Abstract
Despite significant progress in annotating the genome with experimental methods, much of the regulatory noncoding genome remains poorly defined. Here we assert that regulatory elements may be characterized by leveraging local epigenomic signatures where specific transcription factors (TFs) are bound. To link these two features, we introduce IMPACT, a genome annotation strategy that identifies regulatory elements defined by cell-state-specific TF binding profiles, learned from 515 chromatin and sequence annotations. We validate IMPACT using multiple compelling applications. First, IMPACT distinguishes between bound and unbound TF motif sites with high accuracy (average AUPRC 0.81, SE 0.07; across 8 tested TFs) and outperforms state-of-the-art TF binding prediction methods, MocapG, MocapS, and Virtual ChIP-seq. Second, in eight tested cell types, RNA polymerase II IMPACT annotations capture more cis-eQTL variation than sequence-based annotations, such as promoters and TSS windows (25% average increase in enrichment). Third, integration with rheumatoid arthritis (RA) summary statistics from European (N = 38,242) and East Asian (N = 22,515) populations revealed that the top 5% of CD4+ Treg IMPACT regulatory elements capture 85.7% of RA h2, the most comprehensive explanation for RA h2 to date. In comparison, the average RA h2 captured by compared CD4+ T histone marks is 42.3% and by CD4+ T specifically expressed gene sets is 36.4%. Lastly, we find that IMPACT may be used in many different cell types to identify complex trait associated regulatory elements.
Keywords: transcription, binding, heritability, polygenic, epigenomics, CD4+, T cells, rheumatoid, arthritis, autoimmune
Introduction
Transcriptional regulation is the foundation for many complex biological phenotypes, from gene expression to disease susceptibility. However, the complexity of gene regulation, controlled by more than 1,600 human transcription factors (TFs)1 influencing some 20,000 protein coding genes, has made functional annotation of the regulome difficult. Tens of thousands of genomic annotations have been experimentally generated, enabling the success of unsupervised methods such as chromHMM2 and Segway3 to identify global chromatin patterns that better characterize genomic function. However, linking specific regulatory processes to these identified patterns is challenging. Furthermore, although genome-wide association studies (GWASs) have identified ∼10,000 trait-associated variants across hundreds of polygenic traits,4 most variants lie in noncoding regulatory regions with uncertain function.
With continually increasing numbers of genomic annotations generated from high-throughput experimental assays, in silico functional characterization of variants has growing potential. These assays include genome-wide open chromatin, histone mark, and RNA expression profiling, each separately possible at the single-cell level. Initially contributed by genomic consortia, such as ENCODE5 and Roadmap,6 these assays have become more common as easy-to-implement protocols have been developed, thereby contributing to the growing rate of genomic annotation generation.
Recently, integration of datasets, particularly those indicating regulatory elements, with GWAS data has successfully led to the identification of categories of disease-driving variants enriched for genetic heritability (h2).7, 8, 9 Such regulatory annotations identify active promoters and enhancers through open chromatin or histone mark occupancy assays in a cell type of interest.7, 8, 10, 11, 12, 13 However, these annotations include both cell-type-specific and -nonspecific elements, the latter of which may affect a wide range of cellular functions that are not necessarily intrinsic to disease-driving cell states. Therefore, we hypothesized that the identification of regulatory elements specifically driving functional states would help us to not only better characterize regulatory elements genome-wide, but also better capture polygenic h2 of complex traits and diseases. Once the most enriched classes of regulatory elements are recognized, then it may become possible to generate biologically founded mechanistic hypotheses.
Here, we introduce IMPACT (inference and modeling of phenotype-related active transcription), a diversely applicable genome annotation strategy to predict cell-state-specific regulatory elements. We take a two-step approach to define IMPACT regulatory elements. First, we choose a single key TF, known to regulate a cell-state-specific process, and then identify binding motif sites genome-wide, distinguishing between those that are bound and unbound using genomic occupancy identified by ChIP-seq in the corresponding cell state. Here, the term “cell state specific” refers to the observed experimental binding sites of a key TF, which itself may not be entirely cell state specific, assayed in the target cell state. Second, IMPACT predicts TF occupancy at binding motif sites by aggregating and performing feature selection on 503 cell-type-specific epigenomic features and 12 sequence features in an elastic net logistic regression model. The IMPACT model framework can easily be expanded to accommodate thousands of epigenomic annotations and is amenable to increasing rates of data generation. From this regression we learn a TF binding chromatin profile, which IMPACT uses to probabilistically annotate the genome at nucleotide resolution. We refer to high scoring regions as cell-state-specific regulatory elements (Figure 1). With this approach, we aim to better pinpoint sites of causal variation of gene expression and polygenic trait heritability by modeling trait-driving cell-state-specific regulatory processes.
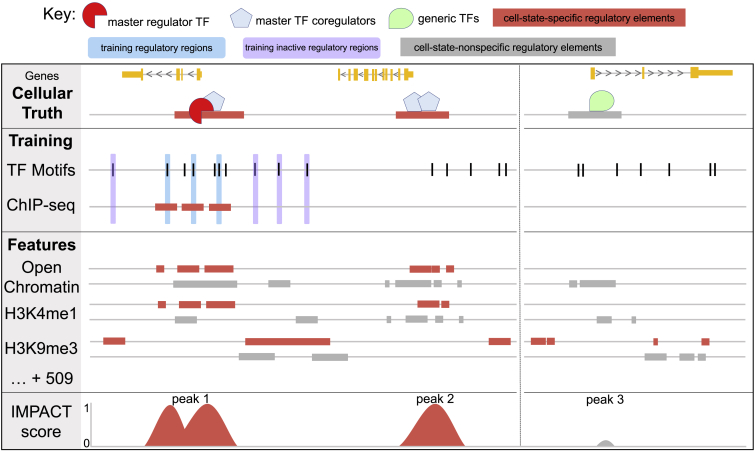
Figure 1.
IMPACT: A Genome Annotation Strategy to Identify Cell-State-Specific Regulatory Elements
IMPACT learns a chromatin profile of cell-state-specific regulation, distinguishing master TF (red) regulatory elements (TF-bound motif sites, blue) from inactive regulatory elements (unbound motif sites, purple). Here, cell-state-specific open chromatin and cell-state-specific H3K4me1 are strong predictors of cell-state-specific regulatory elements. Cell-state-nonspecific open chromatin and nonspecific H3K4me1 are less informative, marking all types of regulatory elements, while H3K9me3 strongly implicates inactive regulatory elements. IMPACT should re-identify regulatory elements marked by master TF binding (peak 1) and those with similar chromatin profiles, presumably sites of related cell-state-specific processes (peak 2). IMPACT should not predict regulation at cell-state-nonspecific elements (peak 3), such as promoters of housekeeping genes.
Material and Methods
Statistical Methods
IMPACT Model
We build a model that predicts TF binding on a motif site by learning the epigenomic profiles of the TF binding sites. We use logistic regression to model the log odds of TF binding on a motif site, or putative binding site, based on a linear combination of the effects of the j epigenomic or sequence features (Table S1), where is an intercept:
where is a value defining some relationship between feature j and the motif site and p is the probability of TF binding at the motif site. From the log odds, which ranges from negative to positive infinity, we compute the probability of TF binding, ranging from 0 to 1:
We use a logistic regression framework with elastic net regularization implemented by the cv.glmnet R (v.1.0.143) package,14 in which optimal are fit according to the following objective function,
where Y represents the binary vector indicating TF bound or unbound motif sites, X is a matrix defining the feature characterization of each motif site, and is the mix term between the ridge (L2), , and lasso (L1), , penalties, where 0 1. We find that no significantly outperforms the others (Figure S1). Therefore, we select = 0.5 to make a compromise between sparsity and information content; enforcing sparsity with lasso performs feature selection thereby helping to avoid overfitting. However, excessive feature selection may remove important information. We use elastic net regularization for two reasons: (1) our model has a large number of features (N > 500) that may result in overfitting if feature selection is not performed (L1 penalty) and (2) the L2 penalty makes the objective function convex, with one stable solution.
Training IMPACT
For each cell state that we model, we train IMPACT to distinguish cell-state-specific regulatory from non-specific or inactive regulatory regions based on cell-state-specific binding of a single key TF. For training, we define the cell-state-specific regulatory class as TF-bound motif sites and the non-specific or inactive regulatory class as unbound motif sites. To define TF-bound motif sites, we use HOMER15 (v.4.8.3) to scan TF ChIP-seq peaks for k-mers with a sequence similarity score, computed from the PWM (position weight matrix) across k nucleotides, that is greater than or equal to the TF binding motif detection threshold, empirically determined by HOMER. Specifically, this log-odds detection threshold is equal to the maximum achievable log odds (computed from the PWM) minus an empirically derived acceptable degree of mismatches. A detailed description of this calculation may be found in the HOMER documentation (Web Resources). We have observed that at most three well-tolerated nucleotide mismatches are permitted for every ten nucleotides. HOMER then scans the genome to assess whether a putative motif site exceeds the detection threshold. The motif-specific detection threshold for each TF used in this study can be found in Table S2. To test how sensitive our selection of training data and genomic annotation is to this parameter, we iterated over multiple motif detection thresholds ranging from lenient to strict (Figure S2). We observe that small changes in the motif log-odds detection threshold lead to modest changes in the proportion of peaks with a detectable motif. For example, decreasing the threshold by 0.5 leads to an increase of at most 10% of ChIP-seq peaks with a detectable motif, and increasing the threshold by 0.5 leads to a decrease of at most 12% of ChIP-seq peaks with a detectable motif. Regarding genomic annotation, we used IFN-G, the quintessential target gene of the TF T-BET, to demonstrate how IMPACT regulatory element probabilities changed in this locus as a result of changing the motif detection threshold (Figure S2). For the following thresholds 4, 5, 6, 6.2 (0.5 lower than the default T-BET detection threshold), 6.7 (default threshold), 7.2 (0.5 greater than the default threshold), 8, and 9, we find that IMPACT regulatory element probabilities do not significantly vary over the IFN-G locus, suggesting that IMPACT genomic annotation is not sensitive to the motif detection threshold parameter.
For each ChIP-seq peak with at least one motif match, we retain only the coordinates of the highest scoring motif match to use in our training set. This ensures that each instance of a bound motif site is in a separate ChIP-seq peak, which avoids double counting ChIP-seq peaks. In terms of training a logistic regression model, this helps to ensure that no two motif instances are in overlapping or proximal genomic coordinates and may be considered independent. We randomly select 1,000 TF-bound motif sites in each training instance.
To define unbound motif sites, we use the genome-wide TF motif scan performed above and select motif matches that do not overlap the corresponding TF ChIP-seq peaks. Then, we retain the genomic coordinates of these matches. We randomly select 10,000 unbound motif sites in each training instance. We select 1,000 bound motif sites and 10,000 unbound motif sites for the following two reasons. First, of all tested TFs, the smallest dataset contained just over 1,000 bound motif sites. Therefore, to uniformly train IMPACT models across TFs, we required the same number of bound motif sites be used in each instance. Second, for the purpose of genome-wide regulatory annotation, we attempt to make our training data represent hypothesized genome-wide regulatory proportions. To this end, we arbitrarily required ten times as many unbound motif sites as bound motif sites to reflect an approximate genome-wide ratio of non-regulatory to regulatory elements, respectively.16, 17 For the purposes of benchmarking IMPACT against state-of-the-art methods, we assessed each model’s performance on the same sets of motif sites.
IMPACT is trained to distinguish TF-bound motif sites from unbound motif sites by their epigenomic and sequence feature characterization. We build a feature matrix by reporting overlap of an annotation and a motif site with a value of 1 and no overlap with a value of 0. Each feature characterization is represented twice in the model, first with respect to local regions and second with respect to distal regions. In the local case, for each motif site and for each feature, we quantify direct positional overlap. In the distal case, we quantify feature overlap with a distal nucleotide relative to the motif site. We reason that although a motif site may not directly overlap a particular feature, such as a promoter, it may be informative to know that there is one nearby. For example, we might look 1,000 nucleotides away from the motif site and report feature overlap at either the upstream or downstream position with a single value of 1 or overlap at neither with a value of 0. After parameter optimization, we set this distance value to 1,000 nucleotides (Figure S1). We do not use absolute distance between annotation and motif site to characterize our feature space in the interest of computational efficiency with specific regard to nucleotide-based genome-wide annotation. Furthermore, IMPACT prediction performance for no TF is significantly improved by using the absolute distance feature characterization strategy (all p > 0.60) (Figure S3).
We note that using motif site-centric gold standards has multiple advantages over predicting TF binding on entire ChIP-seq peak regions. First, using motif sites serves as a quality control for pioneer TF ChIP-seq data, in which case we know the TF is interacting directly with the DNA. Second, it provides an intuitive interpretation for binary labeling as a motif site may be either bound or unbound. Such binary interpretation is not applicable to ChIP-seq peaks, which can each implicate hundreds of nucleotides. Rather than a TF binding uniformly throughout the peak, it is more likely that the ChIP-seq signal is coming from a smaller region of TF binding within the peak, making the use of motif sites an attractive strategy to better localize the signal. Third, it provides TF specificity by focusing on sequences within the peak that only the TF of interest may interact with, whereas within the coordinates of one ChIP-seq peak multiple TFs may be binding. We also observed that on average IMPACT predicts TF binding significantly better when using motif site-centric gold standards according to the AUPRC performance metric (0.18 average increase in AUPRC; all Student’s t test p < 8.3e−36, except for TCF7L2) (Figure S3). Moreover, we find that IMPACT regulatory element probabilities are significantly higher (all p < 0.05, Student’s t test) at nucleotides located in both a motif site and a ChIP-seq peak (Figure S3), suggesting that motif sites provide a non-redundant layer of regulatory information beyond ChIP-seq peak signal. These results suggest that IMPACT’s ability to score motif sites with higher regulatory potential might be used as a strategy to perform quality control on ChIP-seq peaks.
To train the elastic net logistic regression model, we partition the sets of TF-bound and -unbound motif sites by randomly sampling 80% of each set, to be used for 10-fold cross validation (CV), in which these subsets are further partitioned into 90%/10% train/test. The remaining 20%, completely unseen by the CV and not overlapping with the initial 80%, is used as a validation set. In this binary classification problem, probabilistic outputs from the logistic regression are made binary by applying thresholds in the CV. The threshold vector is a sequence from 0 to 1, with resolution of 0.0025, resulting in 401 applied thresholds. We applied IMPACT genome-wide to assign nucleotide-resolution cell-state-specific regulatory element probabilities, using the model learned from the elastic net logistic regression CV.
Interpreting IMPACT Regulatory Element Probabilities
Genome-wide, IMPACT evaluates each nucleotide’s regulatory element potential with respect to a particular TF/cell-state pair and assigns a probability to each nucleotide. In order to understand these probabilities, we compare their distribution across the bound motif site class and the unbound class. As expected, we observe significantly higher IMPACT predictions at TF-bound motif sites compared to unbound motif sites (all p < 1e−3, Student’s t test); unbound motif sites have regulatory probabilities near 0 (Figure S4). This separation informs the interpretation of genome-wide predictions: truly inactive/non-specific regulatory elements are expected to have predicted values close to 0, rather than an arbitrary or uninterpretable non-zero decision boundary.
cis-eQTL Causal Variation Enrichment
We computed a genome-wide enrichment of cis-eQTL causal association across various functional annotations. To this end, we gathered gene-based cis-window summary statistics. Then, for each gene and for each annotation, an enrichment was calculated explicitly as:
where g is the gene, a is the annotation, N is the number of variants within annotation a, M is the number of variants outside annotation a, i is the ith variant, j is the jth variant, and is the chi-square statistic of the association between gene g and SNP i or j. We then computed genome-wide standard errors by block jackknifing the genome into 200 adjacent bins and computed a distribution of enrichment values when leaving one bin out at a time.7 This strategy is designed to prevent the genes of any one region of the genome from dominating the enrichment statistic. Furthermore, we used a permutation strategy to establish a null distribution. To this end, we randomly permuted the chi-square associations in the cis-window of each gene 1,000 times, while matching on 50 LD bins across the cis-window, and recomputed the enrichment with each of the functional annotations. We estimated enrichment significance based on how extreme our result was compared to the permutation distributions.
Partitioning Heritability with S-LDSC
We apply S-LDSC7 (stratified linkage disequilibrium [LD] score regression) (v.1.0.0), a method developed to partition polygenic trait heritability by one or more functional annotations, to quantify the contribution of IMPACT cell-state-specific regulatory annotations to 42 complex traits. We annotate common SNPs (MAF ≥ 0.05) with regulatory element probabilities based on cell-state-specific IMPACT models. Then, we run S-LDSC once on the annotated SNPs to compute population-specific LD scores and again to quantify the complex trait heritability captured by our IMPACT annotations. Here, the two statistics we use to evaluate how well our annotations capture causal variation are enrichment and standardized effect size (∗).
If acj is the value of annotation c for SNP j, we assume the variance of the effect size of SNP j depends linearly on the contribution of each annotation c:
where is the per-SNP contribution from one unit of the annotation to heritability. To estimate , S-LDSC estimates the marginal effect size of SNP j in the sample from the chi-square GWAS statistic :
Considering the expectation of and following the derivation from Gazal et al.,9
where N is the sample size of the GWAS, is the LD score of SNP j with respect to annotation c, and is the true, e.g., population-wide, genetic correlation of SNPs j and k. We define enrichment of an annotation as the proportion of heritability explained by the annotation divided by the average value of the annotation across the M common (MAF 0.05) SNPs. Enrichment may be computed for binary or probabilistic annotations according to the equation below, where is the h2 explained by SNPs in annotation c:
Since is not comparable between annotations or traits, 9 is defined as the per-annotation standardized effect size, or the proportionate change in per-SNP h2 associated with a one standard deviation increase in the value of the annotation. is a function of the standard deviation of the annotation c, sd(c), the trait-specific SNP-heritability estimated by LDSC, , and the total number of reference common SNPs used to compute , M = 5,961,159 in Europeans (EUR) and 5,469,053 in East Asians (EAS):
∗ captures the unique contribution of an annotation to capturing h2 in the S-LDSC model, conditional on other provided annotations. Specifically, a ∗ of 0 means that the annotation does not change per-SNP h2, a strongly negative ∗ means that membership to the categorical annotation decreases per-SNP h2, and a strongly positive ∗ means that membership to the annotation increases per-SNP h2. The significance of ∗ is computed based on a test of how different from 0 the ∗ is. We emphasize that enrichment does not quantify effects that are unique to a given annotation, whereas ∗ does. When conditionally comparing two annotations, say A and B, in a joint S-LDSC model, both annotations may have similar enrichments if they are highly correlated. However, the ∗ for the annotation with greater true causal variant membership will be larger and more significantly positive. Previous work has reported that the threshold for impactful values of ∗| is approximately 0.24.9
Each S-LDSC analysis conditions IMPACT annotations on 69 baseline annotations, a subset of the 75 annotations referred to as the baseline-LD model;9 we removed 6 annotations including T cell enhancers, since IMPACT T cell-state annotations are likely correlated. The 69 annotations consist of 53 cell-type-nonspecific annotations,7 which include histone marks and open chromatin, 10 MAF bins, and 6 LD-related annotations9 to assess whether functional enrichment is cell type specific and to control for the effect of MAF and LD architecture. Consistent inclusion of MAF- and LD-associated annotations in the baseline model is the standard recommended practice of using S-LDSC.
Fine-Mapped RA Posterior Probability Enrichment in IMPACT Regions
For each of 20 chosen RA-associated loci,18 we computed the enrichment of posterior probabilities in the top 1% of cell-state-specific IMPACT regulatory elements. For each RA-associated locus l, we define
where is the posterior causal probability of SNP i, such that i belongs to the top 1% of the cell-state-specific IMPACT annotation c and is the number of SNPs in locus l for which we previously computed a posterior probability.18 The denominator represents the null hypothesis that each SNP in a locus is equally causal. We computed the average of these enrichment values over the 20 RA-associated loci. We assessed significance based on comparison to 10,000 permutation distributions, designed by computing an average enrichment value over these 20 loci, in which random posterior probabilities (of the same quantity ) were selected.
Data
Genome-wide Annotation Data
We obtained publicly available genome-wide epigenomic annotations including ATAC-seq, DNase-seq, FAIRE-seq, HiChIP, polymerase and elongation factor ChIP-seq, and histone modification ChIP-seq assayed in hematopoietic, adrenal, brain, cardiovascular, gastrointestinal, skeletal, and other cell types for the GRCh37 (hg19) assembly (Table S1). Sequence annotations, downloaded from UCSC, include Phastcons conservation, exons, introns, intergenic regions, 3′ UTR (untranslated region), 5′ UTR, promoter-TSS (transcription start site), TTS (transcription termination site), and CpG islands. For benchmarking IMPACT against MocapG,19 MocapS,19 and Virtual ChIP-seq,20 we additionally acquired corresponding cell-type-specific open chromatin and gene expression where applicable (Table S3). For models trained on Pol II ChIP-seq, we removed Pol II and elongation factor ChIP-seq feature tracks from the feature library before running IMPACT.
TF ChIP-Seq Data
We determined genome-wide TF occupancy from publicly available ChIP-seq (Table S4) of 13 key regulators (T-BET,21, 22 GATA3,23 STAT3,24 FOXP3,25 STAT5,25 IRF5,26 IRF1,27 CEBPB,28 PAX5,29 REST,5 RXRA,5 HNF4A,30 TCF7L25) assayed in primary cell states which they have been observed to regulate: Th1, Th2, Th17, Treg cells, macrophages, monocytes, monocytes, B cells, fetal brain cells, brain cells, liver cells, and pancreatic cells, respectively. We additionally acquired ChIP-seq of RNA polymerase II in peripheral blood/lymphocytes, fibroblasts, stomach, liver, left ventricle heart, sigmoid colon, pancreas, and CD4+ T cells.5, 22, 31 All ChIP-seq peaks were called by macs32 (v.1.4.2 20120305) (all p < 1e−5).
cis-eQTL Data
We acquired SNP-level summary statistics from three independent studies. First, we obtained data from 3,754 peripheral blood samples33 in which 7,025 unique genes had measurements. As some genes were represented by several array probes, we retained only summary statistics on one probe, selected randomly, per gene. Second, we obtained data from GTEx V7 (Web Resources) in the following six cell types with the number of samples listed in parentheses: transformed fibroblasts (300), stomach (237), liver (153), left ventricle of heart (272), sigmoid colon (203), and pancreas (220). On average across these cell types, approximately 22,000 genes had measurements in the GTEx data. Third, we obtained eQTL data from CD4+ T cells in East Asian individuals (N = 103) with expression measurements for 20,107 genes.34 For each gene, we truncated the genome-wide summary statistics to a cis window of 1 Mb upstream and downstream of the gene TSS.
Genome-wide Association Data Used in S-LDSC Analyses
We collected RA GWAS summary statistics35 for 38,242 European individuals, combined case and control subjects, and 22,515 East Asian individuals, comprised of 4,873 RA-affected case subjects and 17,642 control subjects.36 We estimated total genome-wide polygenic RA h2 to be about 18% for EUR and 21% for EAS. We further collected 41 other complex trait summary statistics.9, 37, 38 Reference SNPs, used to estimate European LD scores, were the set of 9,997,231 SNPs with minor allele count greater than or equal to five in a set of 659 European samples from phase 3 of 1000 Genomes Projects.39 The regression coefficients were estimated using 1,125,060 HapMap3 SNPs and heritability was partitioned for the 5,961,159 reference SNPs with MAF ≥ 0.05. Reference SNPs, used to estimate East Asian LD scores, were the set of 8,768,561 SNPs with minor allele count greater than or equal to five in a set of 105 East Asian samples from phase 3 of 1000 Genomes Projects.39 The regression coefficients were estimated using 1,026,051 HapMap3 SNPs and heritability was partitioned for the 5,469,053 reference SNPs with MAF ≥ 0.05. Frequency and weight files (1000G EUR phase3, 1000G EAS phase3) are publicly available and may be found in the Web Resources.
Fine-Mapped RA Causal Variation
Previous work from our group aimed to define the most likely causal RA variant for each locus harboring a genome-wide significant variant,18 identified by a GWAS of 11,475 European RA-affected case subjects and 15,870 control subjects.40 To this end, causal posterior probabilities were computed with the approximate Bayesian factor (ABF), assuming one causal variant per locus. The posteriors were defined as:
where i is the ith variant and n is the total number of variants in the locus. As such, the ABF over all variants in a locus sum to 1.
Results
IMPACT Accurately Predicts Transcription Factor Binding
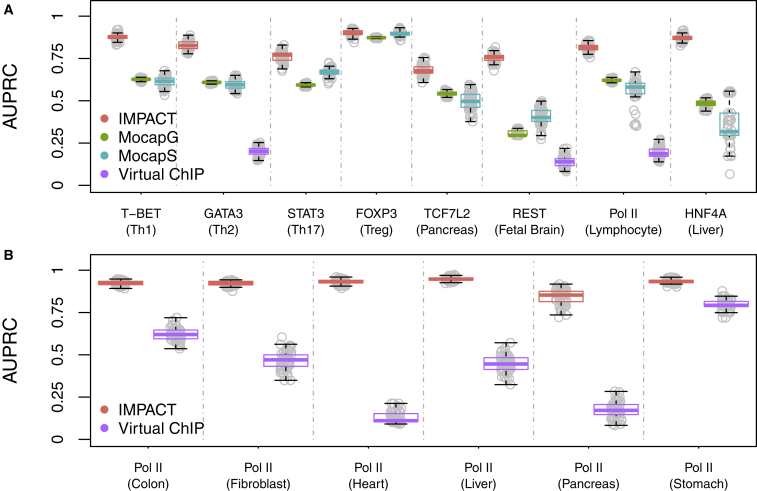
The IMPACT model assumes that cell-state-specific TF binding sites and related regulation may be characterized by a quantitative epigenomic signature. If this is true, IMPACT might predict cell-state-specific genome-wide TF occupancy with high accuracy, which has proven to be a challenging task (see ENCODE-DREAM challenge in Web Resources), leading to a diverse set of TF binding prediction strategies.19, 20, 41, 42 To test this model assumption, we used IMPACT to predict regulatory elements based on experimental binding identified via ChIP-seq of eight tested TFs assayed in eight different cell states: T-BET, GATA3, STAT3, FOXP3, REST, HNF4A, TCF7L2, and RNA polymerase (Pol) II in CD4+ Th1 (T helper 1), CD4+ Th2, CD4+ Th17, CD4+ Treg (T regulatory), fetal brain, liver, pancreatic, and lymphocytic cells, respectively5, 21, 22, 23, 24, 25, 30 (see Material and Methods). We observe that IMPACT predicts TF occupancy with high accuracy across eight tested TFs. The average area under the precision-recall curve (AUPRC) over 50 random sampling trials is 0.81 (SE 0.07), computed via 10-fold cross validation on 80% of data, with AUPRC evaluated on the withheld 20% (Figure 2A). We additionally evaluate IMPACT using Matthew’s correlation coefficient (MCC), mean MCC 0.70 (SE 0.08), and show full precision-recall curves (Figure S5). Next, we compared IMPACT TF binding prediction performance to several recent state-of-the-art methods MocapG,19 MocapS,19 and Virtual ChIP-seq.20 Briefly, MocapG is an unsupervised TF binding prediction method that models “cut counts” from cell-type-specific open chromatin (DNase-seq) with negative binomial distributions. MocapS is a supervised sparse logistic regression approach that predicts TF binding using cell-type-specific DNase-seq cut count modeling from MocapG, TF footprint scores from the same DNase-seq data, conservation scores, GC content, CpG island information, sequence mappability scores, and distance to nearest TSS. Virtual ChIP-seq is a multi-layer perception that predicts TF binding, which similarly uses conservation, cell-type-specific DNase-seq, but also leverages cell-type-specific gene expression from RNA-seq and TF-specific ChIP-seq data over a range of cell types and cell lines. While benchmarking, each method had access to the same training and testing data to ensure fair comparison. We observe that on average, across the eight tested TFs, IMPACT outperforms all three methods: AUPRC IMPACT > MocapG (all p < 1.5e−16, Student’s t test; 0.23 average increase in AUPRC), IMPACT > MocapS (all p < 5.4e−30, except for FOXP3 [p = 0.15]; 0.24 average increase in AUPRC), IMPACT > Virtual ChIP-seq (all p < 8.5e−98; 0.62 average increase in AUPRC) (Figure 2A). We note that using Virtual ChIP-seq we were only able to predict binding for GATA3, REST, and Pol II due to data limitations. In light of this, we predicted Pol II binding in six additional cell types: sigmoid colon, fibroblast, left ventricle heart, liver, pancreas, and stomach. We observed that on average, IMPACT outperforms Virtual ChIP-seq according to the AUPRC (all p < 4.9e−38, Student’s t test; 0.48 average increase in AUPRC) (Figure 2B). We additionally used MCC as a metric to compare TF binding prediction performance, in which IMPACT also on average outperforms the competing methods (all p < 2.0e−39 for MocapG, 0.30 average increase in MCC; all p < 1.2e−22 for MocapS, 0.29 average increase in MCC; all p < 1.2e−77 for Virtual ChIP-seq, 0.49 average increase in MCC), with the following exceptions: MCC FOXP3 IMPACT < MocapG (p < 4.3e−18), MocapS (p < 3.9e−19) (Figure S6).
Figure 2.
IMPACT Outperforms State-of-the-Art TF Binding Prediction Methods
(A) IMPACT outperforms MocapG, MocapS, and Virtual ChIP-seq in predicting cell-state-specific TF binding across 8 TFs, illustrated by AUPRCs on the same training and testing data across 50 trials, with the exception of the MocapS model for FOXP3.
(B) Prediction of Pol II binding in 6 cell types reveals that IMPACT outperforms Virtual ChIP-seq.
Genome-wide IMPACT Regulatory Annotations
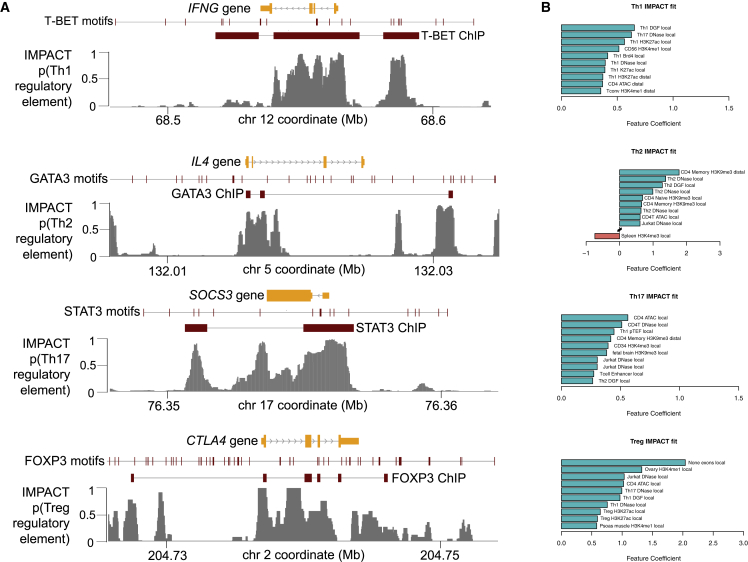
For each of the eight tested TFs, we created genome-wide IMPACT regulatory annotations. Focusing on the four CD4+ T cell-state IMPACT annotations, we illustrate that IMPACT regulatory element probabilities vary dynamically within TF ChIP-seq peaks near canonical CD4+ T cell-state genes. This reflects the high-resolution information that is gained by integrating hundreds of epigenomic and sequence annotations (Figures 3A and S7). Furthermore, we observe that the most heavily weighted features from the logistic regression, indicating TF binding, include cell-state-specific open chromatin and activating histone modifications, as expected (Figure 3B). When training on entire ChIP-seq peaks rather than motif sites within peaks, top weighted features generally have less relevant cell-state specificity (Figure S8), possibly due to high correlation of ChIP-seq signal between CD4+ T cell states. For example, most regulatory elements across CD4+ T cell states may be near similar target genes. While the motif site regions used to train each model are TF specific, independent, and non-overlapping, we still observe relatively high correlations between CD4+ T cell-state IMPACT annotations compared to the epigenomic annotations used to train the models (Figure S9).
Figure 3.
IMPACT Genome-wide Regulatory Tracks
(A) Cell-state-specific regulatory element IMPACT predictions for canonical target genes of T-BET, GATA3, STAT3, and FOXP3.
(B) Highly weighted features of Th1, Th2, Th17, and Treg IMPACT annotations.
The IMPACT epigenomic feature library contains 515 features across many cell and assay types but the importance of annotation categories is not immediately clear. To this end, we systematically removed categories of annotations and retrained TF/cell-state models (Figure S10). First, we observed that TF binding predictive performance significantly decreases upon removal of cell-type-specific features for four of seven TFs (all p < 8.1e−4, Student’s t test). For the three TFs with no significant decrease in performance, this result suggests that presence of annotations from biologically similar cell states may be sufficient to train a high-performing IMPACT model, without requiring annotations specifically assayed in the target cell state. Second, using just histone modification tracks resulted in significantly decreased performance on average (all p < 6.3e−06), while using just open chromatin tracks led to decreased performance for five of seven tested TFs (all p < 2.1e−05) and did not significantly affect the performance for STAT3 and FOXP3. Third, we observed significantly lower performance when restricting to cell-type-specific H3K4me1 (all p < 2.0e−13), except for STAT3 where we observe significantly higher performance (p < 2.5e−44), suggesting that using cell-type-specific features only are generally less informative than a diversity of cell types and assay types. Fourth, we observed that using only cell-type-specific open chromatin results in significantly lower performance for T-BET, TCF7L2, and HNF4A (all p < 4.9e−17), while, for GATA3 and REST, performance improved (both p < 4.5e−3); no comparison could be made for STAT3 or FOXP3 because there were no Th17 or Treg open chromatin annotations to begin with. From this, we learn that integration of diverse cell types and assays generally leads to improved predictive performance. In the case of GATA3, STAT3, and REST, where the use of only cell-type-specific annotations resulted in improved performance over the canonical IMPACT model, such models may overfit to training data and misrepresent true TF binding patterns genome-wide. Therefore, further assessment is necessary, specifically involving training and testing across multiple datasets from the same cell type, which was not possible in this study due to scarcity of primary cell TF ChIP-seq data.
Improved Enrichment of Gene Expression Causal Variation
We developed IMPACT to model regulation specific to a functional cell state, the most general of which may be active cellular transcription. Expression quantitative trait loci (eQTLs) are genetic variations that modulate transcription.43 Most cis-eQTLs map to TSS and promoter annotations, and more rarely to the 5′ UTR.44 We hypothesized that an IMPACT annotation tracking active transcription, trained on RNA polymerase (Pol) II binding sites, would capture cis-eQTL causal variation better than the most strongly enriched canonical eQTL-related annotations.
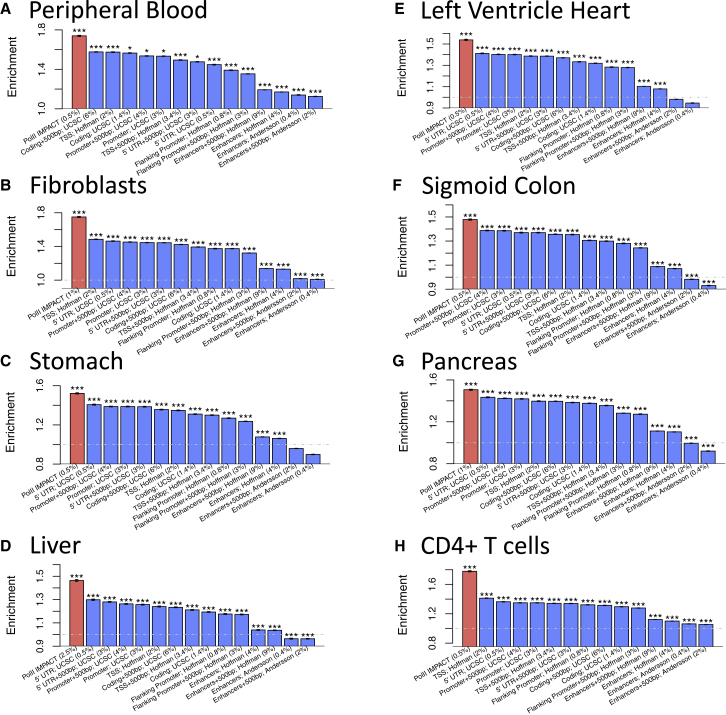
We obtained SNP-level summary statistics from three independent sources: first, from a large and previously published eQTL analysis on 3,754 peripheral blood samples;33 second, from GTEx V7 across 6 tissue types (average sample size = 231), i.e., transformed fibroblasts, stomach, liver, left ventricle heart, sigmoid colon, and pancreas; and third, from a CD4+ T cell eQTL analysis on 103 East Asian individuals.34 We then used IMPACT to annotate SNPs tested in the eQTL analysis with RNA Pol II specific regulatory element probabilities, separately for each tissue or cell type. In this analysis, we were limited by the availability of Pol II ChIP-seq, for which there is an abundance of tissue-specific data but rarely more specific cell-type-level data. While tissues may contain many different cell types, we expect IMPACT to learn an epigenomic signature as general or as specific as the training data provided. For the peripheral blood IMPACT annotation, we combined sites of Pol II binding in both T cells and B cells, the predominant cell populations of peripheral blood, and trained a single IMPACT model. Next, we computed a genome-wide enrichment (see Material and Methods) of chi-square cis-eQTL association statistics, averaged over all genes with at least one significant eQTL, across Pol II IMPACT, Pol II ChIP-seq, and several sequenced-based annotations, such as TSS windows, promoters, and enhancers. We observed that on average Pol II IMPACT across all cell types was more enriched for chi-square association than the Pol II ChIP-seq used for training (paired t test p < 7.7e−4, 7.3% average increase in enrichment). To compute this, we thresholded each Pol II ChIP-seq dataset by peak score, considering 11 uniformly spaced cutoffs, ranging from highest-scoring ChIP-seq peaks to lowest-scoring, while still significant, peaks. To directly compare with IMPACT, we appropriately thresholded each Pol II IMPACT annotation by matching on size, e.g., the genome-wide proportion of SNPs annotated (Figure S11). We also computed enrichment for IMPACT at five other annotation size thresholds (0.5%, 1%, 2.5%, 5%, and 10%), which resulted in larger enrichments than achievable by any thresholding of the ChIP-seq data. Furthermore, we observe that Pol II IMPACT captures more chi-square association than sequenced-based functional annotations (Student’s t test p < 4.8e−4) with the highest performing IMPACT annotations providing a 25% average increase in enrichment over the sequenced-based annotations (Figure 4). For each of the eight tissues or cell types tested, the most enriched Pol II IMPACT annotation outperformed all sequenced-based functional annotations. Specifically, in peripheral blood, Pol II IMPACT introduced a 1.7× enrichment (permutation p < 1e−3), corresponding to a 24% average increase in enrichment compared to the tested sequence-based functional annotations. Similarly, for transformed fibroblasts, Pol II IMPACT introduced a 1.7× enrichment (30% increase); for stomach, a 1.5× enrichment (22% increase); for liver, a 1.5× enrichment (25% increase); for left ventricle heart, a 1.5× enrichment (22% increase); for sigmoid colon, a 1.5× enrichment (22% increase); for pancreas, a 1.5× enrichment (18% increase); and for CD4+ T cells, a 1.8× enrichment (41% increase).
Figure 4.
Pol II IMPACT Captures cis-eQTL Causal Variation Better than Sequence-Based Annotations across Eight Cell and Tissue Types
Enrichment of cis-eQTL chi-square association values with Pol II IMPACT annotations, created for peripheral blood (A), fibroblasts (B), stomach (C), liver (D), left ventricle heart (E), sigmoid colon (F), pancreas (G), and CD4+ T cells (H), highlighting top performing IMPACT annotation compared to enrichments of sequence-based functional annotations. Values in parentheses after annotation name are the average annotation value across all common variants, e.g., the effective size of the annotation. ∗ denotes permutation p < 0.05, ∗∗ permutation p < 0.01, ∗∗∗ permutation p < 0.001. Intervals at the top of each bar represent the 95% confidence interval of the enrichment estimate.
Improved Capture of Rheumatoid Arthritis Causal Variation
We previously hypothesized that IMPACT annotations of pathogenic cell states would more precisely capture polygenic trait h2, compared to regulatory annotations that don’t resolve cell states. Testing this hypothesis requires a polygenic trait with a well-studied disease-driving cell type. Genetic studies of rheumatoid arthritis (RA), an autoimmune disease that attacks synovial joint tissue leading to permanent joint damage and disability,45 have suggested a critical role by CD4+ T cells.7, 8, 11, 12, 46, 47, 48, 49, 50 However, CD4+ T cells are extremely heterogeneous: naive CD4+ T cells may differentiate into memory T cells, and then into effector T cells including Th1, Th2, and Th17 and T regulatory cells, requiring the action of a limited number of key transcription factors (TFs): T-BET or STAT4, GATA3 or STAT6, STAT3 or RORt, FOXP3 or STAT5, respectively.51 As these CD4+ T effector cell states contribute to RA risk,7, 11, 49 we hypothesized that CD4+ T cell-state-specific IMPACT regulatory element annotations would better capture RA h2 than annotations that generalize CD4+ T cells and ignore the differential functionality of effector cell states.
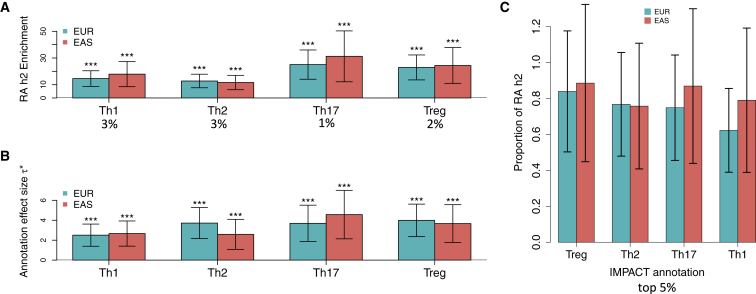
To this end, we built IMPACT annotations in four CD4+ T cell states: Th1, Th2, Th17, and Treg. We then integrated S-LDSC7 with publicly available European (EUR, N = 38,242)7, 35 and East Asian (EAS, N = 22,515)36 RA GWAS summary statistics to partition the common SNP h2 of RA. We use two metrics to evaluate how well our IMPACT annotations capture RA h2: enrichment and per-annotation standardized effect size, ∗ (see Material and Methods). Briefly, enrichment is defined as the proportion of h2 divided by the genome-wide proportion of SNPs in the annotation, and ∗ is defined as the proportionate change in per-SNP h2 associated with a one standard deviation increase in the value of the annotation.9
We observe that each CD4+ T cell-state-specific IMPACT annotation is significantly enriched with RA h2 in both EUR and EAS populations (average enrichment = 20.05, all p < 1.9e−04, Figure 5A, Table S5). Furthermore, we find that ∗ is significantly positive for all CD4+ T IMPACT annotations separately conditioned on the cell-type-nonspecific baseline-LD annotations (all p < 2.1e−03, Figures 5B and S12), supporting the CD4+ T cell-specific role in RA. We then selected the top 5% of regulatory SNPs according to each CD4+ T IMPACT annotation and find that all the CD4+ T cell-state annotations explain a large proportion of RA h2, but the Treg annotation explains the greatest proportion, capturing 85.7% (SE 19.4%, enrichment p < 1.6e−5) of RA h2 meta-analyzed between both EUR and EAS populations (Figure 5C). Furthermore, we observe that the top 9.8% of CD4+ Treg IMPACT regulatory elements, consisting of all SNPs with a non-zero annotation value, capture 97.3% (SE 18.2%, enrichment p < 7.6e−7) of RA h2 in EUR. This powerful result is the most comprehensive explanation for RA h2, to our knowledge, to date.
Figure 5.
CD4+ T Cell-State IMPACT Annotations Are Strongly Enriched for RA Heritability
(A) Enrichment of RA h2 in CD4+ T IMPACT for EUR and EAS populations. Values below cell states are the average annotation value across all common (MAF ≥ 0.05) SNPs, e.g., the effective size of the annotation.
(B) Standardized annotation effect size (∗) of each annotation separately conditioned on annotations from the baseline-LD model.
For (A) and (B), ∗∗∗p < 0.001.
(C) Proportion of total causal RA h2 explained by the top 5% of SNPs in each IMPACT annotation.
For all panels, intervals at the top of each bar represent the 95% confidence interval.
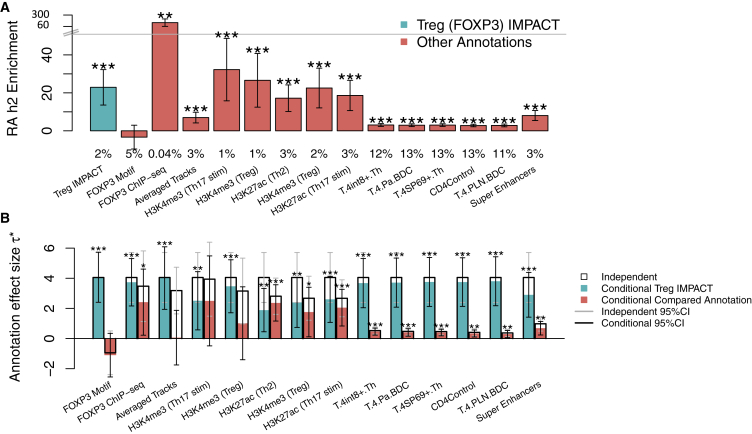
We then assessed whether CD4+ T IMPACT annotations offered improved enrichments of RA h2 compared to canonical CD4+ T cell functional annotations, using S-LDSC and EUR RA summary statistics (Figures 6A and S13). Here, we highlight our comparison of the CD4+ Treg IMPACT annotation to FOXP3 binding motif sites, genome-wide FOXP3 ChIP-seq, the “Averaged Tracks” annotation, which assigns each SNP a value proportional to the number of overlapping IMPACT epigenomic features, the five largest ∗ CD4+ T cell-specific histone mark annotations,7 the five largest ∗ CD4+ T cell-specifically expressed gene sets,8 and CD4+ T cell super enhancers.52 We observe that the CD4+ Treg IMPACT annotation (enrichment = 22.9, SE 4.8, p < 5.2e−08) is significantly more enriched (p < 0.05) for RA h2 than the FOXP3 motif site annotation (enrichment not significantly different from 0), the “Averaged Tracks” annotation (enrichment = 7.0, SE 1.4), all CD4+ T cell-specifically expressed gene sets (average enrichment = 2.9, SE 0.8), and CD4+ T cell super enhancers (enrichment = 8.1, SE 1.3). On the other hand, the FOXP3 ChIP-seq annotation (enrichment = 173.3, SE 58.3), which is used to train the CD4+ Treg IMPACT model, is more strongly enriched (p < 0.05) for RA h2 than the CD4+ Treg IMPACT annotation itself. We additionally created functional annotations representing the overlap of TF ChIP-seq with TF motif sites, as such a combination might improve the enrichment observed for TF ChIP-seq alone. However, these annotations are very small (average annotation size = 0.004% of SNPs) and resulted in non-significant enrichments in the S-LDSC framework. Finally, we observe that all compared CD4+ T cell histone mark annotations are similarly enriched for RA h2, relative to the CD4+ Treg IMPACT annotation (23.4× on average compared to 22.9×, respectively). We note that the average RA h2 captured by these CD4+ T histone mark annotations, ranging in size from 1% to 3% of SNPs, is 42.3%, and the average RA h2 captured by these CD4+ T specifically expressed gene set annotations, ranging in size from 11% to 13% of SNPs, is 36.4%. In terms of total RA h2 explained by a single annotation, these values pale in comparison to the 85.7% of RA h2 captured by the top 5% of SNPs in the Treg IMPACT annotation.
Figure 6.
CD4+ Treg IMPACT Annotation Significantly Captures RA Heritability Conditional on Strongly Enriched CD4+ T Cell Regulatory Annotations
(A) RA h2 enrichment of the CD4+ Treg IMPACT annotation and compared T cell functional annotations. Values below cell states represent the effective size of the annotation. From left to right, we compare Treg IMPACT to genome-wide FOXP3 motif sites, FOXP3 ChIP-seq, the “Averaged Tracks” annotation, which assigns each SNP a value proportional to the number of overlapping IMPACT epigenomic features, the top five cell-type-specific histone modification annotations,7 in terms of independent ∗, the top five cell-type-specifically expressed gene sets (Web Resources),8 in terms of independent ∗, and T cell super enhancers.52
(B) CD4+ Treg IMPACT annotation standardized effect size (∗, teal) conditional on other T cell-related functional annotations (coral).∗ for independent analyses are denoted by the top of each black bar, as a reference for the conditional analyses, denoted by the top of each colored bar.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. For both panels, intervals at the top of each bar represent the 95% confidence interval.
Next, in order to quantify annotation-specific effects of capturing RA h2, we computed the per-annotation standardized effect size, ∗, of each annotation from the previous analysis, conditioned on baseline-LD annotations. We then separately conditioned each CD4+ T cell-state IMPACT annotation jointly on the compared annotations and baseline-LD annotations. Larger and more significantly positive ∗ identifies the annotation that better captures RA h2. We observe that the∗ of both CD4+ Treg and Th2 IMPACT annotations are larger and more significantly positive (all Treg ∗ > 1.9, p < 5.0e−3; all Th2 ∗ > 1.7, p < 0.01) than compared T cell annotations, excluding H3K27ac in Th2 cells, illustrated by taller teal bars than coral bars (Figures 6B and S13). Here, we specifically highlight the CD4+ Treg IMPACT annotation; although the FOXP3 ChIP-seq annotation was more strongly enriched for RA h2 than CD4+ Treg IMPACT, the ∗ of the IMPACT annotation is larger and more significantly positive. Overall, these results suggest that IMPACT annotates areas of concentrated RA h2 that other T cell regulatory annotations do not.
IMPACT Annotation Effect Sizes across 42 Polygenic Traits
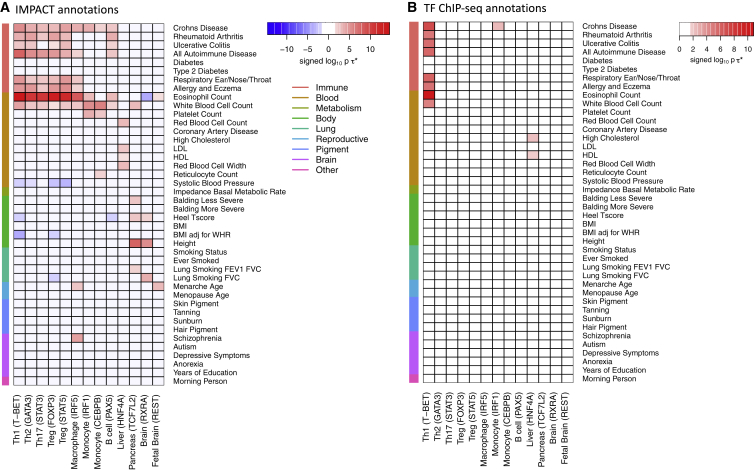
We next applied our CD4+ T IMPACT annotations to 41 additional polygenic traits9, 37, 38 and observed consistently significantly positive per-annotation standardized effect sizes, ∗, for immune-mediated traits, such as Crohn, “all autoimmune disease,” respiratory ear/nose/throat, and “allergy and eczema” (mean ∗ = 3.2; all p < 5.9e−4, p < 1.9e−5, p < 3.6e−3, p < 1.7e−3, respectively), and for several blood traits, eosinophil and white blood cell counts (mean ∗ = 2.5; all p < 1.6e−11, p < 0.02, respectively), but not for non-immune-mediated traits (Figure 7A, Table S6). We then created several different cell-state-specific IMPACT annotations targeting h2 in a range of traits, and we highlight a few examples. For a liver IMPACT annotation, trained on HNF4A5 (hepatocyte nuclear factor 4A), ∗ is positive for liver-associated traits30, 53 LDL and HDL (mean ∗ = 2.0; p < 0.02, p < 1.2e−3, respectively). For a macrophage IMPACT annotation, trained on IRF5,26 ∗ is positive for some immune-mediated and blood traits (mean ∗ = 2.8, all p < 8.2e−3) and intriguingly also for schizophrenia (∗ = 0.9, p < 4.9e−5), supported by studies implicating a putative MHC association.54 Finally, for a CD4+ Treg IMPACT annotation, trained on STAT5,25 an alternative key TF for Treg cells, the values of ∗ across all traits resemble that of FOXP3. This suggests that IMPACT is capturing RA polygenic h2 by annotating loci important to Treg function, rather than TF-specific loci. To ensure that IMPACT annotations were an improvement over the original ChIP-seq used to train each model, we compute ∗ across the same 42 traits for annotations created from the training TF ChIP-seq data (Figure 7B). We observe fewer significant effect sizes, with the exception of stronger ∗ in the T-BET ChIP-seq compared to the T-BET (Th1) IMPACT annotation, first identified in the conditional analysis in Figure S13. Overall, this suggests that IMPACT is a promising strategy to identify complex trait-associated regulatory elements across a range of cell states.
Figure 7.
IMPACT Cell-State-Specific Regulatory Element Annotation Effect Sizes across 42 Polygenic Traits
(A) Signed log10 p values of ∗ for 42 traits across 13 cell-state-specific IMPACT annotations, capturing h2 in distinct sets of complex traits, shown by significantly positive ∗. Each IMPACT annotation is described by its target cell state and key TF used for training in parentheses.
(B) Signed log10 p values of ∗ for 42 traits across annotations representing the TF ChIP-seq used to train the corresponding IMPACT annotations. ChIP-seq annotations are described by the cell state in which the particular TF (in parentheses) was assayed.
Color shown only if p value of ∗ < 0.025 after multiple hypothesis correction.
A Priori Functional Characterization of Variants
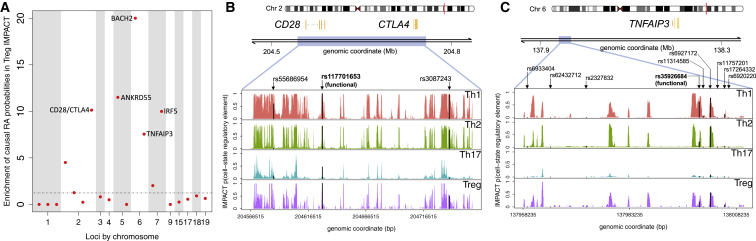
We next hypothesized that improved genomic annotation provided by IMPACT might inform functional variant fine-mapping. Using a GWAS of 11,475 European RA-affected case subjects and 15,870 control subjects,40 an independent study from the European RA summary statistics used in our h2 analyses, our group recently fine-mapped a subset of 20 RA risk loci, each with a manageable number of putatively causal variants, and created 90% credible sets of these SNPs.18 We computed the enrichment of fine-mapped causal probabilities across these 20 loci in the top 1% of our CD4+ T cell-state-specific IMPACT annotations (see Material and Methods). We found that the Treg annotation is significantly enriched (2.87, permutation p < 1.8e−02) while other annotations are not (Table S7). The Treg IMPACT annotation may thus be useful to prune putatively causal RA variants. Furthermore, we observe uniquely high Treg enrichment in the BACH2 and IRF5 loci (16.2 and 8.1, respectively, Figure 8A), suggesting that putatively causal SNPs in these loci may function in a Treg-specific context.
Figure 8.
IMPACT A Priori Identifies Variants with Measured Functionality
(A) Enrichment of posterior probabilities of putatively causal RA SNPs in the top 1% of SNPs with CD4+ Treg regulatory element probabilities highlights the BACH2, ANKRD55, CTLA4/CD28, IRF5, and TNFAIP3 loci.
(B and C) IMPACT regulatory element probabilities (black) at putatively causal SNPs with experimentally validated differential enhancer activity (bolded) and other 90% credible set SNPs (unbolded)18 at two RA-associated loci, CTLA4/CD28 and TNFAIP3.
In the same study, our group observed both differential binding of CD4+ T nuclear extract via EMSA and differential enhancer activity via luciferase assays at two credible set SNPs, narrowing down the list of putatively causal variants in the CD28/CTLA4 and TNFAIP3 loci.18 We observed that both variants with functional activity were located at high probability IMPACT regulatory elements, suggesting that IMPACT may be used to narrow down credible sets to reduce the amount of experimental follow up. First, at the CD28/CTLA4 locus, IMPACT predicts high probability regulatory elements across the four CD4+ T cell states at the functional SNP rs117701653 and lower probability regulatory elements at other credible set SNPs rs55686954 and rs3087243 (Figure 8B). Second, at the TNFAIP3 locus, we observe high probability regulatory elements at the functional SNP rs35926684 and other credible set SNP rs6927172 (Figure 8C) and do not predict regulatory elements at the other seven credible set SNPs. The CD4+ Th1-specific regulatory element at rs35926684 suggests that this SNP may alter gene regulation specifically in Th1 cells and hence, we suggest any functional follow-up be done in this cell state. Fewer than 11% of the credible set SNPs in the other 18 fine-mapped loci have high IMPACT cell-state-specific regulatory element probabilities (Figures S14–S33).
Discussion
In summary, we assume that cell-state-specific regulation may be characterized by an epigenomic signature that may be captured by the cell-state-specific binding sites of a single key TF. To this end, we designed IMPACT to predict cell-state-specific regulatory elements based on epigenomic and sequence profiles of experimental cell-state-specific TF binding by performing a logistic regression on 515 such features. We specifically chose not to employ a deep learning approach in order to retain interpretability of learned annotation weights. Knowledge of which epigenomic or sequence feature annotations are most informative for predicting transcriptional regulation, which varies among cell states, can guide where experimental assay resources might be invested to learn more about the regulome.
We demonstrated the versatility of IMPACT as a genome annotation strategy with several compelling applications. First, we observed that the robust epigenomic footprint of TF binding sites allows for accurate binding prediction. Furthermore, IMPACT outperformed three state-of-the-art methods, MocapG, MocapS, and Virtual ChIP-seq, which use a compendium of sequence-based, open chromatin, and gene expression annotations to predict cell-state-specific TF binding. We believe that this increased predictive power comes from the way in which IMPACT learns which genomic annotations are correlated with TF binding, without knowledge of the cell type or cell state of interest. This is contrary to the compared methods where cell-type-specific DNase-seq or ATAC-seq must be provided as a reference. Moreover, IMPACT provides epigenomic annotations from a wide variety of cell types and assay types which provide complimentary information. We note that we restrict binding prediction to motif sites for each TF in a given cell type. Moreover, validation in a completely independent ChIP-seq dataset was not possible due to the scarcity of primary cell TF ChIP-seq data.
Second, using Pol II IMPACT annotations, for eight tested tissue and cell types, we more precisely captured causal variation of gene expression than by using Pol II ChIP-seq and sequence-based annotations. Our results argue that Pol II IMPACT regions better localize active promoter and proximal regulatory regions driving eQTLs than the compared canonical genomic annotations, which may be less specific due to their larger sizes and restrictive binary characterization. This suggests that IMPACT may be more effective at prioritizing causal SNP variation when fine-mapping eQTLs. These results also argue that the biological basis of eQTLs are related to Pol II binding regions, which is a refinement over previous observations that eQTL causal variation is concentrated near and around TSS and promoter regions.
Third, we more precisely captured causal variation of complex traits. Our CD4+ T IMPACT annotations capture more RA h2 than most canonical CD4+ T cell regulatory annotations. Our findings further reinforce that IMPACT annotations, as an aggregation of hundreds of regulatory annotations, are more informative than single annotations. This is exemplified by the finding that FOXP3 ChIP-seq is strongly enriched for RA h2; and, while this annotation was used as training data for IMPACT, the CD4+ Treg IMPACT annotation captured more RA h2, evident by a larger, more significant annotation effect size, ∗, in the joint analysis. Furthermore, we showed that CD4+ T cell IMPACT annotations explain similar proportions of RA heritability in both European and East Asian populations, suggesting that biological mechanisms driving RA may have similar genetic and regulatory bases in these two populations. We also demonstrated that our approach is generalizable to other trait-driving cell types by showing significantly positive ∗ of IMPACT annotations for 21 of 42 tested complex traits. In particular, CD4+ T IMPACT annotations also captured significant h2 of autoimmune and immune-mediated traits, which is expected given the central role of CD4+ T cells to the immune system and perhaps shared genetic architecture of these traits. We find that h2 of intuitively brain-related traits such as schizophrenia, anorexia, and autism is not captured by brain IMPACT annotations, perhaps suggesting that more complex, cross-cell-type regulatory networks are core to the genetic risk of these traits. Rather, brain IMPACT annotations capture h2 of traits such as menarche age, smoking, and height. We note that we targeted specific polygenic traits using a priori knowledge of the cell states that were most likely to be driving causal biology. To better refine or inform the choice of relevant cell type, we recommend integrating IMPACT with previously published approaches, such as RolyPoly,55 which prioritizes cell types with respect to a particular trait, based on linking single-cell gene expression to GWAS summary statistics. We note that S-LDSC analyses exclude the major histocompatibility complex due to its extremely high gene density and outlier LD structure, which is thought to be the strongest contributor to RA disease h2.56 However, our work supports the notion that there is an undeniably large amount of RA h2 located outside of the MHC.
Lastly, we demonstrated that IMPACT may identify functional variants a priori and suggest the relevant cell-state contexts in which these functional variants may act. We note that disease-relevant IMPACT functional annotations may be integrated with existing functional fine mapping methods, like PAINTOR57 or CAVIARBF,58 to assign causal posterior probabilities to variants.
We recognize several important limitations to our work. First, we have not experimentally validated the activity of any of our predicted regulatory elements. Second, predicted regulatory elements are limited to genomic regions that have been epigenetically assayed. Third, IMPACT as presented in this study is limited to cell states in which ChIP-seq of a key TF has been performed. Furthermore, some TFs are key regulators in more than one cell type or cell state, which should not compromise the cell state specificity of the learned IMPACT annotation. We note that cell state specificity is not gained from the TF itself, but from the unique binding patterns of the TF in a modeled cell state. For example, the CD4+ T cell TFs, for which we create IMPACT annotations, are also key regulators in analogous cell states of ILCs (innate lymphoid cells).59 Under the assumption that these key TFs regulate different sets of genes in the analogous cell states, cell-state-specific IMPACT annotations learned from, for example, T-BET in CD4+ Th1s should be distinguishable from an annotation learned for T-BET in ILC1s. Due to the lack of functional data on ILCs, we were not able to test this claim. However, as more cell-state and cell-type data are generated, especially on more fine-resolution cellular populations, better regulatory annotations may be produced. Moreover, these new functional annotations might nominate other or more precise cellular populations, compared to the ones considered in this study, for explaining polygenic trait heritability and capturing fine-mapped causal variation. While we highlight strong enrichments of IMPACT models trained on CD4+ T cell TFs, especially FOXP3, we acknowledge that it is certainly possible that other cell types and factors play important roles that we have not explored in this study. Fourth, S-LDSC heritability analyses results may be sensitive to the size of the annotation and we recommend enforcing reasonably large annotation sizes, for example at least 0.1% of the genome (Figure S34). In light of these limitations, IMPACT is an emerging strategy for identifying trait-associated regulatory elements and generating hypotheses about the cell states in which variants may be functional, motivating the need to develop therapeutics that target specific disease-driving cell states.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank Eric Lander and Jesse Engreitz for helpful discussions. This work is supported in part by funding from the National Institutes of Health(NHGRI T32 HG002295, UH2AR067677, 1U01HG009088, U01 HG009379 [A.L.P./S.R. U01], and 1R01AR063759) and the Doris Duke Charitable Foundation grant #2013097.
Published: April 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.012.
Contributor Information
Alkes L. Price, Email: aprice@hsph.harvard.edu.
Soumya Raychaudhuri, Email: soumya@broadinstitute.org.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
1000G Phase 3 LD scores, CD4+ T cell specifically expressed genes (binary functional annotations): https://data.broadinstitute.org/alkesgroup/LDSCORE/
ENCODE-DREAM in vivo Transcription Factor Binding Site Prediction Challenge, https://www.synapse.org/#!Synapse:syn6131484/wiki/402026 (accessed: 16th April 2017)
GTEx Portal, https://gtexportal.org/home/
Immgen.tsv, https://gist.github.com/nachocab/3d9f374e0ade031c475a
IMPACT GitHub repository, https://github.com/immunogenomics/IMPACT
RA EUR summary statistics, http://plaza.umin.ac.jp/yokada/datasource/software.htm
RA EAS summary statistics, http://jenger.riken.jp/en/result
S-LDSC tutorial and instructions, https://github.com/bulik/ldsc
Supplemental Data
References
- 1.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.-R., Lareau C., Shoresh N., Brainstorm Consortium Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazal S., Finucane H.K., Furlotte N.A., Loh P.-R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farh K.K.-H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J.H., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickrell J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 15.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponting C.P., Hardison R.C. What fraction of the human genome is functional? Genome Res. 2011;21:1769–1776. doi: 10.1101/gr.116814.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. Defining functional DNA elements in the human genome. PNAS USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westra H.-J., Martínez-Bonet M., Onengut-Gumuscu S., Lee A., Luo Y., Teslovich N., Worthington J., Martin J., Huizinga T., Klareskog L. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet. 2018;50:1366–1374. doi: 10.1038/s41588-018-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Yu B., Carriero N., Silva C., Bonneau R. Mocap: large-scale inference of transcription factor binding sites from chromatin accessibility. Nucleic Acids Res. 2017;45:4315–4329. doi: 10.1093/nar/gkx174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karimzadeh M., Hoffman M.M. Virtual ChIP-seq: Predicting transcription factor binding by learning from the transcriptome. bioRxiv. 2018 doi: 10.1186/s13059-022-02690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soderquest K., Hertweck A., Giambartolomei C., Henderson S., Mohamed R., Goldberg R., Perucha E., Franke L., Herrero J., Plagnol V. Genetic variants alter T-bet binding and gene expression in mucosal inflammatory disease. PLoS Genet. 2017;13:e1006587. doi: 10.1371/journal.pgen.1006587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertweck A., Evans C.M., Eskandarpour M., Lau J.C.H., Oleinika K., Jackson I., Kelly A., Ambrose J., Adamson P., Cousins D.J. T-bet activates Th1 genes through mediator and the super elongation complex. Cell Rep. 2016;15:2756–2770. doi: 10.1016/j.celrep.2016.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustafsson M., Gawel D.R., Alfredsson L., Baranzini S., Björkander J., Blomgran R., Hellberg S., Eklund D., Ernerudh J., Kockum I. A validated gene regulatory network and GWAS identifies early regulators of T cell-associated diseases. Sci. Transl. Med. 2015;7:313ra178. doi: 10.1126/scitranslmed.aad2722. [DOI] [PubMed] [Google Scholar]
- 24.Tripathi S.K., Chen Z., Larjo A., Kanduri K., Nousiainen K., Äijo T., Ricaño-Ponce I., Hrdlickova B., Tuomela S., Laajala E. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 2017;19:1888–1901. doi: 10.1016/j.celrep.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Schmidl C., Hansmann L., Lassmann T., Balwierz P.J., Kawaji H., Itoh M., Kawai J., Nagao-Sato S., Suzuki H., Andreesen R., FANTOM consortium The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood. 2014;123:e68–e78. doi: 10.1182/blood-2013-02-486944. [DOI] [PubMed] [Google Scholar]
- 26.Wang C., Sandling J.K., Hagberg N., Berggren O., Sigurdsson S., Karlberg O., Rönnblom L., Eloranta M.-L., Syvänen A.-C. Genome-wide profiling of target genes for the systemic lupus erythematosus-associated transcription factors IRF5 and STAT4. Ann. Rheum. Dis. 2013;72:96–103. doi: 10.1136/annrheumdis-2012-201364. [DOI] [PubMed] [Google Scholar]
- 27.Qiao Y., Giannopoulou E.G., Chan C.H., Park S.H., Gong S., Chen J., Hu X., Elemento O., Ivashkiv L.B. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham T.-H., Benner C., Lichtinger M., Schwarzfischer L., Hu Y., Andreesen R., Chen W., Rehli M. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119:e161–e171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 29.Dimitrova L., Seitz V., Hecht J., Lenze D., Hansen P., Szczepanowski M., Ma L., Oker E., Sommerfeld A., Jundt F. PAX5 overexpression is not enough to reestablish the mature B-cell phenotype in classical Hodgkin lymphoma. Leukemia. 2014;28:213–216. doi: 10.1038/leu.2013.211. [DOI] [PubMed] [Google Scholar]
- 30.Gertz J., Savic D., Varley K.E., Partridge E.C., Safi A., Jain P., Cooper G.M., Reddy T.E., Crawford G.E., Myers R.M. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell. 2013;52:25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasowski M., Grubert F., Heffelfinger C., Hariharan M., Asabere A., Waszak S.M., Habegger L., Rozowsky J., Shi M., Urban A.E. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright F.A., Sullivan P.F., Brooks A.I., Zou F., Sun W., Xia K., Madar V., Jansen R., Chung W., Zhou Y.-H. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishigaki K., Kochi Y., Suzuki A., Tsuchida Y., Tsuchiya H., Sumitomo S., Yamaguchi K., Nagafuchi Y., Nakachi S., Kato R. Polygenic burdens on cell-specific pathways underlie the risk of rheumatoid arthritis. Nat. Genet. 2017;49:1120–1125. doi: 10.1038/ng.3885. [DOI] [PubMed] [Google Scholar]
- 35.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 37.Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J.-T., Loh P.-R., Schoech A., Reshef Y., Liu X., O’Connor L. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onengut-Gumuscu S., Chen W.-M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E., Type 1 Diabetes Genetics Consortium Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eyre S., Bowes J., Diogo D., Lee A., Barton A., Martin P., Zhernakova A., Stahl E., Viatte S., McAllister K., Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate. Wellcome Trust Case Control Consortium High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arvey A., Agius P., Noble W.S., Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012;22:1723–1734. doi: 10.1101/gr.127712.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davenport E.E., Amariuta T., Gutierrez-Arcelus M., Slowikowski K., Westra H.-J., Luo Y., Shen C., Rao D.A., Zhang Y., Pearson S. Discovering in vivo cytokine-eQTL interactions from a lupus clinical trial. Genome Biol. 2018;19:168. doi: 10.1186/s13059-018-1560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X., Finucane H.K., Gusev A., Bhatia G., Gazal S., O’Connor L., Bulik-Sullivan B., Wright F.A., Sullivan P.F., Neale B.M., Price A.L. Functional architectures of local and distal regulation of gene expression in multiple human tissues. Am. J. Hum. Genet. 2017;100:605–616. doi: 10.1016/j.ajhg.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Firestein G.S. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 46.Raychaudhuri S., Sandor C., Stahl E.A., Freudenberg J., Lee H.-S., Jia X., Alfredsson L., Padyukov L., Klareskog L., Worthington J. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terao C., Okada Y., Ikari K., Kochi Y., Suzuki A., Ohmura K., Matsuo K., Taniguchi A., Kubo M., Raychaudhuri S. Genetic landscape of interactive effects of HLA-DRB1 alleles on susceptibility to ACPA(+) rheumatoid arthritis and ACPA levels in Japanese population. J. Med. Genet. 2017;54:853–858. doi: 10.1136/jmedgenet-2017-104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terao C., Raychaudhuri S., Gregersen P.K. Recent advances in defining the genetic basis of rheumatoid arthritis. Annu. Rev. Genomics Hum. Genet. 2016;17:273–301. doi: 10.1146/annurev-genom-090314-045919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., Kim H., Stahl E., Plenge R., Daly M., Raychaudhuri S. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am. J. Hum. Genet. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trynka G., Westra H.-J., Slowikowski K., Hu X., Xu H., Stranger B.E., Klein R.J., Han B., Raychaudhuri S. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet. 2015;97:139–152. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J., Yamane H., Paul W.E. Differentiation of effector CD4 T cell populations (∗) Annu. Rev. Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vahedi G., Kanno Y., Furumoto Y., Jiang K., Parker S.C.J., Erdos M.R., Davis S.R., Roychoudhuri R., Restifo N.P., Gadina M. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveira T.V., Maniero F., Santos M.H.H., Bydlowski S.P., Maranhão R.C. Impact of high cholesterol intake on tissue cholesterol content and lipid transfers to high-density lipoprotein. Nutrition. 2011;27:713–718. doi: 10.1016/j.nut.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 54.Mokhtari R., Lachman H.M. The major histocompatibility complex (MHC) in schizophrenia: a review. J. Clin. Cell. Immunol. 2016;7 doi: 10.4172/2155-9899.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calderon D., Bhaskar A., Knowles D.A., Golan D., Raj T., Fu A.Q., Pritchard J.K. Inferring relevant cell types for complex traits by using single-cell gene expression. Am. J. Hum. Genet. 2017;101:686–699. doi: 10.1016/j.ajhg.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonseka C.Y., Rao D.A., Raychaudhuri S. Leveraging blood and tissue CD4+ T cell heterogeneity at the single cell level to identify mechanisms of disease in rheumatoid arthritis. Curr. Opin. Immunol. 2017;49:27–36. doi: 10.1016/j.coi.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kichaev G., Roytman M., Johnson R., Eskin E., Lindström S., Kraft P., Pasaniuc B. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics. 2017;33:248–255. doi: 10.1093/bioinformatics/btw615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen W., McDonnell S.K., Thibodeau S.N., Tillmans L.S., Schaid D.J. Incorporating functional annotations for fine-mapping causal variants in a Bayesian framework using summary statistics. Genetics. 2016;204:933–958. doi: 10.1534/genetics.116.188953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koues O.I., Collins P.L., Cella M., Robinette M.L., Porter S.I., Pyfrom S.C., Payton J.E., Colonna M., Oltz E.M. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell. 2016;165:1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.