) shared by all eudicots (Vision et al., 2000; Bowers et al., 2003; Tang et al., 2008; Barker et al., 2009), and two in monocots (σ and ρ) before the diversification of cereal grains and other grasses (Tang et al., 2010). It is also generally accepted that two ancient WGDs took place in angiosperms (ε) and in seed plants (ζ) (Jiao et al., 2011). For reference, the positions of the published genome polyploidization events are shown in Fig. 1.
Fig. 1.
Phylogenetic relationship between species containing NMCP proteins, reported polyploidization events, and numbers of NMCPs. The phylogenetic relationships were generated using the Taxonomy tool on the NCBI webpage (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) and edited in TreeView software. The reported whole-genome duplication events are marked with asterisks: ζ and ε, possible points of appearance of NMCP1 and NMCP2; γ, point of appearance of NMCP3. (This figure is available in colour at JXB online.)
Our data reveal that the number of NMCPs in a given species usually corresponds to its reported number of polyploidization events, which strongly suggests that the additional NMCP genes were created as a result of WGDs. We observed that the species that underwent a WGD contain at least two NMCP genes, whereas species with no reported WGDs, such as the as liverwort M. polymorpha (Bowman et al., 2017) and Selaginella (Banks et al., 2011), contain only one NMCP. These results are in agreement with those of Li et al. (2016) reporting that angiosperm core gene families (the genes present in all angiosperm genomes, as is the case for NMCPs) are more influenced by WGD than by small-scale gene duplications.
It is also known that some proteins involved in specific functions are more frequently retained after WGDs, such as nuclear proteins (Blanc and Wolfe, 2004) and proteins involved in development or defence (Maere et al., 2005; del Pozo and Ramirez-Parra, 2015). NMCPs are nuclear proteins reported to be involved in plant development (van Zanten et al., 2011; Zhao et al., 2016). According to expression data available in eFP databases (Supplementary Dataset S2) and our previous results (Ciska et al., 2013, 2018), NMCPs are in general expressed at higher levels in meristematic and young tissues, as well as during the activation of plant defence against pathogens (Guo et al., 2017).
Phylogeny of NMCPs and retention of duplicated genes
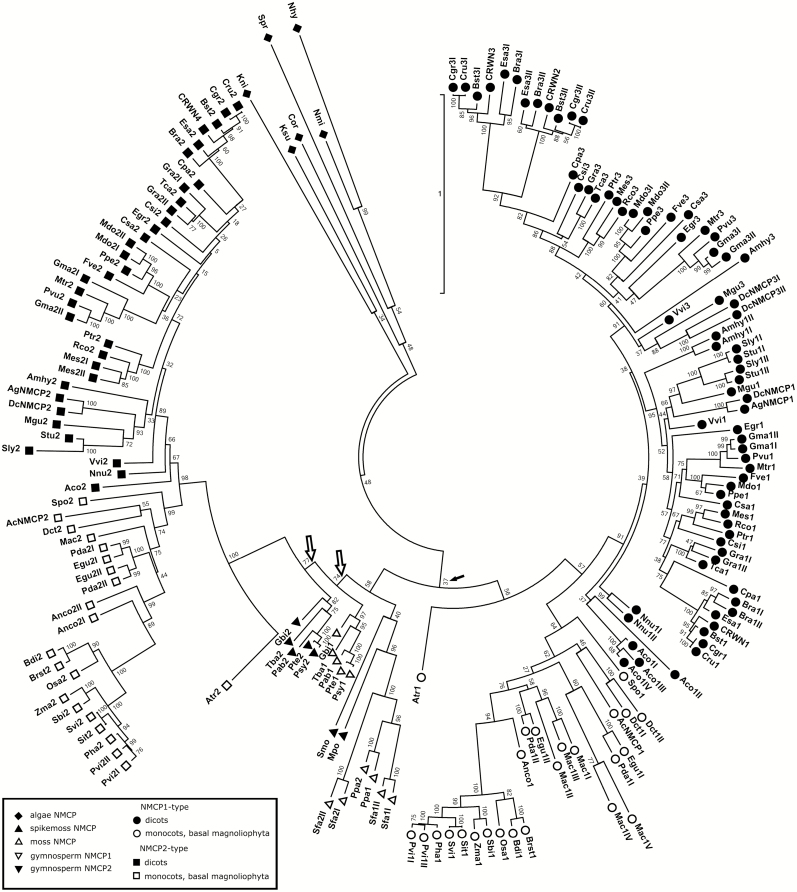
We inferred the phylogenetic relationships between the NMCP homologues using the MEGA7 programme. For this, we aligned the sequences using MUSCLE and generated a Maximum Likelihood phylogenetic tree (Fig. 2; Supplementary Fig. S1). The results obtained were consistent with our previous data and with the classification of angiosperm NMCPs into NMCP1 and NMCP2 types (Ciska et al., 2013, 2018). In this study we incorporate new data from basal and gymnosperm NMCP proteins. In addition, we include new sequences from monocot species, with a focus on those not belonging to the Poaceae (grasses), such as Musa acuminata (banana), Ananas comosus (pineapple), Dendrobium catenatum, Spyrodela polyrhiza, Elaeis guineensis (oil palm), and Phoenix dactylifera (date palm); basal Magnoliophyta (Amborella trichopoda); and the early-diverging dicots Aquilegia coerulea and Nelumbo nucifera (sacred lotus). Our results confirm the origin of NMCPs at least in charophyte algae, in agreement with what was reported by Koreny and Field (2016) and in contrast with the analysis by Poulet et al. (2017), which established the emergence of NMCPs in Bryophyta (mosses and clubmosses).
Fig. 2.
Phylogenetic tree of NMCPs. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The tree with the highest log likelihood is shown. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 176 amino acid sequences. There were a total of 2761 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Reliability of the branches was inferred from bootstrap analyses of 1000 replicates. All positions with less than 95% site coverage were eliminated (i.e. less than 5% alignment gaps, missing data or ambiguous bases were allowed at any position). Filled arrows indicate low bootstrap values for the node including NMCP2 and basal NMCPs. Empty arrows indicate the two nodes of gymnosperm NMCP1 and NMCP2.
As seen on the phylogenetic trees (Fig. 2; Supplementary Fig. S1), algal NMCPs form a separate cluster with long branches, which corresponds to their diverged sequences. NMCPs of mosses, liverwort (M. polymorpha), lycophytes (Selaginella moellendorffii), and gymnosperms cluster with the NMCP2-type proteins (although with low bootstrap values; Fig. 2), which suggests that these NMCPs present more basal features than the quickly diverging NMCP1-type proteins.
Gymnosperms contain two NMCP proteins that form two separate clusters and cluster together with angiosperm NMCP2 proteins and the ‘basal NMCPs’. The fact that the two different NMCP types of gymnosperms, including those in Ginkgo biloba, cluster with NMCP2 proteins (Fig. 2), but not together, as is the case for NMCPs of mosses, clubmosses, or spikemosses, could suggest that they originated in the ζ duplication event that included all seed plants. The observation that the rates of molecular evolution per unit of time in gymnosperms are on average seven times lower than in angiosperms (De La Torre et al., 2017) is in agreement with our phylogenetic analysis, in which gymnosperm NMCPs locate closer to the centre of the phylogenetic tree (Fig. 2; Supplementary Fig. S1), suggesting they evolved more slowly than the sequences located further away from the centre (lengths of the branches are measured by the number of substitutions per site) (Kumar et al., 2016). A similar observation was made for ferns and horsetail (with the exception of E. giganteum). This, together with the fact that gymnosperm and other basal NMCPs cluster together in the phylogenetic analysis, suggests that they diverged more slowly than NMCP1-type proteins and maintained ‘basal NMCP’ features.
Most monocots and the basal angiosperm A. trichopoda (which arose before the separation of monocots and eudicots) contain a single NMCP protein of each type, with the exception of the few species that underwent a recent WGD: Musa acuminata (D’Hont et al., 2012), P. dactylifera (Al-Mssallem et al., 2013), E. guineensis (Singh et al., 2013), and D. catenatum (Zhang et al., 2016).
As previously reported, dicots contain a single NMCP2 gene, except for a few species that underwent a recent WGD event and contain two genes [soybean (Glycine max), apple (Malus domestica), and cassava (Manihot esculenta)]. In contrast, they present one to four genes encoding NMCP1 proteins, depending on the number of WGD events experienced in each case (Fig. 1).
WGDs and the evolution of the NMCP family
WGDs (polyploidization) can be divided into ancient (paleopolyploidy) and recent (neopolyploidy) events. Based on the information available on the WGD events and the phylogenetic relationships in the NMCP family, we can distinguish: (i) species that have never undergone WGD and contain a single NMCP (Charophyta algae, the liverwort M. polymorpha, S. moellendorffii, and other spikemosses); (ii) species that have undergone a recent WGD and retained the duplicates (P. patens, Sphagnum fallax, E. giganteum); (iii) species that in the past underwent an ancient WGD and contain two (seed plants), three (dicots), or four (Brassicaceae) NMCP homologues; and (iv) species that, as well as the ancient WGD event, have undergone a recent polyploidization and contain a variable number of NMCPs depending on the number of genes retained in the genome.
There is evidence for WGDs in the ferns Botrypus virginianus and Sceptridium dissectum, which belong to the Ophioglossales (Clark et al., 2016). In the case of S. dissectum, there is just one NMCP gene. B. virginianus presents two identical NMCPs at consecutive loci, which we named Bvi for the phylogenetic analysis. Consecutive loci suggest tandem duplications that result in two adjacent duplicates on the same chromosome (Wang et al., 2011b). This may suggest that, if ferns have undergone WGD, duplicates were lost over time.
Looking at species that have undergone only a recent WGD, we found two mosses, P. patens and S. fallax, which contain, respectively, two and four NMCP proteins, correlating with the number of reported WGD events (Fig. 1) (Rensing et al., 2008; Devos et al., 2016). A horsetail and lycophytes contain two or three NMCPs that probably were created by WGDs in their lineages (Vanneste et al., 2015; Clark et al., 2016).
Based on genomic data and NMCP phylogeny, we conclude that three NMCP homologues were created in ancient WGDs and posteriorly retained in genomes. A correlation was noted between the long-term survival of WGDs and decisive moments in plant evolution, for example, in the origin and diversification of flowering plants, the transition from woodland to grassland, and mass extinctions (Van de Peer et al., 2017).
The divergence into NMCP1 and NMCP2 proteins could have taken place in one of the two ancient polyploidyzation events (ζ during the evolution of seed plants and ε during the evolution of flowering plants) (Jiao et al., 2011). Taking into account that gymnosperms contain two NMCP homologues, we investigated the possibility that the NMCP1-type and NMCP2-type proteins diverged after the first WGD and were later retained in all seed plants. The fact that G. biloba contains two NMCP homologues that cluster with other gymnosperm NMCP1 and NMCP2 separately seems to support this hypothesis. There is evidence of two WGD events in Pinaceae and cupressophytes (including Taxus sp.) that did not involve G. biloba (Li et al., 2015). Although another study (Guan et al., 2016) suggests that G. biloba could have undergone another round of WGD, the topology of our phylogenetic analysis suggests that gymnosperm NMCP1 and NMCP2 were created in a WGD shared by all gymnosperms, which is probably the ζ WGD event. In contrast, Ruprecht et al. (2017), after revision of phylogenetic data and methodology, shed some doubt about the evidence for two separate WGDs (ζ and ε), and advise caution when interpreting their analysis.
Our results seem to support the hypothesis that NMCP1 and NMCP2 diverged in a WGD shared by all seed plants (ζ). In addition, we propose that if there was another round of WGD in flowering plants (ε), the duplicates were lost over the time, as a basal angiosperm (A. trichopoda) and most monocots contain only one NMCP of each type.
The appearance of NMCP3 coincides with the γ WGD in Gunneridae, which was not shared by N. nucifera and Aquilegia sp. (Van de Peer et al., 2017; Ren et al., 2018), neither of which contain NMCP3 (Fig. 1; Supplementary Fig. S1).
The appearance of the third NMCP1-type protein, a homologue of Arabidopsis thaliana CRWN2/CRWN3, coincides with two duplication events (α and β) in the Brassicaceae (mustard family) not shared by Carica papaya (Ren et al., 2018) (Fig. 2; Supplementary Fig. S1). These WGD events were concurrent with the wave of WGDs that occurred close to the Cretaceous–Paleogene boundary, which was marked by a number of cataclysmic events that resulted in major climate change and led to the extinction of 60–70% of plant and animal species (Vanneste et al., 2014; Van de Peer et al., 2017).
We found a number of angiosperm species that, apart from the ancient WGDs, have recently undergone a recent duplication event, for example, M. acuminata (D’Hont et al., 2012), P. dactylifera (Al-Mssallem et al., 2013), E. guineensis (Singh et al., 2013), D. catenatum (Zhang et al., 2016), Aquilegia (Van de Peer et al., 2017), N. nucifera (Ming et al., 2013), carrot (Daucus carota; Iorizzo et al., 2016), tomato and potato (Solanum sp.; Sato et al., 2012), M. esculenta (Bredeson et al., 2016), Brassica rapa (Wang et al., 2011a), G. max (Blanc and Wolfe, 2004; Schmutz et al., 2010), Gossypium raimondii (Blanc and Wolfe, 2004; Wang et al., 2012), and M. domestica (Velasco et al., 2010), and contain multiple NMCP1-type proteins.
Some species have retained a higher number of NMCP genes of one type than of the other. The favoured type is usually NMCP1, as is the case in D. catenatum, Panicum virgatum, D. carota, M. acuminata, B. rapa, and A. coerulea. In contrast, just a few duplicated NMCP2 genes were preferentially retained (in A. comosus, which underwent σ WGD, and M. esculenta) (Ming et al., 2015). This suggests that NMCP2 is under higher evolutionary (selective) pressure to return to a single copy, as was observed for angiosperm core gene families (Li et al., 2016). In spite of this, a few species seem to have lost all duplicated genes quickly after a WGD event, such as S. polyrhiza (Wang et al., 2014), maize (Zea mays; Schnable et al., 2009), poplar (Populus trichocarpa; Tuskan et al., 2006), and Eucalyptus grandis (Myburg et al., 2014).
A variation in gene retention has been reported before, as in G. max, which retains almost four times as many duplicated gene pairs as Z. mays from recent WGD events that happened at similar times (Liang and Schnable, 2018).
Retention and expression of the duplicated NMCPs
The NMCP family follows the general trend in evolution according to which, during the process of diploidization, genes duplicated in recent WGDs are retained for some time and then lost (Panchy et al., 2016; van de Peer et al., 2017). Species with young polyploids contain a higher number of NMCP genes, such as S. fallax (Devos et al., 2016), E. guineensis (Singh et al., 2013), P. dactylifera (Al-Mssallem et al., 2013), Aquilegia (Van de Peer et al., 2017), B. rapa (Wang et al., 2011a), G. max (Blanc and Wolfe, 2004; Schmutz et al., 2010), G. raimondii (Blanc and Wolfe, 2004; Wang et al., 2012), M. domestica (Velasco et al., 2010), and M. acuminata (D’Hont et al., 2012). The fact that the duplicate is still present in the genome does not necessarily mean that the protein is expressed. Immediately after WGD events, high chromosomal instability is observed (Soltis et al., 2015), and epigenetic modifications are induced to avoid extreme changes in the transcriptome of the polyploid (del Pozo and Ramirez-Parra, 2015). In fact, at least in autotetraploids, the transcriptome is only slightly modified in comparison to the diploids (Stupar et al., 2007; Riddle et al., 2010; Allario et al., 2011; del Pozo and Ramirez-Parra, 2014). Shortly afterwards, a diploidization process begins, which includes loss of duplicate genes, chromosomes, and/or repetitive DNA and gene silencing to return to the diploid genome (Soltis et al., 2015). During the diploidization process, one copy of the duplicated gene is usually lost from the genome through short and medium-sized sequence deletions resulting from non-homologous recombination (Woodhouse et al., 2014). In addition, duplicates inserted into another chromosome are rarely transcribed and translated into proteins due to genomic rearrangements (Walley et al., 2016).
We observed a higher number of NMCPs in comparison to lamins due to recent WGD events, as it is generally recognized that WGD events are much more common in plants than in animals (Mable, 2003; Wertheim et al., 2013). In addition, duplicates from WGDs are retained in angiosperms for a longer time than in mammals (Ren et al., 2018). Up to 35% of vascular plant species are recent polyploids that have not completed the process of diploidization (Mayrose et al., 2011; Barker et al., 2016; Van de Peer et al., 2017). Therefore, there is a high probability that many of the NMCP genes are not expressed and will be lost over time (Panchy et al., 2016).
We investigated the expression of NMCPs on the eFP browser for the following species: A. thaliana, Eutrema salsugineum, G. max, Z. mays, Oryza sativa, Brachypodium distachyon, and P. patens. We found expression data for most NMCPs in these species, with exception of two of the six G. max homologues (Gma3I and Gma2I), suggesting that a single homologue of NMCP2 and NMCP3 proteins is expressed in soybean. In P. patens, both Ppa1 and Ppa2 NMCP genes are expressed and show clear differences in expression patterns (see Supplementary Dataset S2). In G. max, some duplicates (Gma1I and Gma1II) seem to be expressed. We have not found expression data for more polyploid species.
In A. thaliana autotetraploids, genes that are included in gene ontogeny categories of abiotic and biotic stimuli and stress responses are expressed at higher levels than in diploid A. thaliana (Ng et al., 2012), and the levels of transcription of the duplicated genes involved in stress responses seem to depend on environmental conditions (del Pozo and Ramirez-Parra, 2014). As a rule, genes that provide an advantage in adaptation to new environmental conditions, such as better salt tolerance, higher photosynthetic rates, or better resistance to drought and other abiotic stresses, are more often retained in the genome and transcribed (del Pozo and Ramirez-Parra, 2015), as is the case for CRWN1, an A. thaliana homologue of Gma1 involved in plant defence (Guo et al., 2017).
Predicted structure of CCDs and conserved regions
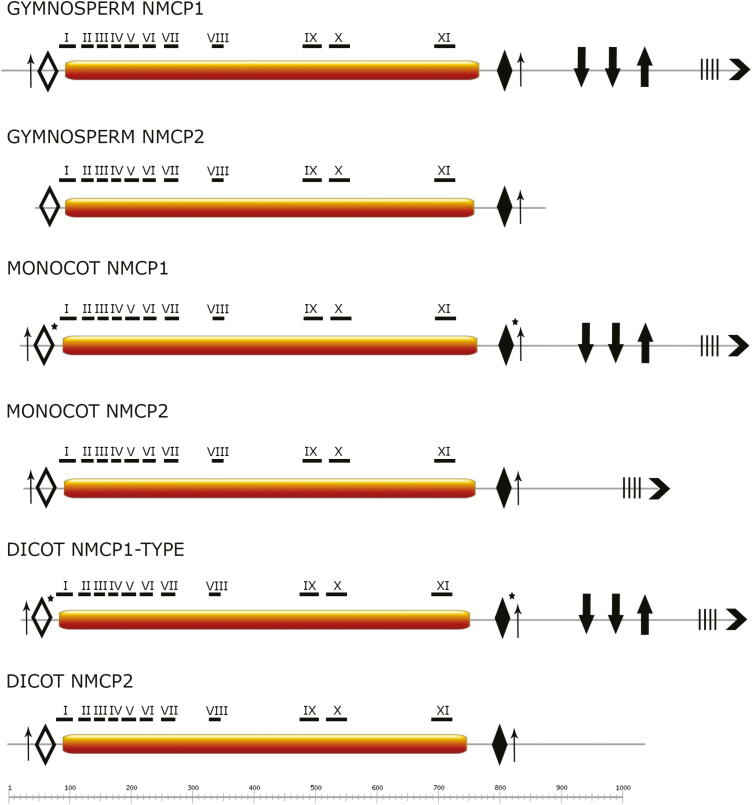
NMCP proteins present a tripartite domain layout, similar to metazoan lamins (Ciska and Moreno Diaz de la Espina, 2013). All obtained protein sequences, including those from basal species, are predicted to contain a central CCD flanked by a short head and a longer tail domains (Figs 3 and 4). The CCDs of all proteins are predicted to dimerize. In a few basal NMCPs, short coiled-coil regions (28–30 amino acids) were also predicted in the tail domain (Fig. 4). We grouped the sequences based on NMCP classification and phylogenetic relationship and searched for conserved features such as total protein length, length of each domain, and the presence of conserved motifs for each group. Mean ±SD values of the total length of the different proteins and of the three domains for each group in Magnoliophyta are included in Table 1.
Fig. 3.
Comparison of NMCP domain layout and the position of conserved motifs across the Spermatophyta. NMCPs present a tripartite domain layout, with a central CCD of highly conserved length (represented by bars) flanked by a short head domain and a longer tail domain. The tail domain is generally longer in NMCP1 than in NMCP2 proteins. Highly conserved motifs I–XI (MEME search) are located along the CCD and concentrated at the N-terminal end; motif I is characteristic for all NMCPs with the exception of a few algal sequences. The head and tail domains also contain highly conserved motifs specific to the NMCP family. Empty diamonds, GGLDEESLERKDRAALJAYI (motif 11 in Supplementary Fig. S2); filled diamonds, SWLRKCASKIF (motif 5 in Supplementary Fig. S2); thin upward arrows, phosphorylation sites; thick downward arrows, nuclear localization signal; thick upward arrows, QTPGEKRYNLRRSTIVNTVA (motif 14 in Fig. S2); four vertical bars, stretch of acidic amino acids; chevron arrows at C-terminus, EDEEPGEASIGKKLWNFLTT (motif 4 in Supplementary Fig. S2). Asterisks indicate conserved regions that were present in most but not all NMCPs analysed. Dicot NMCP1-type includes NMCP1 and NMCP3. (This figure is available in colour at JXB online.)
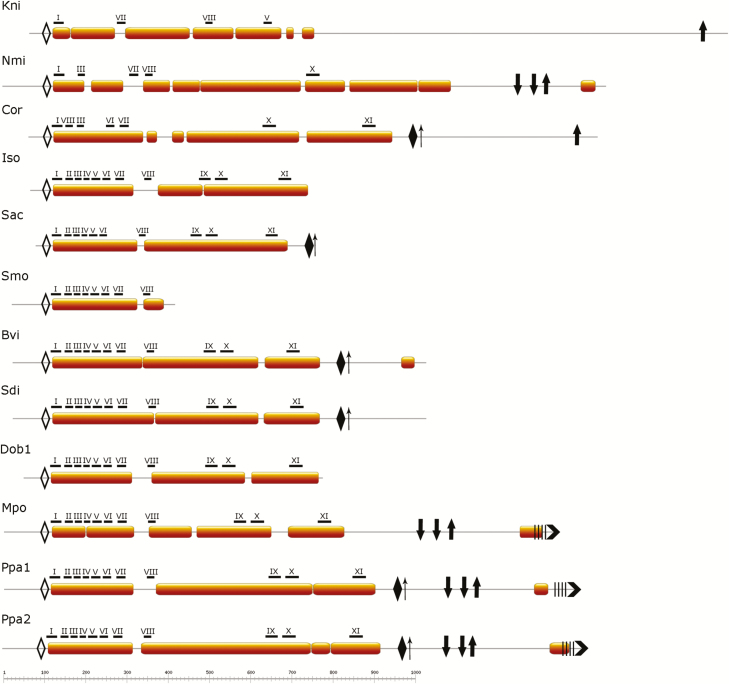
Fig. 4.
Comparison of NMCP domain layout and positions of conserved motifs in selected basal NMCPs. Basal NMCPs also present a tripartite domain layout, with a central CCD (represented by bars) of conserved length flanked by a short head domain and a longer tail domain. Highly conserved motifs I–XI (MEME search) are located along the CCD and concentrated at the N-terminal end. The head and tail domains also contain highly conserved motifs specific for the NMCP family. Empty diamonds, GGLDEESLERKDRAALJAYI (motif 11 in Supplementary Fig. S2); filled diamonds, SWLRKCASKIF (motif 5 in Supplementary Fig. S2); thin upward arrows, phosphorylation sites; thick downward arrows, nuclear localization signal; thick upward arrows, QTPGEKRYNLRRSTIVNTVA (motif 14 in Supplementary Fig. S2); four vertical bars, stretch of acidic amino acids; chevron arrows at C-terminus, EDEEPGEASIGKKLWNFLTT (motif 4 in Supplementary Fig. S2). Kni, Klebsormidium nitens; Nmi, Nitella mirabilis; Cor, Coleochaete orbicularis; Iso, Isoetes sp.; Sac, Selaginella acanthonota; Smo, Selaginella moellendorffii; Bvi, Botrypus virginianus; Sdi, Sceptridium dissectum; Dob, Dendrolycopodium obscurum; Mpo, Marchantia polymorpha; Ppa, Physcomitrella patens. (This figure is available in colour at JXB online.)
Table 1.
Total protein length and lengths of the three protein domains in NMCP families from Magnoliophyta
| Total length | Head | CCD | Tail | |
|---|---|---|---|---|
| Gymnosperm NMCP1-type | 1203±72 aa | 106±17 aa | 673±9 aa | 430±72 aa |
| Gymnosperm NMCP2-type | 832±155 aa | 51±23 aa | 665±30 aa | 60–463 aa |
| Monocot NMCP1-type | 1171±75 aa | 72±35 aa | 674±20 aa | 463±91 aa |
| Monocot NMCP2-type | 1034±77 aa | 68±18 aa | 669±16 aa | 297±70 aa |
| Dicot NMCP1-type | 1169±49 aa | 64±24 aa | 668±16 aa | 435±34 aa |
| Dicot NMCP2-type | 1039±60 aa | 92±30 aa | 657±40 aa | 285±60 aa |
The corresponding mean ±SD values are given for each NMCP-type. NMCP1-type proteins are generally longer than NMCP2-type due to the longer tail domain. aa, Amino acids; CCD, coiled-coil rod domain.
In seed plants, the length of the central CCD of NMCPs is highly conserved (Fig. 3, Table 1), in agreement with our previous observation for angiosperms (Ciska et al., 2018). This feature is crucial in the process of polymerization in higher-order structures (oligo- and polymers), which are essential for the formation and functionality of the lamina (Surkont et al., 2015). Interestingly, in NMCP1 and NMCP2 proteins the total length of the protein and the length of the tail domain are also conserved in angiosperms and gymnosperms, with the exception of a few NMCP2 gymnosperm sequences that seem to be lacking a fragment of the C-terminal end (Rmi2, Fho2; Cde2, Gbi2).
The mosses P. patens and S. fallax, as well as M. polymorpha, have a longer amino acid chain, owing to a longer CCD (between 752 and 888 amino acids) than the typical CCD found in seed plants. Ferns and a horsetail (E. giganteum) also have a slightly longer CCD (714–730 amino acids), with the exception of Egi3, which seems to lack a fragment at the C-terminal end. The homologues in the clubmosses and spikemosses seem to have a variable amino acid chain length (449–846 amino acids), while a variable CCD length (423–730 amino acids) was observed for clubmosses; in spikemosses, a total length of 348–745 amino acids, and CDD length of 292–696 amino acids, is present (Fig. 4).
We investigated the presence of conserved regions in NMCPs using the MEME suite and identified several conserved motifs characterizing the protein family, some of which have previously reported in angiosperms (Ciska et al., 2013). For the general NMCP search we selected 51 NMCP sequences comprising all phylogenetic groups (including algae), which is the maximum number allowed by the MEME suite. The conserved regions were distributed along the whole protein sequences, although the N-terminal end of the CCD and some motifs in close proximity to the CCD in the head and tail domains exhibited especially high conservation in all NMCPs (Figs 3 and 4; Table 2; Supplementary Fig. S2). The conserved motifs I–XI are located along the CCD but are concentrated at the N-terminal end. Motif I (ELYDYQYNMGLLLIEKKEWT) is conserved in all NMCPs and was previously reported to be essential for the localization of DcNMCP1 in the nuclear periphery (Kimura et al., 2014).
Table 2.
Regions conserved among the NMCP protein family
| Pattern | e-value | Domain | |
|---|---|---|---|
| 1. | ELYDYQYNMGLLLIEKKEWT | 1.1e-589 | CCD (I) |
| 2. | ALGVEKQCVADLEKALKEMR | 2.2e-428 | CCD (III) |
| 3. | ELKQEKEKFEKEWELLDEKR | 1.6e-361 | CCD (X) |
| 4. | EDEEPGEASIGKKLWNFLTT | 8.2e-216 | Tail |
| 5. | SWLRKCASKIF | 2.1e-184 | Tail |
| 6. | EREELLRLQSELKZEIDELR | 2.1e-262 | CCD (IX) |
| 7. | LKREQAAHLIALSEAEKREE | 1.7e-274 | CCD (II) |
| 8. | RKLQEVEAREDALRRERLSF | 4.1e-266 | CCD (VI) |
| 9. | WEKKLQEGZERLLEGQRJLN | 1.2e-247 | CCD (VII) |
| 10. | IKKDIEELXVQREKLKEQRE | 1.2e-257 | CCD (XI) |
| 11. | GGLDEESLERKDRAALJAYI | 1.4e-239 | Head |
| 12. | AEXAEVKVTAESKLAZAREL | 1.8e-220S | CCD (IV) |
| 13. | LEAEAKLHAAEALLAEASRK | 1.5e-194 | CCD (V) |
| 14. | QTPGEKRYNLRRSTIVNTVA | 2.5e-184 | Tail |
| 15. | LDKKEQELLLLZEKLASRER | 1.9e-158 | CCD (VIII) |
The regions are ordered from the highest statistical significance of the motif to the lowest (i.e. from the lowest to the highest e-value). The patterns were generated using the MEME standard protein alphabet (standard amino acid symbols; ambiguous symbols: B=N or D; Z=Q or E; J=L or I; X=any amino acid). Bold letters indicate the most conserved residues. The location in the head, CCD (the roman numbers represent position in the CCD; see Fig. 3), or tail domain is given. The conserved motifs in the form of logos are provided in Supplementary Fig. S3.
The head and tail domains also contain highly conserved motifs specific for the NMCP family, mostly located within the longer tail domain. The short head domain contains a highly conserved motif close to the CCD (GGLDEESLERKDRAALJAYI). Conserved motif 5 (SWLRKCASKIF) (Table 2) is present in the tail domain of almost all NMCP proteins, apart from several NMCP3 proteins (including CRWN3), several monocot NMCP1-type proteins (AcNMCP1, in cereals), and M. polymorpha NMCP (Supplementary Fig. S2). This highly conserved short region seems to be essential for the localization of CRWN1 to the nuclear periphery (Guo et al., 2017).
We also identified blocks of conserved motifs in the tail domain that always appear in association. These include one motif in the conserved C-terminal end (EDEEPGEASIGKKLWNFLTT), which is present in all NMCP1-type proteins (including those of gymnosperms and also in most monocot NMCP2-type proteins) and is always preceded by a stretch of acidic amino acids. The QTPGEKRYNLRRSTIVNTVA region, which was also reported to be involved in localization to the nuclear periphery (Kimura et al., 2014), is preceded by two nuclear localization motifs. Gymnosperm and dicot NMCP2-type proteins have shorter tail domains and lack these two conserved regions, with the exception of Pinus taeda and Pinus silvestris NMCP2s, which contain a full-length tail domain, and P. abies NMCP2, which contains the QTPGEKRYNLRRSTIVNTVA region but not the conserved C-terminal end (Fig. 3).
In addition, we identified short minimal consensus phosphorylation motifs Ser/Thr-Pro (S/T-P) for cyclin-dependent kinases (Errico et al., 2010) at the N-terminal end of all NMCPs, with the exception of gymnosperm NMCP2, and an additional CDK site located after the conserved SWLRKCASKIF region.
Although the Charophyta sequences do not present the typical layout of conserved motifs, the MEME search identified several conserved motifs in each sequence (see Fig. 4), including GGLDEESLERKDRAALJAYI in the head and QTPGEKRYNLRRSTIVNTVA in the tail domains. As for the NMCP members in mosses, ferns, and horsetail, many sequences seem to lack the N- or C-terminal end fragments and related conserved regions, although P. patens NMCPs contain the full set of conserved motifs identified by the MEME suite (Fig. 4).
Although in some phylogenetic groups NMCPs seem to have diverged from the typical layout, it is clear that there is a set of conserved regions characterizing the NMCP family that is present from the most basal Charophyta to evolved angiosperms (see Figs 3 and 4). These conserved domains would play important roles in the fundamental functions performed by NMCPs, not only supporting polymerization for the formation of a functional lamina, but also in protein interactions. The latter would facilitate chromatin organization and gene expression in key cellular processes controlled by NMCPs such as germination and plant immunity (Zhao et al., 2016; Guo et al., 2017). This analysis deserves further investigation in the future.
Immunofluorescence localization of P. patens NMCP1 to the lamina
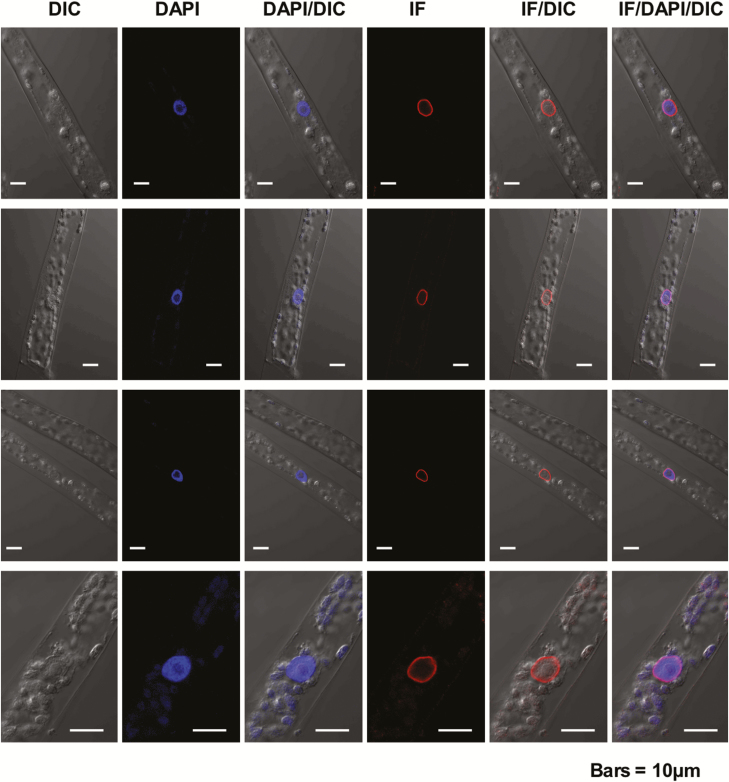
To investigate whether conservation of the predicted secondary structure and conserved domains in basal NMCPs reflects a conserved functionality, we produced an antibody to the NMCP1 protein of the bryophyte P. patens and investigated the localization of the endogenous protein by confocal microscopy. Western blot analysis showed that the antibody is specific for this protein and does not react with PpNMCP2 (Supplementary Fig. S3). The immunofluorescence images revealed that, as reported for NMCPs from angiosperms (Masuda et al., 1997; Ciska et al., 2013; Sakamoto and Takagi, 2013; Ciska et al., 2018), the protein accumulates underneath the nuclear envelope wrapping chromatin (Fig. 5). This localization suggests that, in spite of the different length and organization of the CCD relative to angiosperm NMCPs, this basal NMCP is also attached to the nuclear membrane. These data, together with the conservation of predicted secondary structures, further suggest a functional conservation in basal NMCPs.
Fig. 5.
Localization of Physcomitrella patens NMCP in protonema cells. Confocal sections of four representative protonemata after staining with the anti-PpaNMCP1 antibody, showing the distribution of PpaNMCP1, chromatin staining with DAPI, and the corresponding differential interference contrast (DIC) images. Overlays of the PpaNMCP1 and DAPI stainings and the corresponding DIC images are also shown. PpaNMCP1 localizes at the nuclear periphery wrapping chromatin. Bars=10 µm. (This figure is available in colour at JXB online.)
Conclusions
Our phylogenetic, structural, and specific domain conservation results confirm the origin of the NMCP family of proteins as early as in Charophyte algae, the basal Streptophyta. This origin is placed after branching from Chlorophyta according to the phylogenetic results of Koreny and Field (2016), and updates a previous report establishing the origin of these proteins in mosses (Poulet et al., 2017). Charophyte algae have a single basal NMCP protein, and ancient WGD events resulted in the creation of new homologues of NMCPs during evolution. The WGD of seed plants is in the origin of NMCP1- and NMCP2-type proteins in Spermatophyta, and the WGD of Gunneridae originates the NMCP3 type. Recent WGDs are quite common in plants and result in a higher number of NMCP homologues in certain species; nevertheless, their expression and retention in the genome depends on genomic rearrangements and the functions they fulfil, with the NMCP2 proteins being under higher evolutionary pressure than NMCP1s.
There is a set of structural features characterizing the NMCP family: a tripartite structure with a highly conserved CCD, a short head with a highly conserved region, and a long tail domain with several highly conserved regions. Some of these regions are involved in the proteins’ association to the nuclear periphery, while others may be binding sites for interacting proteins. The regions conserved from Charophyta to angiosperms play fundamental roles in the different functions performed by NMCPs, not only supporting polymerization for the formation of a functional lamina, but also mediating protein interactions facilitating chromatin organization and expression in cellular processes such as germination and plant immunity (Zhao et al., 2016; Guo et al., 2017). The localization of a basal NMCP at the nuclear periphery, along with the conservation of its predicted secondary structure, suggests conservation of functions from basal to more evolved NMCPs, and would date the origin of the NMCP-based lamina at least at the mosses.
Supplementary data
Supplementary data are available at JXB online.
Dataset S1. Accession data and sequences used for the phylogenetic analyses.
Dataset S2. Expression data of different NMCPs from the eFP browser.
Fig. S1. Phylogenetic tree of NMCP proteins (extended version with transcriptomic data).
Fig. S2. Regions conserved in NMCPs generated by MEME search presented as logos.
Fig. S3. Validation of target specificity of the anti-PpNMCP1 antibody using western blot analysis.
Acknowledgements
We thank Dr Fujita, Faculty of Science, Hokkaido University, for kindly providing us with P. patens plants. This work was supported by the Spanish Ministry of Science and Innovation (BFU2010-15900) and CSIC (PIE 201020E019).
References
- Al-Mssallem IS, Hu S, Zhang X, et al. . 2013. Genome sequence of the date palm Phoenix dactylifera L. Nature Communications 4, 2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allario T, Brumos J, Colmenero-Flores JM, Tadeo F, Froelicher Y, Talon M, Navarro L, Ollitrault P, Morillon R. 2011. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. Journal of Experimental Botany 62, 2507–2519. [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ. 1977. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Molecular Genetics and Genomics 154, 87–95. [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, et al. . 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytologist 210, 391–398. [DOI] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME. 2009. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biology and Evolution 1, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsios P, Peter T, Baumann O, Stick R, Meyer I, Gräf R. 2012. A lamin in lower eukaryotes? Nucleus 3, 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B. 2009. Streptophyte algae and the origin of embryophytes. Annals of Botany 103, 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, et al. . 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15. [DOI] [PubMed] [Google Scholar]
- Bredeson JV, Lyons JB, Prochnik SE, et al. . 2016. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotechnology 34, 562–570. [DOI] [PubMed] [Google Scholar]
- Ciska M, Masuda K, Moreno Díaz de la Espina S. 2013. Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. Journal of Experimental Botany 64, 1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciska M, Masuda K, Moreno Díaz de la Espina S. 2018. Characterization of the lamin analogue NMCP2 in the monocot Allium cepa. Chromosoma 127, 103–113. [DOI] [PubMed] [Google Scholar]
- Ciska M, Moreno Diaz de la Espina S. 2013. NMCP/LINC proteins: putative lamin analogs in plants? Plant Signaling & Behavior 8, e26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciska M, Moreno Díaz de la Espina S. 2014. The intriguing plant nuclear lamina. Frontiers in Plant Science 5, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Hidalgo O, Pellicer J, et al. . 2016. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytologist 210, 1072–1082. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury JM, et al. . 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. [DOI] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. 2014. Broken nuclei–lamins, nuclear mechanics, and disease. Trends in Cell Biology 24, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Li Z, Van de Peer Y, Ingvarsson PK. 2017. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Molecular Biology and Evolution 34, 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Ramirez-Parra E. 2014. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant, Cell & Environment 37, 2722–2737. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Ramirez-Parra E. 2015. Whole genome duplications in plants: an overview from Arabidopsis. Journal of Experimental Botany 66, 6991–7003. [DOI] [PubMed] [Google Scholar]
- Delorenzi M, Speed T. 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Cooper ED. 2015. The evolutionary origin of a terrestrial flora. Current Biology 25, R899–R910. [DOI] [PubMed] [Google Scholar]
- Devos N, Szövényi P, Weston DJ, Rothfels CJ, Johnson MG, Shaw AJ. 2016. Analyses of transcriptome sequences reveal multiple ancient large-scale duplication events in the ancestor of Sphagnopsida (Bryophyta). New Phytologist 211, 300–318. [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. 2007. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. The Plant Cell 19, 2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T. 2010. Identification of substrates for cyclin dependent kinases. Advances in Enzyme Regulation 50, 375–399. [DOI] [PubMed] [Google Scholar]
- Fiserova J, Goldberg MW. 2010. Relationships at the nuclear envelope: lamins and nuclear pore complexes in animals and plants. Biochemical Society Transactions 38, 829–831. [DOI] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW. 2009. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. The Plant Journal 59, 243–255. [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Huttenlauch I, Hutchison CJ, Stick R. 2008. Filaments made from A- and B-type lamins differ in structure and organization. Journal of Cell Science 121, 215–225. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. . 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto C, Tamura K, Fukao Y, Shimada T, Hara-Nishimura I. 2014. The novel nuclear envelope protein KAKU4 modulates nuclear morphology in Arabidopsis. The Plant Cell 26, 2143–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K. 2014. Evidence for LINC1-SUN associations at the plant nuclear periphery. PLoS One 9, e93406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. 2015. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annual Review of Biochemistry 84, 131–164. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Medalia O. 2015. Lamins: the structure and protein complexes. Current Opinion in Cell Biology 32, 7–12. [DOI] [PubMed] [Google Scholar]
- Guan R, Zhao Y, Zhang H, et al. . 2016. Draft genome of the living fossil Ginkgo biloba. Gigascience 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Mao X, Zhang H, Zhang Y, Fu M, Sun Z, Kuai P, Lou Y, Fang Y. 2017. Lamin-like proteins negatively regulate plant immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Molecular Plant 10, 1334–1348. [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, et al. . 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications 5, 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorizzo M, Ellison S, Senalik D, et al. . 2016. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nature Genetics 48, 657–666. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. . 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Fujino K, Ogawa K, Masuda K. 2014. Localization of Daucus carota NMCP1 to the nuclear periphery: the role of the N-terminal region and an NLS-linked sequence motif, RYNLRR, in the tail domain. Frontiers in Plant Science 5, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Kuroda C, Masuda K. 2010. Differential nuclear envelope assembly at the end of mitosis in suspension-cultured Apium graveolens cells. Chromosoma 119, 195–204. [DOI] [PubMed] [Google Scholar]
- Kollmar M. 2015. Polyphyly of nuclear lamin genes indicates an early eukaryotic origin of the metazoan-type intermediate filament proteins. Scientific Reports 5, 10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koreny L, Field MC. 2016. Ancient eukaryotic origin and evolutionary plasticity of nuclear lamina. Genome Biology and Evolution 8, 2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, Meyer I, Gräf R. 2012. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Molecular Biology of the Cell 23, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Baniaga AE, Sessa EB, Scascitelli M, Graham SW, Rieseberg LH, Barker MS. 2015. Early genome duplications in conifers and other seed plants. Science Advances 1, e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Defoort J, Tasdighian S, Maere S, Van de Peer Y, De Smet R. 2016. Gene duplicability of core genes is highly consistent across all angiosperms. The Plant Cell 28, 326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Schnable JC. 2018. Functional divergence between subgenomes and gene pairs after whole genome duplications. Molecular Plant 11, 388–397. [DOI] [PubMed] [Google Scholar]
- Mable BK. 2003. Breaking down taxonomic barriers in polyploidy research. Trends in Plant Science 8, 582–590. [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. 2005. Modeling gene and genome duplications in eukaryotes. Proceedings of the National Academy of Sciences, USA 102, 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M. 1997. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Experimental Cell Research 232, 173–181. [DOI] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, et al. . 2014. Data access for the 1000 plants (1KP) project. Gigascience 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP. 2011. Recently formed polyploid plants diversify at lower rates. Science 333, 1257. [DOI] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Liu Y, et al. . 2013. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biology 14, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Wai CM, et al. . 2015. The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics 47, 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mínguez A, Moreno Díaz de la Espina S. 1993. Immunological characterization of lamins in the nuclear matrix of onion cells. Journal of Cell Science 106, 431–439. [DOI] [PubMed] [Google Scholar]
- Mochizuki R, Tsugama D, Yamazaki M, Fujino K, Masuda K. 2017. Identification of candidates for interacting partners of the tail domain of DcNMCP1, a major component of the Daucus carota nuclear lamina-like structure. Nucleus 8, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Diaz de la Espina S, Barthellemy I, Cerezuela MA. 1991. Isolation and ultrastructural characterization of the residual nuclear matrix in a plant cell system. Chromosoma 100, 110–117. [Google Scholar]
- Myburg AA, Grattapaglia D, Tuskan GA, et al. . 2014. The genome of Eucalyptus grandis. Nature 510, 356–362. [DOI] [PubMed] [Google Scholar]
- Ng DW, Zhang C, Miller M, Shen Z, Briggs SP, Chen ZJ. 2012. Proteomic divergence in Arabidopsis autopolyploids and allopolyploids and their progenitors. Heredity 108, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M. 2000. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Research 7, 9–17. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171, 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar V, Poulet A, Détourné G, Tatout C, Vanrobays E, Evans DE, Graumann K. 2016. A novel family of plant nuclear envelope-associated proteins. Journal of Experimental Botany 67, 5699–5710. [DOI] [PubMed] [Google Scholar]
- Poulet A, Probst AV, Graumann K, Tatout C, Evans D. 2017. Exploring the evolution of the proteins of the plant nuclear envelope. Nucleus 8, 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Research 43, D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Wang H, Guo C, Zhang N, Zeng L, Chen Y, Ma H, Qi J. 2018. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Molecular Plant 11, 414–428. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. . 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. [DOI] [PubMed] [Google Scholar]
- Riddle NC, Jiang H, An L, Doerge RW, Birchler JA. 2010. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theoretical and Applied Genetics 120, 341–353. [DOI] [PubMed] [Google Scholar]
- Ruprecht C, Lohaus R, Vanneste K, Mutwil M, Nikoloski Z, Van de Peer Y, Persson S. 2017. Revisiting ancestral polyploidy in plants. Science Advances 3, e1603195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Takagi S. 2013. LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant & Cell Physiology 54, 622–633. [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, et al. . 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. . 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. . 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Singh R, Ong-Abdullah M, Low ET, et al. . 2013. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 500, 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. 2015. Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development 35, 119–125. [DOI] [PubMed] [Google Scholar]
- Stupar RM, Bhaskar PB, Yandell BS, et al. . 2007. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 176, 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkont J, Diekmann Y, Ryder PV, Pereira-Leal JB. 2015. Coiled-coil length: size does matter. Proteins 83, 2162–2169. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. 2008. Synteny and collinearity in plant genomes. Science 320, 486–488. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH. 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proceedings of the National Academy of Sciences, USA 107, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O. 2017. The molecular architecture of lamins in somatic cells. Nature 543, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. . 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K. 2017. The evolutionary significance of polyploidy. Nature Reviews Genetics 18, 411–424. [DOI] [PubMed] [Google Scholar]
- van Zanten M, Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, Koornneef M, Fransz P, Soppe WJ. 2011. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proceedings of the National Academy of Sciences, USA 108, 20219–20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Research 24, 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Sterck L, Myburg AA, Van de Peer Y, Mizrachi E. 2015. Horsetails are ancient polyploids: evidence from Equisetum giganteum. The Plant Cell 27, 1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, et al. . 2010. The genome of the domesticated apple (Malus × domestica Borkh.). Nature Genetics 42, 833–839. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. 2000. The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Walley JW, Sartor RC, Shen Z, et al. . 2016. Integration of omic networks in a developmental atlas of maize. Science 353, 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dittmer TA, Richards EJ. 2013. Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biology 13, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, et al. . 2012. The draft genome of a diploid cotton Gossypium raimondii. Nature Genetics 44, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Wang W, Haberer G, Gundlach H, et al. . 2014. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nature Communications 5, 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. . 2011a. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Tang H, Tan X, Ficklin SP, Feltus FA, Paterson AH. 2011b. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS One 6, e28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim B, Beukeboom LW, van de Zande L. 2013. Polyploidy in animals: effects of gene expression on sex determination, evolution and ecology. Cytogenetic and Genome Research 140, 256–269. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proceedings of the National Academy of Sciences, USA 111, 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GQ, Xu Q, Bian C, et al. . 2016. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Scientific Reports 6, 19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Guan C, Feng J, Liang Y, Zhan N, Zuo J, Ren B. 2016. The Arabidopsis CROWDED NUCLEI genes regulate seed germination by modulating degradation of ABI5 protein. Journal of Integrative Plant Biology 58, 669–678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.