Abstract
Individuals with germline PTEN tumor-suppressor variants have PTEN hamartoma tumor syndrome (PHTS). Clinically, PHTS has variable presentations; there are distinct subsets of PHTS-affected individuals, such as those diagnosed with autism spectrum disorder (ASD) or cancer. It remains unclear why mutations in one gene can lead to such seemingly disparate phenotypes. Therefore, we sought to determine whether it is possible to predict a given PHTS-affected individual’s a priori risk of ASD, cancer, or the co-occurrence of both phenotypes. By integrating network proximity analysis performed on the human interactome, molecular simulations, and residue-interaction networks, we demonstrate the role of conformational dynamics in the structural communication and long-range allosteric regulation of germline PTEN variants associated with ASD or cancer. We show that the PTEN interactome shares significant overlap with the ASD and cancer interactomes, providing network-based evidence that PTEN is a crucial player in the biology of both disorders. Importantly, this finding suggests that a germline PTEN variant might perturb the ASD or cancer networks differently, thus favoring one disease outcome at any one time. Furthermore, protein-dynamic structural-network analysis reveals small-world structural communication mediated by highly conserved functional residues and potential allosteric regulation of PTEN. We identified a salient structural-communication pathway that extends across the inter-domain interface for cancer-only mutations. In contrast, the structural-communication pathway is predominantly restricted to the phosphatase domain for ASD-only mutations. Our integrative approach supports the prediction and potential modulation of the relevant conformational states that influence structural communication and long-range perturbations associated with mutational effects that lead to PTEN-ASD or PTEN-cancer phenotypes.
Keywords: PTEN, germline PTEN mutations, PTEN interactome, molecular dynamics simulation, network proximity analysis, PTEN allostery, conformation dynamics
Introduction
Phosphatase and tensin homolog deleted on chromosome ten (PTEN [MIM: 601728]) is a multi-functional tumor-suppressor gene found to be mutated in the germline of individuals with Cowden syndrome (CS [MIM: 158350]) and related cancer-predisposition syndromes or somatically in a diverse range of solid tumors.1, 2, 3 Individuals, regardless of clinical presentation, with germline PTEN mutations are molecularly diagnosed with PTEN hamartoma tumor syndrome (PHTS). PHTS is typically an autosomal-dominant disorder variably characterized by macrocephaly, hamartomatous overgrowths, and malignant neoplasia, especially of the breast and thyroid.4, 5, 6 The phenotypically diverse clinical manifestations encompassed by PHTS often share overlapping clinical features but have great variability, making clinical outcomes difficult to predict. Paradoxically, approximately one-fifth of PHTS-affected individuals also present with autism spectrum disorder (ASD), raising an intriguing question about how two seemingly disparate clinical outcomes can result from germline mutations in one gene when there are no obvious genotype-phenotype associations.5, 7 Current clinical management guidelines assign enhanced clinical surveillance for component cancers in all individuals with PHTS and a low threshold to evaluate for ASD in children. It would enhance precision practice if clinicians could predict ahead of time which PHTS-affected individual would develop ASD only, cancer only, or both.
PTEN functions as a dual-specificity phosphatase that antagonizes both the PI3K/AKT/mTOR signaling pathway8, 9, 10 and the mitogen-activated protein kinase (MAPK) pathway.11 The critical ability of PTEN to catalyze the dephosphorylation of phosphatidylinositol (3,4,5)-triphosphate (PIP3) restrains pro-growth, pro-survival, and pro-proliferation signaling, guarding against tumorigenesis.10, 12, 13, 14 The regulation of the enzymatic activity and functional diversity of PTEN is complex and involves multiple cellular processes and interactions with other proteins. Hence, deficiency of any of these functions contributes to disease.15, 16, 17 In fact, PTEN structure-function analyses reveal distinctive functional patterns that correlate with variants found in the catalytic active site. For example, there are variants (e.g., p.Gly129Glu) that specifically disrupt the lipid phosphatase activity while leaving the protein phosphatase activity intact and vice versa (p.Tyr138Leu, for example). Additionally, there are variants (e.g., p.Cys124Ser) that completely disrupt the phosphatase core motif, leaving a lipid- and protein-phosphatase-dead protein; this is a mechanism by which PTEN is inactivated in tumors.13, 18, 19, 20 Furthermore, these variants result in specific alterations (i.e., expanding or contracting its depth) within the catalytic pocket, and they affect substrate preference.21
In a recent pilot study, we sought to identify differences in ASD- versus cancer-associated germline PTEN missense variants in silico by investigating putative structural and conformational dynamics.22 Five of six ASD-associated variants showed localized destabilization, contributing to the partial opening of the active site, whereas all six cancer-associated variants showed long-range perturbations that decreased structural stability and increased dynamics across the domain interface, mediating a closed active site.22 Most notable was the identification of an inter-domain disruption with an increase in dynamics across the phosphatase-C2 domain interface in PTEN, and this disruption was observed in both the c.388C>G (p.Arg130Gly) (cancer only) and c.517C>T (p.Arg173Cys) (shared in individuals with both ASD and cancer) mutations, indicating both residue positions play a role in inter-residue signal propagation and are crucial to structural stability.22, 23 These results provide evidence to support the recent identification of pivotal mutational-sites that might serve as key mediating bridges of allosteric communication in PTEN.24 Allosteric propagation results in communication between distinct sites in the protein structure and takes place through dynamic shifts of conformational ensembles.25 The quantitation of key factors that govern structural communication is essential in order to assess signal propagation for predicting the effects of mutations.25 Thus, our pilot observations encouraged further investigation in order to profile conformational changes that mediate long-range effects and allosteric signal propagation and thus reveal separate molecular features contributing specifically to ASD or cancer. Despite the specific nature of PTEN’s functional dynamics, mutation-induced conformational changes that are associated with ASD or cancer and that govern potential allosteric signaling pathways and structural-communication propagation have yet to be investigated at an atomistic level. It is, therefore, essential to determine how ASD- and cancer-associated variants that disrupt PTEN’s long-range communication and inter-domain dynamics give rise to a potential mutation-driven allosteric interface.
In this study, we combined molecular dynamics (MD)-based modeling of residue-interaction networks (RINs) and structural communication to further explore the effects that germline missense variants have on the dynamics of PTEN. We had a particular interest in changes that occur within the active site and the inter-domain interface. Understanding inter-residue characteristics while combining network behavior properties has proven to be a novel approach for studying interactions between proteins,26, 27 protein conformational changes,28 and allosteric communication.29, 30 Moreover, inter-residue network analysis is crucial for identifying functional residues.31 As such, here we investigate the relationship between germline PTEN variants that are involved in both ASD and cancer via network perturbation, subsequent structural-conformational dynamics, and altered inter-residue interactions that coordinate key functional centers of the protein (Figure S1).
Material and Methods
Human Protein-Protein Interactome Network
To build a comprehensive human protein-protein interactome, we assembled data from a total of 18 bioinformatics and systems-biology databases with multiple experimental pieces of evidence. Specifically, we focused on high-quality protein-protein interactions (PPIs) with five types of experimental evidence: (1) binary PPIs tested by high-throughput yeast-two-hybrid (Y2H) systems—we combined binary PPIs tested from two publicly available, high-quality Y2H datasets;32, 33 (2) kinase-substrate interactions in literature-derived low-throughput and high-throughput experiments; (3) literature-curated PPIs identified by affinity purification and then mass spectrometry (AP-MS), Y2H, and literature-derived low-throughput experiments; (4) high-quality PPIs from protein three-dimensional (3D) structures reported in Instruct;34 (5) and a signaling network made by literature-derived low-throughput experiments as annotated in SignaLink2.0.35 The genes were mapped to their Entrez ID on the basis of the NCBI database,36 as well as their official gene symbols based on the GeneCards database.37 Duplicated pairs were removed. In addition, all computationally predicted data, such as evolutionary analysis, gene-expression data, and metabolic associations, were excluded. The resulting updated human interactome that was used in this study includes 351,444 PPIs connecting 17,706 unique proteins. The detailed descriptions are provided in our previous studies.38, 39, 40
Selecting PTEN Germline Mutations for Residue Network and Molecular Dynamics Analyses
This prospective study was performed within the framework of the “Molecular Mechanisms Involved in Cancer Predisposition” protocol (Cleveland Clinic Institutional Review Board approved, IRB# 8458) and conducted with informed consent and in accordance with the World Medical Association Declaration of Helsinki. Germline PTEN missense variants from a series of 138 prospectively accrued individuals were compiled with a number of descriptive variables, which included clinical phenotypes, pathogenicity predictors, other genotype information, and predictors of structural stability. The dataset was evaluated by an in-house, random forest algorithm and further condensed with a principal component analysis (PCA), as described in Smith et al., 2018.22 By inspecting the PCA, we selected a total of 17 missense variants that segregated the most distantly from each other and were associated with ASD and cancer (Table 1). The accession numbers for the PTEN nucleotide and protein reference sequences reported in this paper are GenBank: NM_000314.7 and NP_000305.3, respectively.
Table 1.
Germline PTEN Missense Variants Analyzed
| Nucleotide Changea | PTEN Missense Variantsb |
|---|---|
| ASD-associated variants | |
| c.69A>C | p.Leu23Phe |
| c.194A>G | p.Tyr65Cys |
| c.202T>C | p.Tyr68His |
| c.302T>C | p.Ile101Thr |
| c.365T>G | p.Ile122Ser |
| c.658C>G | p.Leu220Val |
| Cancer-associated variants | |
| c.71A>G | p.Asp24Gly |
| c.275A>C | p.Asp92Ala |
| c.388C>G | p.Arg130Gly |
| c.401T>G | p.Met134Arg |
| c.613A>G | p.Met205Val |
| c.1033C>G | p.Leu345Val |
| ASD or cancer (shared) variants | |
| c.389G>A | p.Arg130Gln |
| c.406T>C | p.Cys136Arg |
| c.464A>G | p.Tyr155Cys |
| c.517C>T | p.Arg173Cys |
| ASD and cancer (co-existing) variants | |
| c.509G>T | p.Ser170Ile |
Reference sequence: GenBank: NM_000314.7
Reference sequence: GenBank: NP_000305.3
Network Proximity Analysis
We comprehensively accessed and assembled PTEN genetic and protein-protein interaction data from BioGRID.41 Collectively, we refer to these genes and proteins as PTEN influencers (Table S1). We collected significantly somatically mutated genes from The Cancer Genome Atlas (TCGA) projects, as described in the previous studies.38, 42 In addition, we collected known cancer-associated genes (germline-mutation-related) from four public databases: the Online Mendelian Inheritance in Man (OMIM) database,43 the HuGE Navigator,44 PharmGKB,45 and the Comparative Toxicogenomics Database (CTD),46 as described in our recent study.39
Here, we used a network proximity measure for quantifying network-based relationships between PTEN influencers and ASD-related genes or cancer genes from the human protein-protein interactome. Specifically, we calculated network proximity between PTEN influencers (A) and ASD-related genes or cancer genes (B) via:
| (1) |
where indicates the shortest distance between proteins within each gene set and , and and represent the shortest distance for proteins within and in the human interactome. To evaluate the significance of the network distance between PTEN influencers and ASD-related genes or cancer genes, we constructed a reference distance distribution corresponding to the expected distance between two randomly selected groups of proteins of the same size and degree (connectivity) distribution as the original PTEN influencers and disease genes (proteins) in the human interactome. This procedure was repeated 10,000 times, on the basis of previous studies.39, 47 We used the mean (μ) and SD (σ) of the reference distribution to calculate a Z score by converting an observed (non-Euclidean) distance to a normalized distance. In addition, a p value was computed on the basis of a permutation test.
Protein-Structure Model Preparation and Molecular Dynamics Simulations
The wild-type (WT) crystal structure of human PTEN protein (PDB: 1D5R) was obtained from the RCSB Protein Data Bank (PDB).12, 48 The tartrate (TLA) molecule was removed, and all calculations were conducted on apo PTEN. The overall three-dimensional structure of PTEN contains deleted segments that consist of unstructured or loosely folded regions of 7 and 49 residues at the N and C termini, respectively, and 24 residues in an intrinsically disordered internal loop (residues 286–309).12 Though only four PTEN mutations (c.3G>T [p.Met1?], c.892C>G [p.Gln298Glu], c.1061C>A [p.Pro354Gln], and c.1066A>G [p.Asn356Asp]) within our PHTS cohort occur within these deleted segments, these mutations were not modeled because they reside within these flexible regions that are likely to appear in multiple distinct conformations. Consequently, these mutations were excluded from molecular dynamics (MD) analysis. The 17 selected PTEN mutant structures were constructed in silico by side-chain replacement with the Visual Molecular Dynamics (VMD) Mutator Plugin 1.349 from the WT PTEN crystal structure file. 17 mutant structure models were generated for each of the mutations (c.69A>C [p.Leu23Phe], c.71A>G [p.Asp24Gly], c.194A>G [p.Tyr65Cys], c.202T>C [p.Tyr68His], c.275A>C [p.Asp92Ala], c.302T>C [p.Ile101Thr], c.365T>G [p.Ile122Ser], c.388C>G [p.Arg130Gly], c.389G>A [p.Arg130Gln], c.401T>G [p.Met134Arg], c.406T>C [p.Cys136Arg], c.464A>G [p.Tyr155Cys], c.509G>T [p.Ser170Ile], c.517C>T [p.Arg173Cys], c.613A>G [p.Met205Val], c.658C>G [p.Leu220Val], and c.1033C>G [p.Leu345Val]).
We performed all MD simulations by using GROMACS 4.6.350 with GROMOS53a6 forcefield51 on (apo) WT PTEN (PDB: 1D5R) and each PTEN mutant structure. Each system was subjected to energy minimization via the steepest descent method, and a series of five overall minimization steps was performed as previously described22 to remove steric clashes and minimize the forces as a result of the mutation that was introduced to the WT structure. Total minimization was carried out until convergence, where the maximum atomic force was <1000 kJ/mol-nm. The minimized structures were then slowly heated from 0 to 300 K over 100 ps and equilibrated for an additional 250 ps. The production simulations were carried out at constant pressure (1 atm), temperature (300 K), and particle number (NPT, isobaric-isothermal ensemble), as previously described.22 The total simulation time for each model was 200 ns, and coordinates were saved every 1 ps.
Clustering Analysis
We utilized the GROMACS clustering analysis tool to explore the conformation heterogeneity in the ensemble of protein structures generated by each molecular-dynamics simulation system. We used the GROMOS clustering algorithm52 with a Cα of 0.20 nm root-mean-square deviation (RMSD) cut-off to determine structurally similar clusters. For each system, we found three dominant clusters holding greater than ∼60% of total protein structures.
Residue-Interaction Network Analysis
The most populated structure from the clustering analysis for both WT PTEN and each mutant PTEN structure was submitted to the Residue Interaction Network Generator (RING)53 to construct the interactive residue-interaction network (RIN). RINs are constructed by considering amino acid residues as nodes, which are connected by non-covalent interactions, and edges are represented by contacts between atoms such as hydrogen bonds, salt bridges, van der Waals force, and π-π interactions. We utilized the RINs for WT PTEN and each mutant PTEN structure to analyze the residue-residue interaction of PTEN, and we visualized them with Cytoscape 3.6.1.54
After the construction of the structure topology, the network was characterized by the evaluation of quantitative descriptors, as previously described.55, 56 On the basis of the variance of the connectivity, the network heterogeneity was used to measure the connection tendency of a network that included hub nodes. Two important centrality measures related to distance and connectedness are betweenness and closeness, respectively. The betweenness centrality Bk of a node k is the number of times that a node is included in the shortest path between each pair of nodes, normalized by the total number of pairs, and it is defined as:
| (2) |
where N denotes the number of shortest paths between two nodes. The betweenness centrality of a node reflects the amount of control that this node exerts over the interactions of other nodes in the network.57 The closeness centrality Ck of a node is the reciprocal of the average shortest path length, which is calculated as follows:
| (3) |
where the descriptor of the node distance di is normalized by dividing into the number of vertex distances (x − 1). Closeness centrality is a measure of how quickly information spreads from a given node to other reachable nodes in the network.57 We evaluated the nodes by computing estimation plots58 for both betweenness and closeness centrality to determine the difference of means (Δ) on an effect size. Subsequently, we further assessed the nodes by calculating Z score values versus the corresponding residues, and we chose a cutoff threshold of the absolute value of 2 to represent residue peaks in the centrality profiles as potential mediating sites of structural communication and allosteric regulation.
Protein-Structure Network and Global Metapath Analysis
Long-range communication and allosteric networks were characterized by a mixed protein-structure network (PSN) and an elastic network model-normal mode analysis (ENM-NMA) approach previously applied to investigate structural and allosteric communication pathways.30, 59, 60 On the basis of early work established by Vishveshwara et al., the PSN is constructed from the atomic coordinates of residues, which represent the nodes of the network.26, 61 Two nodes are connected by an edge if the percentage of the interaction between them is greater than or equal to a given interaction strength cut-off:
| (4) |
where Iij is the interaction percentage of nodes i and where j is the number of side-chain atom pairs within a given cut-off (4.5 Å), and Ni and Nj are, respectively, the normalization factors (NF) for residues i and j; the NF account for the difference in size of different nodes and their propensity to make the maximum number of contacts with other nodes in protein structures.
The mixed PSN-ENM approach was recently implemented to predict salient structural-communication pathways in biomolecular systems. The underpinning of WebPSN59 lies with a protein structure graph (PSG) and searches for all of the shortest communication pathways between user-specified residues. A PSG defines amino acids as nodes and the non-covalent interactions among them as links. Such graphs are useful for identifying clusters of residues that stabilize the protein structure and protein-protein interfaces.26, 62 The network topology of PSGs depends on the cut-off of the interaction strength between the residues that are used in the constructed graph. The building of the PSG is carried out by means of the PSN algorithm. The strength of interaction between residues i and j (Iij) is evaluated as a percentage, then Iij is calculated for all node pairs. The interaction strength Imin was chosen (Table S2), and any residue pair for which Iij ≥ Imin is considered to be interacting and hence connected to the PSG. This graph-based approach computes network features (e.g., nodes, hubs, and edges) and the shortest communication pathways from MD simulation trajectories.30 In this approach, information on the structural dynamics contributes to the building of the PSG and is utilized in the search for the shortest communication path in which the conformational dynamics contributes in terms of both correlated motions and path occurrence in the trajectories.30
The mixed PSN-ENM method (WebPSN) involves a multi-step process wherein network features (i.e., nodes, hubs, links, etc.) are computed by building a PSG, and the shortest communication pathways for ensembles of structures are acquired from a single, high-resolution structure. The algorithm defines all possible communication paths between selected node pairs and filters the results to the cross-correlation of atomic motions, as derived from ENM-NM. Filtering consists of retaining only the shortest path(s) that contains at least one residue correlated (i.e., with a cross-correlation value ≥0.6) with either one of the two extremes (i.e., the first and last residues in the path). Metapaths made of the most recurrent nodes and links in the path pool (i.e., global metapaths) infer a coarse picture of the structural communication in the considered system. In detail, metapaths are made of nodes ≥5% of the considered path pool (i.e., ‘frequent nodes’) and of links, satisfying both the conditions of being present in one of the paths and of connecting ‘frequent nodes.’ All global metapaths were visualized with VMD 1.9.3.49
Results
Network Proximity of PTEN Influencers of ASD and of Cancer
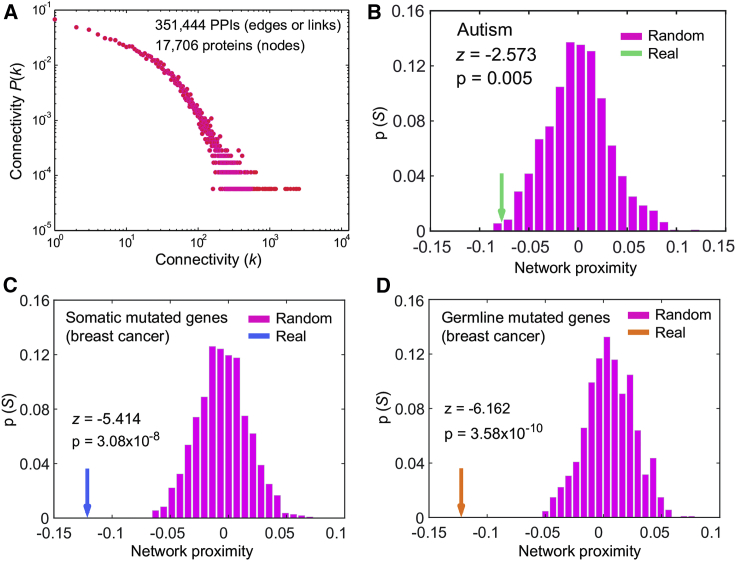
Cells govern biological functions via complex networks, such as in the human protein-protein interactome.38 Perturbations to the human interactome often drive cells to various phenotypic states, such as tumorigenesis and neurological diseases.38 To determine whether germline PTEN variants lead to ASD and/or to cancer via network perturbation, we measured the network-based relationship of PTEN influencers to ASD compared to their relationship to cancer within the human protein-protein interactome network model (Figure 1A). We integrated PTEN genetic interactions and PTEN protein-protein interactions as PTEN influencers (see Material and Methods). To examine PTEN network effects, we used a network-proximity measure that quantifies the network-based relationship between PTEN influencers and known disease genes/proteins in the human protein-protein interactome (see Material and Methods). Specifically, we calculated a Z score for quantifying the significance of the shortest path lengths between PTEN influencers and proteins associated with ASD or cancer from the human interactome. We found a significant network proximity between PTEN influencers and ASD-related genes from the human protein-protein interactome (Z = −2.573, p = 0.005, Figure 1B), indicating stronger network perturbations by PTEN in ASD.
Figure 1.
Significant Network Proximity of PTEN Influencers to Known ASD- or Cancer-Associated Genes within the Human Protein-Protein Interactome Network Model
(A) Connectivity distribution of the human interactome that was used in this study.
(B) A significant network proximity of PTEN influencers with known ASD-associated genes within the human interactome.
(C and D) Significant network proximities of PTEN influencers with known somatic mutant genes (C) or with known breast cancer germline mutant genes (D) within the human interactome. The network proximity analyses of PTEN influencers with more than 10 different other cancer types are provided in Table S1.
In addition, we found significant network proximity between PTEN influencers and cancer in both somatically mutated genes and germline-related genes. For example, PTEN influencers show closer network distance in both breast cancer somatic mutant genes (Z = −5.414, p = 3.08 × 10−8, Figure 1C) from TCGA project and breast cancer germline-associated genes (Z = −6.162, p = 3.58 × 10−10, Figure 1D) in breast cancer compared to the same number of random gene sets with similar (degree of) connectivity in the human interactome. Furthermore, we found a significant network proximity of PTEN influencers with more than 10 different cancer types as well (Table S1). These observations of network proximity are consistent with previous studies that found that PTEN variants can involve both cancer and ASD. In order to understand the functional consequences of how PTEN variants perturb network effects further, we next turned to investigate the structural-network perturbations by using several selected PTEN germline variants found in both ASD and cancer.
Critical Hub Residues Identified by Residue-Interaction Network Analysis in PTEN Mutants Observed in ASD versus Cancer
To address the functional consequences from mutation-driven structural-network perturbations, we utilized a RIN to test our hypothesis that variants associated with ASD and/or cancer might act as a driving force that orchestrates structural communication and allosteric regulation. Previous studies suggest that protein-structure topologies produce small-world networks in which a high local connectivity of residue nodes could be balanced by a smaller number of long-range interactions, giving rise to a high degree of interaction cooperativity.31, 63, 64, 65 The RIN is described as a protein structural graph represented as a network topology map that provides a robust framework for investigating allosteric communications within a protein.26, 62, 66 The network topology map is comprised of nodes (residues) connected by edges (residue interactions) that determine the shortest communication paths between nodes. We computed network-centrality measures (degree, betweenness, and closeness) to identify critical nodes that extend through the entire network topology and participate in both structural communication and allosteric signal propagation.57
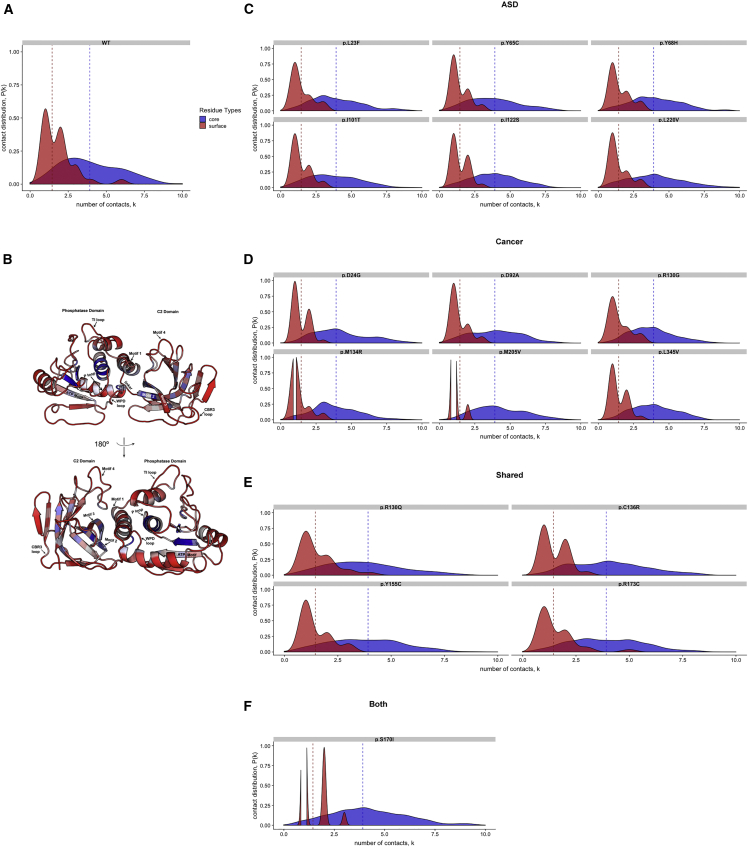
RINs have been utilized to identify hubs (nodes), which have a high degree of connectivity (e.g., first-degree neighbors) and are crucial for stability and signal propagation throughout a network.67 Thus, we initially computed the degree of connectivity to identify the crucial node(s) that are vital for communication and signal propagation. RIN models have demonstrated that small-world allosteric networks are characterized by high-degree connectivity and a small amount of long-range connectivity, whereby long-range protein communication is determined by specific residue clusters playing critical roles in the transmission of functional signals.63, 68, 69 Therefore, in order to distinguish critical residue hubs that play a key role in small-world communication, we calculated the residue depth (RD) and the connectivity distribution by computing the differences between core and surface residues for both WT PTEN and each mutant PTEN structure associated with ASD and/or cancer (Figures 2A–2D and S2A–S2D). RD distinguishes between exposed and buried residues, quantifies the distance of a residue from the bulk solvent, and also correlates with protein-protein interaction hotspots.70 We define the core residues as those residing at depths greater than 4Å, in accordance with previous work.71
Figure 2.
Degree Density Distribution of Core and Surface Residues in ASD- and Cancer-Associated Mutations
Density distribution on core and surface residues for (A) WT PTEN, (B) three-dimensional PTEN structure, (C) ASD only, (D) cancer only, (E) mutations shared across both phenotypes, and (F) one mutation with co-existing ASD and cancer. Core residues encompass two key regions: (1) the active site (residues 32–35; residues overlapping P loop, ATP B-binding motif, and pα4 loop, residues 122–138) and (2) inter-domain (motif 1, residues 169–180; motif 2, residues 250–259, and motif 3, residues 264–276). The core (blue) and surface (red) residues are mapped within the three-dimensional structure of PTEN (inset).
Comparison of the depth distribution of ASD-associated mutations (c.69A>C [p.Leu23Phe], c.194A>G [p.Tyr65Cys], c.202T>C [p.Tyr68His], c.302T>C [p.Ile101Thr], c.365T>G [p.Ile122Ser], and c.658C>G [p.Leu220Val]) demonstrates low connectivity with surface residues when compared to WT PTEN (Figure 2A). Cancer-associated mutations (c.71A>G [p.Asp24Gly], c.275A>C [p.Asp92Ala], c.388C>G [p.Arg130Gly], c.401T>G [p.Met134Arg], c.613A>G [p.Met205Val], and c.1033C>G [p.Leu345Val]) demonstrate stronger connectivity for core residues, highlighting them as critical hubs for signal propagation (Figure 2B). Moreover, we see a notable loss of surface-residue interactions, further demonstrating that the core residues are key players in PTEN structural communication. The mean difference between the core and surface connectivity was larger among cancer-associated variants; however, this difference was not statistically significant (p = 0.31). Additionally, when we compared area under the curve (AUC) distributions across phenotype categories, there were no statistically significant differences. We also observed a loss in connectivity of the surface residues for variants that occur in individuals with ASD, as well as unrelated individuals with cancer (but with no single individual having both ASD and cancer) when compared to either variants associated only with ASD or variants associated only with cancer (Figure 2C). The PTEN c.509G>T (p.Ser170Ile) mutation associated with concurrent ASD and cancer phenotypes in a single individual showed effects similar to cancer (only) variants (Figure 2D). Though the high RD of cancer-associated variants reveals potential regions (P loop, residues 123–131; motif 1, residues 169–180; and CBR3 loop, residues 260–269) prone to mediate allosteric effects, no significant differences were seen when compared to ASD-associated variants (Figures S2A–S2D). Overall, our results demonstrate that in comparison to ASD-associated variants, cancer-associated variants demonstrate stronger connectivity for core residues, identifying them as critical hubs for signal propagation. Additionally, we see a loss of surface-residue interactions, further demonstrating that the core residues are key players in PTEN structural communication and underscoring the extent to which the cancer-associated RIN is perturbed at long distances.
Structural Communication in PTEN’s Inter-domain Region Influences Heightened Allosteric Communications Specific to Cancer
In order to further distinguish critical residue hubs that play a key role in potential allosteric regulation and long-range structural communication, we computed two critical, quantitative-centrality network parameters, betweenness and closeness centrality. Nodes with high betweenness values control the flow of topological information in a network,72 whereas nodes with high closeness values play a principal part in the transmission of information to all other residues in the network.57 Nodes with large betweenness and closeness values have been shown to lie in critical regions in proteins, and they are typically binding free energy hotspots or located in the vicinity of hotspots.24, 31, 64, 73 We therefore posit that allosteric communication is effectively propagated by way of highly conserved residues that are within the active site and inter-domain region and that exhibit a significantly higher betweenness compared to the network average. Moreover, our recent studies reveal these regions are prone to mutations that thermodynamically destabilize the structure.22
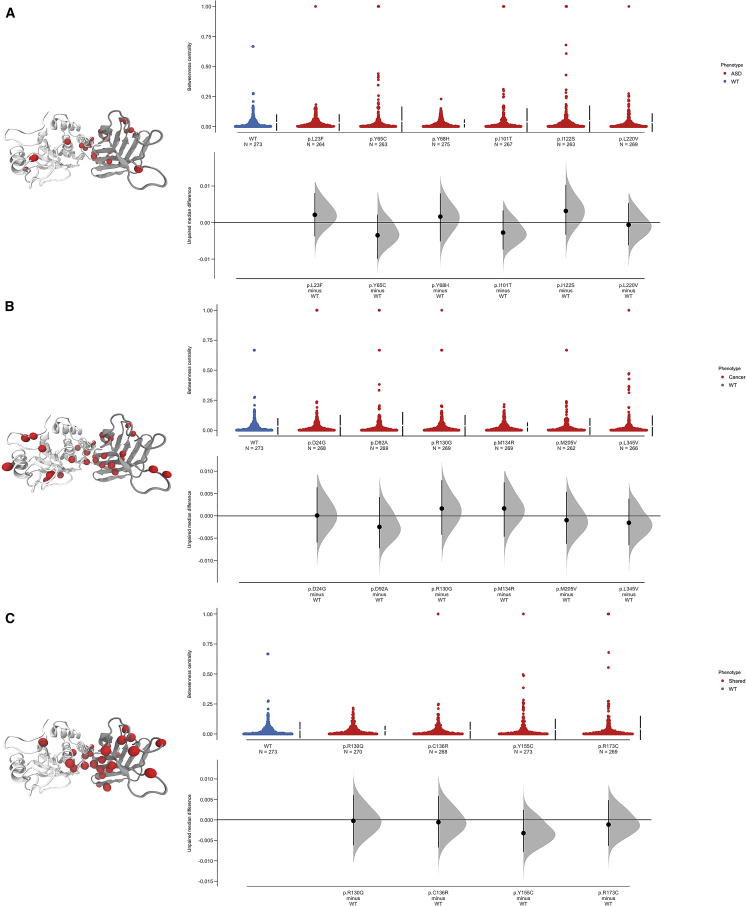
In order to investigate the role of specific residues within ASD- versus cancer-associated PTEN mutations in allosteric communication, we computed betweenness centrality for all residues of the PTEN mutants and assessed the difference between the median betweenness centrality of a given mutant versus wild-type PTEN and Z scores versus the corresponding residues (Figures 3A–3C, S3, and S4A–S4D). All of the 95% confidence intervals for the mutants compared to the wild-type networks include 0, indicating that, as networks, there is little difference in betweenness centrality between the mutants and wild-type structures (Figures 3A–3C and S3). Because of the lack of striking, network-level differences, we computed betweenness Z scores to identify the specific nodes in the mutant networks with high betweenness (Figures S4A–S4D). Residue nodes with a high betweenness in ASD-associated variants are enriched in both the phosphatase and C2 domains, but they are predominantly distributed across the inter-domain motifs for cancer-associated variants (Figures S4A and S4B). Interestingly, the overall betweenness profile across both ASD and cancer phenotypes revealed prominent peaks related to highly conserved and critically functional residues. The most notable residues with high Z score values (>2) for ASD-associated variants were nodes that lie in the N-terminal region (Arg47 and Asn48), inter-domain interface (motif 1, Pro169, Ser170, Gln171, Arg173, and Tyr174; motif 2, Cys250, Gly251, Val255, and Glu256; motif 3, Trp274, and Asn276; and motif 4, Leu325, and Asn329), as well as one node (Lys260) in the CBR3 loop (Figures S4A and S4B).
Figure 3.
Residue-based Betweenness Centrality Estimation Plot Profiles for ASD- and Cancer-Associated PTEN Germline Mutations
Dynamics-based analysis of betweenness network centrality for (A) ASD only, (B) cancer only, and (C) mutations shared across both phenotypes. The gray-filled curve indicates the complete Δ distribution given the observed data. In-line with the median of each group, the Δ is indicated by the black circle. The 95% confidence interval of Δ is illustrated by the vertical black line. Significant betweenness centrality peaks were mapped to the three-dimensional PTEN structure for each phenotype (insets).
The nodes with significant absolute Z score values for the cancer-associated variants are also located in the N-terminal region (Gln17, Glu18, Asp24, Tyr46, Arg55, Tyr76, Gln87, and Asn94) in addition to the active-site residue Arg130. In contrast to the ASD-associated variants, cancer-associated variants also have nodes that clustered more within the inter-domain motifs (motif 1, Pro169, Arg173, Tyr174, and Tyr178; motif 2, Glu256; motif 3, Met270, His272, Trp274, Val275, and Asn276; and motif 4, Leu325, Ala328, and Asn329), indicating heightened structural communication across both domains and long-range perturbation allosteric effects (Figure S4B). Germline mutations that can result in ASD or in cancer show similar centrality peaks compared to cancer-associated variants where nodes cluster more within the inter-domain motifs, most particularly motif 1 (motif 1, Pro169, Ser170, Arg173, and Tyr174; motif 2, Lys254 and Glu256; motif 3, His272 and Trp274; and motif 4, Lys327) (Figure S4C). Interestingly, the c.509G>T (p.Ser170Ile) mutation observed in the single individual with both ASD and cancer demonstrated no peaks beyond the defined Z score threshold (Figure S4D).
To explore structural communication in the ASD- versus cancer-associated variants, we next computed closeness centrality. First, we assessed the difference in closeness centrality at the network level, finding that two ASD-associated mutations (c.194A>G [p.Tyr65Cys] and c.658C>G [p.Leu220Val]), two cancer-associated mutations (c.613A>G [p.Met205Val] and c.1033C>G [p.Leu345Val]), and one shared mutation (c.464A>G [p.Tyr155Cys]) had significant differences compared to the wild type (Figures S5A and S5B). Subsequent to these findings, we examined closeness centrality at the residue level by computing Z scores for each node (Figures S6A–S6D). Nodes with significant closeness values in ASD-associated variants were enriched in both the phosphatase and C2 domains; however, they were more predominant in nodes in the CBR3 loop (Gln261, Asn262, Met264, Leu265, Lys266, and Lys267) of the C2 domain (Figure S6A). In contrast, cancer-associated variants demonstrate nodes with significant closeness Z scores across both domains, most specifically in a rather large area of the N-terminal region, the ATP-B binding motif (Lys60, Ala72, and Arg74), TI loop (Arg161, Asp162, Lys163, Lys164, and Thr167), and the inter-domain motifs motif 2 (Gly251, Glu256, and Pro258), motif 3 (Met264, Leu265, Lys266, Lys267, and Phe272), and motif 4 (Thr321, Asp324, Lys327, and Lys330) (Figure S6B), demonstrating that the inter-domain region is a key site for structural-signal propagation. This is consistent with the significant betweenness residue peaks located within the inter-domain motifs corresponding to global structural communication (Figure S4B), further highlighting this region as a hub for long-range allosteric communication. The variants that can result in ASD or cancer phenotypes also demonstrate similar features with cancer (only)-associated variants, in that significant absolute Z scores were seen across both domains. However, several nodes in the N-terminal region had twice the absolute Z scores compared to WT PTEN and ASD-associated variants (Figure S6C), indicating that long-range perturbations across both domains affect allosteric regulation. Most notably, the c.389G>A (p.Arg130Gln) and c.406T>C (p.Cys136Arg) mutations have significant absolute Z scores that are lowered within the inter-domain interface (residues 150–200) compared to WT PTEN.
This might be, in part, due to the structure compensating for an increase in thermodynamic stability and thus exhibiting a more rigid inter-domain region.22 In contrast, the c.464A>G (p.Tyr155Cys) and c.517C>T (p.Arg173Cys) mutations demonstrate significant closeness values within the C2 domain, whereas WT does not, and this is also in line with our previous results, which exhibit a greater, more flexible (destabilized) inter-domain region (Figure S6C). Similar to the cancer-associated variants, the c.509G>T (p.Ser170Ile) mutation seen in both ASD and cancer reveals significant closeness Z scores across both domains, more specifically in nodes within the phosphatase domain (Thr78 and Val85) and the TI loop (Lys164 and Thr167), demonstrating that these nodes are key sites for structural signal propagation (Figure S6D).
Global Conformational Changes Influence Structural Communication and Define Functional Residues of Allosteric Regulation in ASD and Cancer
In order to determine how mutation-induced conformational dynamics modulates structural communication and allosteric propagation, we investigated the global conformational changes induced by ASD- versus cancer-associated variants. We therefore analyzed the conformational effects of a set of 17 independent, all-atom molecular dynamics (MD) simulations conducted for WT PTEN (apo), ASD-, and cancer-associated variants. Within the active site, a conformational change occurred, and this change involved the P loop (residues 123–130) and WPD loop (residues 88–98) forming a more “open” active site and displacing catalytic residues Asp92 and Arg130 in the ASD-associated variants and a “closed” active site in the cancer-associated variants (Figure S7). The positioning of these two residues is essential for their interaction with the PIP3 lipid substrate and the overall function of PTEN.
To obtain a detailed map of the critical hubs influenced by global conformational changes and vital to the structural communication within PTEN, a RIN comparison was carried out on the open and closed forms of the ASD- versus cancer-associated variants. The crucial hubs within the active-site loops (WPD, P, and TI loops) are defined as nodes that are highly connected (>4 edges, interactions).29 In comparing the connectivity distribution throughout the entire structure of PTEN in ASD- versus cancer-associated variants, we see a difference in connectivity in critical functional loops and motifs (P loop, residues 123–131; ATP-A binding motif, residues 122–136; CBR3 loop, residues 260–269; and motif 1 residues, 169–180) within the phosphatase domain compared to the C2 domain (Figure S8 and Table 2). This might be, in part, due to cooperative conformational dynamics formed by conserved residues within the active site and inter-domain regions, suggesting rapid signal propagation through a small network of core residues.
Table 2.
Inverse Correlation in Degree Connectivity of Phosphatase Domain Catalytic Loops and C2 Domain Inter-domain Motifs
| Phosphatase Domain | C2 Domain | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalytic loops | ATP motifs | Inter-domain motifs | |||||||||||||
| WPD | P | TI | ATP A | ATP B | CBR3 | Motif 1 | Motif 2 | Motif 3 | Motif 4 | Linker | |||||
| Wild type | 78 | 61 | 41 | 117 | 91 | 29 | 128 | 83 | 86 | 49 | 17 | ||||
| p.Leu23Phe | ASD | 85 | 30∗ | 37 | 82∗ | 100 | 31 | 122 | 82 | 105∗ | 66∗ | 25 | |||
| p.Tyr65Cys | 76 | 56 | 48 | 103 | 76 | 22∗ | 120 | 97 | 102∗ | 46 | 24 | ||||
| p.Tyr68His | 81 | 54 | 31 | 90∗ | 84 | 31 | 96∗ | 94 | 104∗ | 60∗ | 17 | ||||
| p.Ile101Thr | 69 | 49∗ | 40 | 90∗ | 91 | 20∗ | 137 | 61 | 62 | 49 | 22∗ | ||||
| p.Ile122Ser | 66 | 47∗ | 42 | 94∗ | 100 | 36∗ | 99∗ | 80 | 91 | 55 | 19 | ||||
| p.Leu220Val | 80 | 43∗ | 38 | 99 | 81 | 34 | 118 | 75 | 94 | 39 | 20∗ | ||||
| p.Asp24Gly | Cancer | 72 | 48∗ | 41 | 95 | 83 | 41∗ | 110 | 91 | 99 | 65∗ | 20∗ | |||
| p.Asp92Ala | 57∗ | 40∗ | 40 | 79∗ | 71∗ | 19∗ | 104 | 70 | 79 | 70∗ | 11∗ | ||||
| p.Arg130Gly | 76 | 40∗ | 39 | 87∗ | 74 | 25 | 102∗ | 78 | 86 | 62∗ | 20∗ | ||||
| p.Met134Arg | 77 | 37∗ | 45 | 89∗ | 102 | 46∗ | 105 | 105∗ | 99 | 51 | 10∗ | ||||
| p.Met205Val | 98∗ | 59 | 57∗ | 108 | 88 | 23∗ | 121 | 89 | 80 | 49 | 20∗ | ||||
| p.Leu345Val | 67 | 56 | 49 | 99 | 97 | 43∗ | 91∗ | 80 | 107∗ | 59∗ | 21∗ | ||||
| p.Arg130Gln | Shared | 81 | 56 | 48 | 101 | 105 | 51∗ | 84∗ | 91 | 103∗ | 71∗ | 23∗ | |||
| p.Cys136Arg | 64 | 67 | 44 | 116 | 72∗ | 16∗ | 106 | 76 | 79 | 41 | 19 | ||||
| p.Tyr155Cys | 90 | 43∗ | 21∗ | 96 | 81 | 29 | 105 | 79 | 89 | 60∗ | 18 | ||||
| p.Arg173Cys | 71 | 44∗ | 27∗ | 89∗ | 103 | 51∗ | 97∗ | 104∗ | 102∗ | 55 | 13∗ | ||||
| p.Ser170Ile | Both | 107∗ | 49∗ | 52∗ | 105 | 77 | 27 | 124 | 95 | 112∗ | 66∗ | 24∗ | |||
| Residues Key | |||||||||||||||
| WPD loop: residues 88–98 | ATP A binding motif: residues 122–136 | motif 1: residues 169–180 | motif 4: residues 321–334 | ||||||||||||
| P loop: residues 123–131 | ATP B binding motif: residues 60–73 | motif 2: residues 250–259 | Linker: residues 185–191 | ||||||||||||
| TI loop: residues 160–171 | CBR3 loop: residues 260–269 | motif 3: residues 264–276 | |||||||||||||
17 germline missense PTEN mutations associated with ASD only or cancer only, mutations shared across both phenotypes (shared), and one mutation with co-existing ASD and cancer (both). Inversely correlated regions are marked with an asterisk, indicating either a 20% increase or decrease compared to the WT.
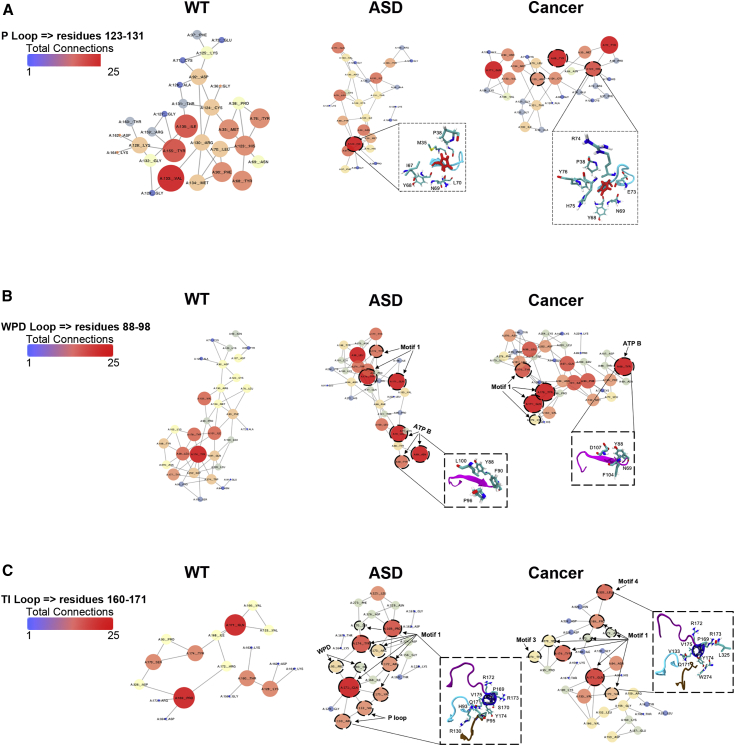
Distinct differences in connectivity distribution reveal changes in hub residues within the three active-site loops in ASD- versus cancer-associated variants. Within the P loop, cancer-associated variants had a slight decrease in connectivity with residue His123, going from nine total connected interactions to seven, compared to ASD-associated variants (Figure 4A).12 Residue His123 is essential for P loop conformation and participates in a critical π-π stacking interaction with Tyr76 and Phe37. Changes in connectivity that disrupt these interactions would extend the pβ5-strand residues before the P loop, leading to marked conformational changes in the WPD loop. The cancer-associated variants also demonstrate more interactions between His123 and residues Tyr68 and Tyr76, which lie within the ATP-B binding motif. The identification of these P loop hub residues, in addition to residue Gln171, reveals salient structural communication specific to cancer-associated variants among the P loop, ATP-B binding motif, and inter-domain region. Interestingly, residue Arg130 is a critical hub residue in cancer-associated variants. Mutations at this position seen in both cancer (only)-associated variants and variants that can result in ASD or cancer phenotypes in different individuals reveal an increase in structural stability, exhibiting a more rigid inter-domain as previously described.22 Moreover, within the RIN model, Arg130 participates in a structural-communication pathway between proximal and distal sites (e.g., Arg130-Met134-Asn94-Gln171). The interaction of Arg130 with these proximal and distal sites expands the stiffness of the network and therefore leads to a more closed active site for cancer-associated variants. The changes in connectivity within the WPD loop reveal that ASD-associated variants have more hub residues in the ATP-B binding motif, from 41 to 54 total connected interactions, compared to cancer-associated variants (Figure 4B).
Figure 4.
Residue Interaction Connectivity in Catalytic Loops of ASD- and Cancer-Associated Mutations
Connectivity in (A) the P loop (residues 123–131), (B) the WPD loop (residues 88–98), and (C) the TI loop (residues 160–171). The size and color of the nodes within the residue-interaction network indicate the importance of the hub node in the network, revealing it as a key player in structural communication (e.g., expansion in size and progression toward red color indicate increasing connectivity).
Interestingly, mutation-induced conformational changes associated with PTEN-ASD also reveal an increase in degree connectivity in motifs 3 and 4 within the inter-domain region, further indicating the consequential effects of conformational changes on long-range communication. In contrast to ASD-associated variants, the loss of structural communication between the WPD loop hub residues and the ATP-B binding site generally occurs in cancer-associated variants. This apparent loss of inter-residue communication reveals the induced conformation changes associated with PTEN-cancer mutations, resulting in an increased inter-residue signal propagation in motif 1 residues. Moreover, because of changes in connectivity in the ATP-B binding motif in ASD- versus cancer-associated variants, we would expect ATP binding to be more favorable in ASD-associated variants compared to WT.
Connectivity differences in the TI loop reveal that ASD-associated variants have more hub residues (from 25 total connected interactions to 30) within the WPD loop compared to cancer-associated variants (Figure 4C). Moreover, ASD-associated variants reveal a structural-communication pathway that participates in proximal hub sites, whereas the structural-communication pathway in cancer-associated variants participates in both proximal and distal hub sites. This structural rearrangement seen in cancer-associated variants can be attributed to global conformations of the TI loop (Figure S7).
A Global Metapath of Communication Reveals Distinct, Long-range Communication Pathways Associated with ASD and Cancer
To identify a specific global metapath of communication within the structure of ASD- and cancer-associated variants, a mixed protein structure network (PSN) and elastic network model (ENM) approach was employed.30, 59, 60 ASD- versus cancer-associated-mutation-induced conformational dynamics exert differential impact on structural communication, and this impact can be examined by representing PTEN structures as an elastic network of interacting residues. We utilized the PSN analysis to deduce the effects that mutations have on the native PTEN fingerprint and to identify key hub residues that govern allosteric regulation within the long-range communication pathway. A detailed comparison of the communication paths between ASD- versus cancer-associated variants allows us to identify distinct, long-range, salient communication pathways.
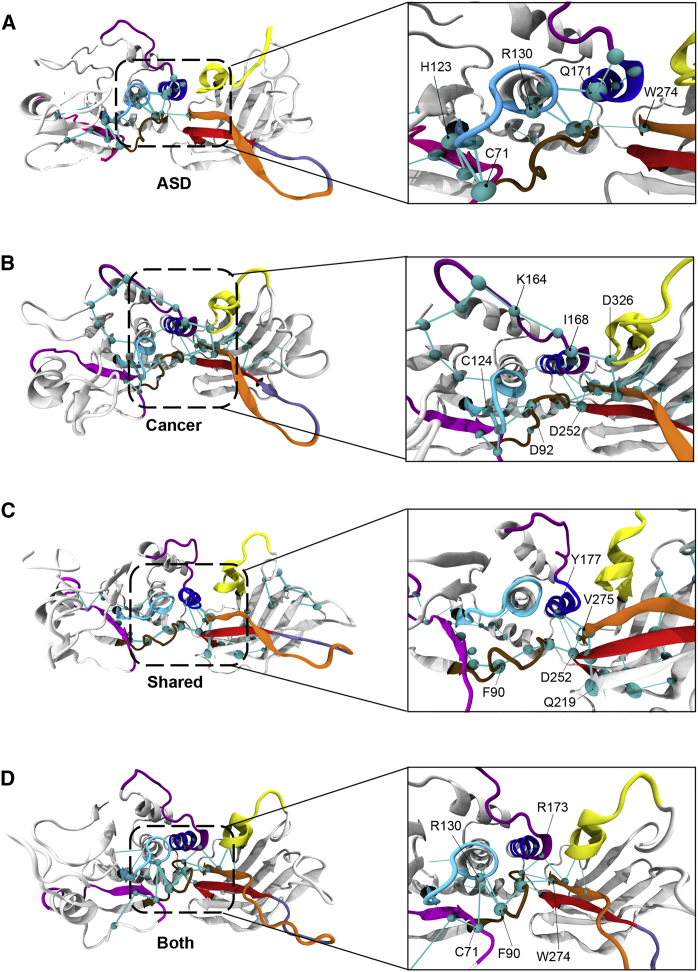
Our results reveal that the ASD-associated global metapath has a distinguishable pathway that illustrates the global metapath traversing through nodes in the WPD loop (residues 88–98) of the active site, moving into the core of the protein (e.g., motif 1), and spreading to residue Trp274 of motif 3 within the C2 domain (Figure 5A). Most notably, the global metapath is predominantly restricted to the phosphatase domain within the ASD-associated variants. Conversely, the cancer-associated global metapath extends from the N-terminal region of the protein structure and distinctly spreads across the inter-domain interface (Figure 5B). Interestingly, cancer-associated variants demonstrate a more clustered pathway concentrated in their core (inter-domain interface).
Figure 5.
Global Metapath of Communication in ASD- and Cancer-Associated Mutations
The figure shows (A) ASD only, (B) cancer only, (C) mutations shared across both phenotypes, and (D) one mutation resulting in co-existing ASD and cancer. The metapath outlines critical nodes where the spheres (cyan) are centered on C-alpha atoms and the diameter is proportional to the number of edges made by the node.
The global metapaths involving the mutations that can be associated with either ASD or with cancer (c.389G>A [p. Arg130Gln], c.406T>C [p.Cys136Arg], c.464A>G [p.Tyr155Cys], and c.517C>T [p.Arg173Cys]), as well as the c.509G>T (p.Ser170Ile) variant seen in both ASD and cancer in a single individual, had the active-site loops involved in their pathways. Most notable was the WPD loop (residues 88–98), which contains the acidic Asp92 that participates in catalysis (Figures 5C and 5D). These pathways share several other features that consist of hub residues containing both ATP-binding sites (residues 60–73 and 122–136) and motif 3 (residues 264–276), which governs flexibility of the CBR3 loop (residues 260–269). The minimal involvement of the inter-domain region can be attributed to the fact that half of these mutations (c.389G>A [p.Arg130Gln], c.464A>G [p.Tyr155Cys], and c.517C>T [p.Arg173Cys]) are in the central core of the protein; therefore, there is a decrease in the normal interactions required for long-range communication. This further emphasizes the role of the highly conserved Arg130 and Arg173 positions, both in structural stability and allosteric communication, because they are integral hub residues participating in both open and closed conformational states.
Overall, the global metapath of each mutant PTEN phenotype highlights the inter-domain interface as a crucial region that participates both in the stability of the core and in the overall dynamics of the protein (Figure 5 and Table 3). Our results indicate that cancer-associated variants are distinct in that they possess a global metapath communication pathway that propagates across the entire protein structure and comprises critical inter-domain hub nodes that govern allosteric communication (Figure 5B).
Table 3.
Summary of Main Findings in Germline PTEN Missense Mutations Associated with ASD Versus Mutations Associated with Cancer
| Autism Spectrum Disorder | Cancer |
|---|---|
| open active site | closed active site |
| significant network centrality peaks in N-terminal and inter-domain interface | significant network centrality peaks across both domains and inter-domain interface |
| phosphatase-restricted structural communication pathway | long-range structural communication pathway – nodes in active-site and inter-domain region |
Discussion
To date, there have been no reports aimed at unraveling the human interactome of genes/proteins associated with both ASD and cancer. Moreover, inter-residue, long-range structural communication, as well as allosteric regulation, has not been previously reported in ASD (only)- or cancer (only)-associated germline PTEN missense variants. As such, we integrated network-proximity analysis performed on the human interactome, molecular-dynamics simulations, and RIN analysis to investigate the relationship of functional dynamics, structural-communication pathways, and long-range allosteric regulation within ASD-associated (PTEN-ASD) versus cancer-associated germline PTEN (PTEN-cancer) missense variants. Our results provide new insight into a possible PTEN allostery mechanism that governs salient structural-communication pathways and perturbations in the human protein-protein interactome network model. We demonstrate that the PTEN influencers share significant overlap with the ASD and cancer interactomes, providing network-based evidence that PTEN is a crucial player in the biology of both ASD and cancer. We identified altered, inter-residue interactions that coordinate key functional centers that might govern long-range allosteric regulation, and we found distinct differences between the ASD- and cancer-associated PTEN germline variants (Figure S1 and Table 3). Our results highlight the critical role of mutation-induced conformational dynamics associated with PTEN-ASD or PTEN-cancer missense variants and offer novel perspectives in long-range allosteric regulation, and they are an important step in predicting effects of germline PTEN missense variants.
Our dynamics-based network approach, in which we utilized the results of MD simulations, allowed us to determine the depth and distribution of highly connected residues in ASD- versus cancer-associated PTEN germline variants. In comparison to ASD-associated variants, cancer-associated variants demonstrate more connectivity for core residues, identifying them as critical hubs for signal propagation (Figure 2). This might be due, in part, to compensation from decreased thermostability and deleterious conformational changes.74 We have shown that cancer-associated germline PTEN variants tend to be highly destabilizing, affecting the entirety of the three-dimensional structure.22 Additionally, a loss of surface-residue interaction further revealed that the core residues are key players in small-world communication, underscoring the extent to which the RIN can be perturbed, even at long distances (Figure 2 and Table 3). Such long-distance communication through residues can affect catalysis, as seen in missense mutations and double mutations >15 Å from the active site.75, 76, 77 Moreover, it is well established that mutations that introduce residues that increase the molecular volume decrease structural stability, as we previously demonstrated for cancer-associated mutations.22, 78
The central finding of our study is that distinct structural-communication-pathway signatures exist for ASD-associated compared to cancer-associated variants. The specific signature for cancer-associated variants is governed by critical nodes, which include the highly conserved active-site loops and inter-domain region. We found that global features that differentiate critical nodes include degree connectivity, significant betweenness and closeness centrality that mediate structural communication, and allosteric signal propagations. Our results revealed significant centrality peaks related to critical functional residues across both ASD versus cancer phenotypes (Figures S4 and S6). Interestingly, the c.509G>T (p.Ser170Ile) mutation, occurring in an individual with both ASD and cancer, demonstrated no significant peaks for betweenness centrality (Figure S4D and Table 3). This might be due, in part, to the fact that this residue position is located within both the active-site TI loop and a rich-hydrogen-bond network of the inter-domain region of PTEN, and this position diminishes long-range communication and allosteric effects. In fact, a missense mutation at this position leads to decreased dynamics across the inter-domain region and the CBR3 loop.22 Mutations in position Ser170 have been previously reported in multiple individuals with cancer,12, 79 further demonstrating the importance of this position as a hub for allosteric regulation. Moreover, Ser170 was found to be generally intolerant to mutation, whereby most changes induce a decrease in the steady-state protein abundance, emphasizing the importance this position plays in maintaining the structural integrity of WT PTEN.80 This finding further highlights this position as a long-range-allosteric-communication hub that is specific to cancer-associated variants (Figures S4A–S4D and S6A–S6D). In fact, possible allosteric pathways involving the inter-domain region were found to correspond with regions that most efficiently propagate signals in response to external perturbations, as demonstrated by Verkhivker (2018),24 whereby mutations in these positions associate with severe phenotypes.
Our results demonstrate that global conformational changes within the active site correspond with significant network-centrality values, indicating that the structural rearrangement of key mediating centers is enriched by residues located in the active-site loops, CBR3 loop, and inter-domain interface (Figures S4A–S4D and S7). Differences seen in connectivity reveal changes in hub residues within the three active-site loops (WPD, P, and TI loops) in ASD- versus cancer-associated variants (Figure 4 and Table 3). Mutations in residues within this catalytic region might give rise to global changes that shift the conformational dynamics related to ASD compared to cancer. Because allosteric effects involve the release of strain energy through fluctuation and changes in van der Waals contacts and hydrogen bonds,81 signal propagation is more efficient in a tightly packed environment with high atomic density.82 Therefore, mutation-induced conformational dynamics seen in cancer-associated variants lead to a more packed and closed active-site conformation, eliciting an allosteric effect induced by long-range perturbations from the active site to the inter-domain. Furthermore, conformational changes associated with PTEN-cancer variants that affect connectivity in the WPD loop demonstrate a structural-communication loss that results in an increased inter-residue signal propagation in motif 1 residues in the ATP-B binding site. In contrast, PTEN-ASD variants exhibit conformational changes that generally restore direct communication between the ATP-B binding region and both motifs 3 and 4. These changes, occurring within the inter-domain region, might be functionally related to the opening of the WPD and P loops. Our findings on mutation-induced conformational dynamics that lead to an open versus closed active site seen in ASD- and cancer-associated variants, respectively, are also in agreement with functional studies by Spinelli et al. (2015). These studies demonstrate effects of allelic variants in vitro that that lead to the partial loss of function of PTEN enzymatic activity and seem to be preferentially associated with ASD, whereas catalytically inactive mutants lead to a more aggressive phenotype.20
The highly conserved active-site loops, despite their obvious central role in catalysis, have been subject to minimal scrutiny in dynamic cross-talk with a possible allosteric regulation site.22, 23, 24 However, our RIN analysis highlights how different conformations within the active-site loops mediate long-range interactions between the active site, N-terminal region, ATP-B binding site, CBR3 loop, and the inter-domain region, and thus might play an active role in allostery. In fact, it has been shown that PI(4,5)P2 binds to N-terminal residues 6–15, thereby inducing an allosteric conformational change that activates PTEN.83 In addition, the variant p.Arg15Ala occurs in human cancers and is reported to reduce PTEN activity.83 Moreover, the CBR3 loop facilitates membrane binding, which results in conformational changes that occur in the active site and leads to the assumption that there is an allosteric activation mechanism in PTEN (Figure 4).83, 84 A more recent study utilizing neutron scattering combined with all-atom MD simulations revealed the importance of the CBR3 loop in membrane and active-site interactions with phosphatidylinositide substrates [PI(4,5)P2 and PI(3,4,5)P3].85 These inositolphosphates bind more strongly to the protein in the absence of PIPs bound within the membrane.85 Additionally, a cluster of contacts of accumulated PI(4,5)P2 was observed at the C2 domain. This cluster of contacts populated the CBR3 loop and the cleft between the phosphatase and C2 domains (inter-domain). In a recent but separate study, this same region was implicated to be rather significant for the binding of a cluster of 5-HT2C receptor α-helical 3L4F-F1 peptide conformations.86 Interestingly, previous studies have revealed that the binding of an aptamer that corresponds to residues 31–43 induces a slight conformational change in PTEN and exposes both adjacent ATP-binding regions; thus, it establishes the existence of a possible allosteric mechanism not previously described for this tumor suppressor.87, 88 These results further implicate the importance of the ATP-B binding region and CBR3 loop in the inter-domain dynamics, as well as the communication pathway. Overall, conformational changes associated with PTEN-ASD variants affect connectivity in the ATP-B binding region. This change in connectivity induces direct communication between motifs 3 and 4, and this communication might be related to CBR3 regulation and the functional opening of the WPD and P loops. In contrast, conformational changes that affect connectivity in the WPD loop and are associated with PTEN-cancer variants demonstrate a loss of structural communication in the ATP-B binding site, and this loss results in an increased inter-residue signal propagation in motif 1 residues. These mutation-induced displacements in the active-site pocket elicit long-range, inter-residue propagation that governs potential allosteric regulation and plays a key role in modulating the catalytic function of PTEN that leads to ASD or cancer.
A unique aspect of this work is the utilization of elastic-network and protein-structure-network modeling to identify global metapaths of communication within the structure of ASD- and cancer-associated variants (Figure 5 and Table 3). We demonstrate that the inter-domain region acts as a hinge point for collective motions, where hub behavior and structural communication play a central role in differentiating the regulatory actions associated with ASD- and cancer-associated phenotypes. Our results revealed specific pathways of structural communication where the cancer-associated global metapath distinctly spreads across the inter-domain interface, whereas the ASD-associated global metapath is predominantly restricted to the phosphatase domain. The inter-domain region is highlighted as a potential hub that is implicated in allosteric regulation, whereby nodes cross-talk with the adjacent active site, suggesting that these regions might be integral for the coordination of global structural changes.
Overall, the network perturbation and long-range structural-communication effects of specific mutations on the PTEN structure, elucidated through our present study, provide clues into salient structural pathways that lead to ASD or to cancer (Table 3). Furthermore, our observations begin to give hints as to mutant PTEN’s allosteric druggability, which might actually be different for ASD versus cancer outcomes. An understanding of the allosteric modulation of PTEN offers rich and diverse opportunities to further guide clinical management, including targeted therapy, for individuals with ASD and cancer. In doing so, we might successfully optimize conformational states for PTEN function, either by stabilizing active conformations or destabilizing inactive conformations. Our results indicate that the inter-domain region of PTEN forms a potential basis for allosteric communication, which offers potential clinical utility as an alternative strategy for effective targeting of this region to overcome the clinical morbidities associated with ASD and cancer. Our current observations highlight the importance of utilizing multiple approaches, such as network perturbations, biophysical simulations, and RINs, to understand the effects of specific mutations on PTEN structure and infer possible mechanisms and clinical potentials for allostery in helping to predict ahead of time whether a specific PTEN variant will have a high likelihood of being associated with ASD or with cancer risk, or both. Our observations provide considerable insight into the crucial aspects related to novel structural-communication and network perturbations that contribute to allosteric modulation and, essentially, the identification of the disease-specific molecular features that contribute to autism or to cancer. This will provide a basis for empiric studies that will include understanding the targeting of allosteric differences between PTEN-ASD and PTEN-cancer and validating in vivo that, indeed, different tactics might be required in PTEN-ASD-associated allosteric alterations compared to those of PTEN-cancer.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank all the individuals with PHTS and PTEN-ASD and their families, whose participation was crucial to making this study possible. We thank Dr. Lamis Yehia for critical review of the manuscript. This study was funded, in part, by the National Cancer Institute grant P01CA124570, the National Institute of Neurological Disorders and Stroke (NINDS) grant U54NS092090, the Ambrose Monell Foundation PTEN-Switch Grant, and a grant allocation of computing time from the Ohio Supercomputing Center (PCCF0020) [all to C.E.]. I.N.S. is funded, in part, by the Ambrose Monell Cancer Genomic Medicine Fellowship and the National Institutes of Health (NIH) T32 5T32CA59366-22. C.E. is an American Cancer Society Clinical Research Professor and the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic.
Published: April 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.009.
Web Resources
BioGRID, https://thebiogrid.org
CTD, http://ctdbase.org
GeneCards, https://www.genecards.org
HuGE Navigator, https://phgkb.cdc.gov/PHGKB/hNHome.action
OMIM, http://www.omim.org
PharmGKB, https://www.pharmgkb.org
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
WebPSN, http://webpsn.hpc.unimore.it
Supplemental Data
References
- 1.Nelen M.R., van Staveren W.C., Peeters E.A., Hassel M.B., Gorlin R.J., Hamm H., Lindboe C.F., Fryns J.P., Sijmons R.H., Woods D.G. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum. Mol. Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 2.Nelen M.R., Padberg G.W., Peeters E.A., Lin A.Y., van den Helm B., Frants R.R., Coulon V., Goldstein A.M., van Reen M.M., Easton D.F. Localization of the gene for Cowden disease to chromosome 10q22-23. Nat. Genet. 1996;13:114–116. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 3.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 4.Eng C. PTEN: One gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 5.Tan M.H., Mester J., Peterson C., Yang Y., Chen J.L., Rybicki L.A., Milas K., Pederson H., Remzi B., Orloff M.S., Eng C. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am. J. Hum. Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan M.H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin. Cancer Res. 2012;18:400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers M.P., Stolarov J.P., Eng C., Li J., Wang S.I., Wigler M.H., Parsons R., Tonks N.K. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc. Natl. Acad. Sci. USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stambolic V., Suzuki A., de la Pompa J.L., Brothers G.M., Mirtsos C., Sasaki T., Ruland J., Penninger J.M., Siderovski D.P., Mak T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 10.Maehama T., Dixon J.E. PTEN: A tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 11.Gu J., Tamura M., Yamada K.M. Tumor suppressor PTEN inhibits integrin- and growth factor-mediated mitogen-activated protein (MAP) kinase signaling pathways. J. Cell Biol. 1998;143:1375–1383. doi: 10.1083/jcb.143.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.O., Yang H., Georgescu M.M., Di Cristofano A., Maehama T., Shi Y., Dixon J.E., Pandolfi P., Pavletich N.P. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez-Escudero I., Oliver M.D., Andrés-Pons A., Molina M., Cid V.J., Pulido R. A comprehensive functional analysis of PTEN mutations: Implications in tumor- and autism-related syndromes. Hum. Mol. Genet. 2011;20:4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 14.Georgescu M.M. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1:1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamguney T., Stokoe D. New insights into PTEN. J. Cell Sci. 2007;120:4071–4079. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
- 16.Salmena L., Carracedo A., Pandolfi P.P. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Leslie N.R., Foti M. Non-genomic loss of PTEN function in cancer: Not in my genes. Trends Pharmacol. Sci. 2011;32:131–140. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X.C., Piccini A., Myers M.P., Van Aelst L., Tonks N.K. Functional analysis of the protein phosphatase activity of PTEN. Biochem. J. 2012;444:457–464. doi: 10.1042/BJ20120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papa A., Wan L., Bonora M., Salmena L., Song M.S., Hobbs R.M., Lunardi A., Webster K., Ng C., Newton R.H. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spinelli L., Black F.M., Berg J.N., Eickholt B.J., Leslie N.R. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J. Med. Genet. 2015;52:128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson L., Maccario H., Perera N.M., Yang X., Spinelli L., Tibarewal P., Glancy B., Gray A., Weijer C.J., Downes C.P., Leslie N.R. Suppression of cellular proliferation and invasion by the concerted lipid and protein phosphatase activities of PTEN. Oncogene. 2010;29:687–697. doi: 10.1038/onc.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith I.N., Thacker S., Jaini R., Eng C. Dynamics and structural stability effects of germline PTEN mutations associated with cancer versus autism phenotypes. J. Biomol. Struct. Dyn. 2019;37:1766–1782. doi: 10.1080/07391102.2018.1465854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith I.N., Briggs J.M. Structural mutation analysis of PTEN and its genotype-phenotype correlations in endometriosis and cancer. Proteins. 2016;84:1625–1643. doi: 10.1002/prot.25105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verkhivker G.M. Biophysical simulations and structure-based modeling of residue interaction networks in the tumor suppressor proteins reveal functional role of cancer mutation hotspots in molecular communication. Biochim. Biophys. Acta, Gen. Subj. 2019;1863:210–225. doi: 10.1016/j.bbagen.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Nussinov R., Tsai C.J., Ma B. The underappreciated role of allostery in the cellular network. Annu. Rev. Biophys. 2013;42:169–189. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinda K.V., Vishveshwara S. A network representation of protein structures: Implications for protein stability. Biophys. J. 2005;89:4159–4170. doi: 10.1529/biophysj.105.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu G., Zhou J., Yan W., Chen J., Shen B. The topology and dynamics of protein complexes: Insights from intra- molecular network theory. Curr. Protein Pept. Sci. 2013;14:121–132. doi: 10.2174/1389203711314020004. [DOI] [PubMed] [Google Scholar]
- 28.Fang Y.P., Ma D.C., Li M.L., Wen Z.N., Diao Y.B. Investigation of the proteins folding rates and their properties of amino acid networks. Chemometr Intell Lab. 2010;101:123–129. [Google Scholar]
- 29.Vishveshwara S., Ghosh A., Hansia P. Intra and inter-molecular communications through protein structure network. Curr. Protein Pept. Sci. 2009;10:146–160. doi: 10.2174/138920309787847590. [DOI] [PubMed] [Google Scholar]
- 30.Raimondi F., Felline A., Seeber M., Mariani S., Fanelli F. A mixed protein structure network and elastic network model approach to predict the structural communication in biomolecular systems: The PDZ2 domain from tyrosine phosphatase 1E as a case study. J. Chem. Theory Comput. 2013;9:2504–2518. doi: 10.1021/ct400096f. [DOI] [PubMed] [Google Scholar]
- 31.Amitai G., Shemesh A., Sitbon E., Shklar M., Netanely D., Venger I., Pietrokovski S. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 2004;344:1135–1146. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 32.Rolland T., Taşan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 34.Meyer M.J., Das J., Wang X., Yu H. INstruct: A database of high-quality 3D structurally resolved protein interactome networks. Bioinformatics. 2013;29:1577–1579. doi: 10.1093/bioinformatics/btt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fazekas D., Koltai M., Türei D., Módos D., Pálfy M., Dúl Z., Zsákai L., Szalay-Bekő M., Lenti K., Farkas I.J. SignaLink 2 - A signaling pathway resource with multi-layered regulatory networks. BMC Syst. Biol. 2013;7:7. doi: 10.1186/1752-0509-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coordinators N.R., NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(D1):D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 38.Cheng F., Jia P., Wang Q., Lin C.C., Li W.H., Zhao Z. Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol. Biol. Evol. 2014;31:2156–2169. doi: 10.1093/molbev/msu167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng F., Desai R.J., Handy D.E., Wang R., Schneeweiss S., Barabási A.L., Loscalzo J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018;9:2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng F., Liu C., Lin C.C., Zhao J., Jia P., Li W.H., Zhao Z. A gene gravity model for the evolution of cancer genomes: A study of 3,000 cancer genomes across 9 cancer types. PLoS Comput. Biol. 2015;11:e1004497. doi: 10.1371/journal.pcbi.1004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O’Donnell L. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J., Cheng F., Zhao Z. Tissue-specific signaling networks rewired by major somatic mutations in human cancer revealed by proteome-wide discovery. Cancer Res. 2017;77:2810–2821. doi: 10.1158/0008-5472.CAN-16-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W., Gwinn M., Clyne M., Yesupriya A., Khoury M.J. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 45.Hernandez-Boussard T., Whirl-Carrillo M., Hebert J.M., Gong L., Owen R., Gong M., Gor W., Liu F., Truong C., Whaley R. The pharmacogenetics and pharmacogenomics knowledge base: Accentuating the knowledge. Nucleic Acids Res. 2008;36:D913–D918. doi: 10.1093/nar/gkm1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis A.P., King B.L., Mockus S., Murphy C.G., Saraceni-Richards C., Rosenstein M., Wiegers T., Mattingly C.J. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng F., Kovács I.A., Barabási A.L. Network-based prediction of drug combinations. Nat. Commun. 2019;10:1197. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45(D1):D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 50.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 51.Oostenbrink C., Villa A., Mark A.E., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 52.Daura X., Gademann K., Juan B., Seebach D., van Gunsteren W.F., Mark A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. 1999;38:236–240. [Google Scholar]
- 53.Piovesan D., Minervini G., Tosatto S.C. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw315. W367-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonchev D., Buck G.A. From molecular to biological structure and back. J. Chem. Inf. Model. 2007;47:909–917. doi: 10.1021/ci7000617. [DOI] [PubMed] [Google Scholar]
- 56.Doncheva N.T., Assenov Y., Domingues F.S., Albrecht M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 2012;7:670–685. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 57.Borgatti A.P.E., M. G. A grapth-theoretic perspective on centrality. Soc. Networks. 2006;28:466–484. [Google Scholar]
- 58.Ho J., Tumkaya T., Aryal S., Choi H., Claridge-Chang A. Moving beyond P values: Everyday data analysis with estimation plots. bioRxiv. 2018 doi: 10.1038/s41592-019-0470-3. [DOI] [PubMed] [Google Scholar]
- 59.Seeber M., Felline A., Raimondi F., Mariani S., Fanelli F. WebPSN: A web server for high-throughput investigation of structural communication in biomacromolecules. Bioinformatics. 2015;31:779–781. doi: 10.1093/bioinformatics/btu718. [DOI] [PubMed] [Google Scholar]
- 60.Fanelli F., Felline A., Raimondi F., Seeber M. Structure network analysis to gain insights into GPCR function. Biochem. Soc. Trans. 2016;44:613–618. doi: 10.1042/BST20150283. [DOI] [PubMed] [Google Scholar]
- 61.Kannan N., Vishveshwara S. Identification of side-chain clusters in protein structures by a graph spectral method. J. Mol. Biol. 1999;292:441–464. doi: 10.1006/jmbi.1999.3058. [DOI] [PubMed] [Google Scholar]
- 62.Brinda K.V., Vishveshwara S. Oligomeric protein structure networks: Insights into protein-protein interactions. BMC Bioinformatics. 2005;6:296. doi: 10.1186/1471-2105-6-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atilgan A.R., Akan P., Baysal C. Small-world communication of residues and significance for protein dynamics. Biophys. J. 2004;86:85–91. doi: 10.1016/S0006-3495(04)74086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.del Sol A., O’Meara P. Small-world network approach to identify key residues in protein-protein interaction. Proteins. 2005;58:672–682. doi: 10.1002/prot.20348. [DOI] [PubMed] [Google Scholar]
- 65.del Sol A., Fujihashi H., Amoros D., Nussinov R. Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vijayabaskar M.S., Vishveshwara S. Interaction energy based protein structure networks. Biophys. J. 2010;99:3704–3715. doi: 10.1016/j.bpj.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Böde C., Kovács I.A., Szalay M.S., Palotai R., Korcsmáros T., Csermely P. Network analysis of protein dynamics. FEBS Lett. 2007;581:2776–2782. doi: 10.1016/j.febslet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 68.Daily M.D., Upadhyaya T.J., Gray J.J. Contact rearrangements form coupled networks from local motions in allosteric proteins. Proteins. 2008;71:455–466. doi: 10.1002/prot.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daily M.D., Gray J.J. Allosteric communication occurs via networks of tertiary and quaternary motions in proteins. PLoS Comput. Biol. 2009;5:e1000293. doi: 10.1371/journal.pcbi.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan K.P., Nguyen T.B., Patel S., Varadarajan R., Madhusudhan M.S. Depth: A web server to compute depth, cavity sizes, detect potential small-molecule ligand-binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt503. W314-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baysal C., Atilgan A.R. Relaxation kinetics and the glassiness of proteins: The case of bovine pancreatic trypsin inhibitor. Biophys. J. 2002;83:699–705. doi: 10.1016/S0006-3495(02)75201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freeman L.C. A set of measures of centrality based upon betweenness. Sociometry. 1977;40:35–41. [Google Scholar]
- 73.Di Paola L., De Ruvo M., Paci P., Santoni D., Giuliani A. Protein contact networks: An emerging paradigm in chemistry. Chem. Rev. 2013;113:1598–1613. doi: 10.1021/cr3002356. [DOI] [PubMed] [Google Scholar]
- 74.Nussinov R., Tsai C.J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 75.Wang L., Tharp S., Selzer T., Benkovic S.J., Kohen A. Effects of a distal mutation on active site chemistry. Biochemistry. 2006;45:1383–1392. doi: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isogai S., Deupi X., Opitz C., Heydenreich F.M., Tsai C.J., Brueckner F., Schertler G.F., Veprintsev D.B., Grzesiek S. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature. 2016;530:237–241. doi: 10.1038/nature16577. [DOI] [PubMed] [Google Scholar]
- 77.Doshi U., Holliday M.J., Eisenmesser E.Z., Hamelberg D. Dynamical network of residue-residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. Proc. Natl. Acad. Sci. USA. 2016;113:4735–4740. doi: 10.1073/pnas.1523573113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajasekaran N., Naganathan A.N. A self-consistent structural perturbation approach for determining the magnitude and extent of allosteric coupling in proteins. Biochem. J. 2017;474:2379–2388. doi: 10.1042/BCJ20170304. [DOI] [PubMed] [Google Scholar]
- 79.Marsh D.J., Dahia P.L., Zheng Z., Liaw D., Parsons R., Gorlin R.J., Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 80.Matreyek K.A., Starita L.M., Stephany J.J., Martin B., Chiasson M.A., Gray V.E., Kircher M., Khechaduri A., Dines J.N., Hause R.J. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 2018;50:874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gardino A.K., Villali J., Kivenson A., Lei M., Liu C.F., Steindel P., Eisenmesser E.Z., Labeikovsky W., Wolf-Watz M., Clarkson M.W., Kern D. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nussinov R., Tsai C.J., Csermely P. Allo-network drugs: Harnessing allostery in cellular networks. Trends Pharmacol. Sci. 2011;32:686–693. doi: 10.1016/j.tips.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Campbell R.B., Liu F., Ross A.H. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 84.Redfern R.E., Redfern D., Furgason M.L.M., Munson M., Ross A.H., Gericke A. PTEN phosphatase selectively binds phosphoinositides and undergoes structural changes. Biochemistry. 2008;47:2162–2171. doi: 10.1021/bi702114w. [DOI] [PubMed] [Google Scholar]
- 85.Nanda H., Heinrich F., Lösche M. Membrane association of the PTEN tumor suppressor: Neutron scattering and MD simulations reveal the structure of protein-membrane complexes. Methods. 2015;77-78:136–146. doi: 10.1016/j.ymeth.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anastasio N.C., Gilbertson S.R., Bubar M.J., Agarkov A., Stutz S.J., Jeng Y., Bremer N.M., Smith T.D., Fox R.G., Swinford S.E. Peptide inhibitors disrupt the serotonin 5-HT2C receptor interaction with phosphatase and tensin homolog to allosterically modulate cellular signaling and behavior. J. Neurosci. 2013;33:1615–1630. doi: 10.1523/JNEUROSCI.2656-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moncalero V.L., Costanzo R.V., Perandones C., Radrizzani M. Different conformations of phosphatase and tensin homolog, deleted on chromosome 10 (PTEN) protein within the nucleus and cytoplasm of neurons. PLoS ONE. 2011;6:e18857. doi: 10.1371/journal.pone.0018857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lobo G.P., Waite K.A., Planchon S.M., Romigh T., Nassif N.T., Eng C. Germline and somatic cancer-associated mutations in the ATP-binding motifs of PTEN influence its subcellular localization and tumor suppressive function. Hum. Mol. Genet. 2009;18:2851–2862. doi: 10.1093/hmg/ddp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.