Abstract
Phosphoglucomutase 1 (PGM1) encodes the metabolic enzyme that interconverts glucose-6-P and glucose-1-P. Mutations in PGM1 cause impairment in glycogen metabolism and glycosylation, the latter manifesting as a congenital disorder of glycosylation (CDG). This unique metabolic defect leads to abnormal N-glycan synthesis in the endoplasmic reticulum (ER) and the Golgi apparatus (GA). On the basis of the decreased galactosylation in glycan chains, galactose was administered to individuals with PGM1-CDG and was shown to markedly reverse most disease-related laboratory abnormalities. The disease and treatment mechanisms, however, have remained largely elusive. Here, we confirm the clinical benefit of galactose supplementation in PGM1-CDG-affected individuals and obtain significant insights into the functional and biochemical regulation of glycosylation. We report here that, by using tracer-based metabolomics, we found that galactose treatment of PGM1-CDG fibroblasts metabolically re-wires their sugar metabolism, and as such replenishes the depleted levels of galactose-1-P, as well as the levels of UDP-glucose and UDP-galactose, the nucleotide sugars that are required for ER- and GA-linked glycosylation, respectively. To this end, we further show that the galactose in UDP-galactose is incorporated into mature, de novo glycans. Our results also allude to the potential of monosaccharide therapy for several other CDG.
Keywords: glycosylation, galactose, PGM1-CDG, CDG, tracer metabolomics, nucleotide sugars, central carbon metabolism, mitochondria
Introduction
Congenital disorders of glycosylation (CDG) are a subgroup of inborn errors of metabolism (IEMs) affecting one of the most crucial post-translational modifications of proteins: glycosylation.1 Glycosylation is the biochemical process of activating sugars, assembling them into oligosaccharides, and attaching them to proteins via N-, O-, or lipid-linkages. As such, the process plays crucial roles in both the stability and functionality of the target molecules2 (Figures 1A–C).
Figure 1.
Glycosylation
(A) N-linked glycosylation requires the nucleotide sugars CMP-sialic acid, GDP-fucose, GDP-mannose, UDP-glcNAc, UDP-galactose, and UDP-glucose; all are synthesized through a number of connected pathways with close connections to PGM1 activity.
(B and C) Glycan construction commences in the endoplasmic reticulum (ER) (B), and it is completed in the Golgi apparatus (GA) (C). Transferrin is used as an example of a mature glycoprotein.
More than 100 different types of CDG are currently described, and they present with extremely wide phenotypic variability.3 The most common form of CDG is caused by the group of N-glycosylation defects; these CDG present with decreased glycan occupancy on mature protein as a result of either the impaired synthesis and transfer of glycans in the endoplasmic reticulum (ER; CDG type I) or the impaired synthesis of glycans in the Golgi apparatus (GA; CDG type II). Glycosylation begins with the biosynthesis of nucleotide sugars (the building blocks of glycans, e.g., UDP-glucose, UDP-galactose, and GDP-mannose, etc. [Figure 1A]). Biosynthesis is followed by nucleotide-sugar transport (Figures 1B and 1C), oligosaccharide synthesis in the ER (Figure 1B), and finally glycan maturation in the GA (Figure 1C). The most immediate therapeutic potential exists in targeting the first step (nucleotide sugar biosynthesis). Specifically, different monosaccharides, such as mannose, fucose, or galactose, have been trialed as dietary supplements for bypassing underlying metabolic defects and replenishing the essential glycosylation precursor products (see overview in Witters et al.4).
One of the few CDG for which a tailored treatment exists is PGM1-CDG, and the treatment takes the form of oral D-galactose therapy (further referred to as galactose therapy).4
PGM1-CDG (OMIM: 614921),5, 6 primarily classified as glycogen storage disorder (GSD XIV), is a multi-systemic CDG that is caused by mutations in PGM1 (GenBank: NM_002633.2, MIM: 171900), which encodes the phosphoglucomutase 1 enzyme (PGM1, EC: 5.4.2.2). This metabolic defect leads to abnormal N-glycan synthesis in both the ER and the GA, as well as decreased galactosylation of truncated glycans (combined defects characteristic of both CDG types I and II).4, 6 The underlying pathomechanism of the complex biochemical defect in PGM1-CDG is not yet understood.
PGM1-deficient individuals show severe liver and muscle impairment, hypoglycemia, endocrine and coagulation problems, and cardiomyopathy.6 Nutritional interventions such as frequent feeding, high glucose intake, or treatment with 1–3 g/kg/day of raw cornstarch added to the diet, as are applied for other glycogen disorders, do not resolve the symptoms. On the basis of the deficient galactosylation in PGM1 deficiency, we recently trialed individuals with PGM1-CDG on galactose therapy.
This therapy trial led to a significant improvement, including the restoration of glycosylation profiles,6, 7, 8 in biochemical measures collected in studies of affected individuals over the course of an 18-week pilot trial.7 No clinical progression data have been reported, and a longer-term follow-up (1 year) was limited to a single individual on galactose, restricting the ability to assess the efficacy and safety of galactose therapy.7 The reported laboratory data about PGM1-deficient individuals on galactose supplementation are also difficult to interpret because of the history of variable compliance. We hypothesized that deficient PGM1 activity (in glucose-1-P to glucose-6-P) impairs nucleotide-sugar metabolism linked to glucose-1-P, but galactose could bypass the impairment (Figure 1A). Although a preliminary analysis (limited in sample size) in PGM1-CDG fibroblasts did not find that activated sugar pools were depleted, it suggested that galactose could potentially boost levels of nucleotide sugars.7 The data were unable, however, to provide an explanation for the mechanism or the mixed CDG type I-II seen in PGM1-CDG.
Here, we provide detailed evidence, from multiple experiments, that galactose therapy for PGM1 deficiency is not effective through targeting either the regulation of the PGM1 gene or protein or the altered metabolism linked to glucose-6-P. Rather, galactose therapy is effective through the replenishment of galactose-1-P and the nucleotide sugars UDP-glucose and UDP–galactose. In turn, these UDP-hexoses are necessary for the correct ER- and GA-associated glycosylation, respectively (Figures 1B and 1C). Moreover, we show that the biosynthesis of these two nucleotide sugars is metabolically re-wired in the presence of galactose. Critically, we also show that the labeled galactose is incorporated into mature N-glycans on the cell surface, providing evidence of the mode of action of galactose treatment in this therapy for individuals with PGM1-CDG.
The approach further suggests that the direct administration of nucleotide sugars may be a more effective and less onerous form of treatment for affected individuals than galactose therapy. The data also offer new possibilities for developing novel nutritional treatments for related CDG with other defects in nucleotide-sugar metabolism and transport. Importantly, this study also highlights the use of tracer metabolomics as an underexploited tool in the investigation of metabolic disease mechanisms and therapy.
Material and Methods
Ethics
Samples from affected individuals sourced from Tulane University were collected with informed consent and analyzed in accordance with application number IRB 533377-7 (“Screening for congenital disorders of glycosylation in individuals with hypoglycemia”); samples from the individuals sourced from UZ Leuven Hospital were collected with informed consent and analyzed in accordance with ethics application number S58358 (“EURO-CDG-2: Een Europees onderzoeknetwerk gericht op het verbeteren van de diagnostiek en de behandeling van aangeboren glycosylatie defecten”). The analysis of fibroblasts at the UZ Leuven Hospital was in accordance with ethics application number S60206 (“Retrospective metabolomic analysis of archived fibroblasts”).
Study Cohort
13 individuals (P1–13) with a molecular diagnosis of PGM1-CDG are included in this study (refer to Table S1 for their characteristics).
Clinical Data
The clinical studies were performed according to the Tulane IRB protocol #517339, ClinicalTrials.gov NCT02955264. Eight individuals (four previously reported individuals: P2,6 P7,9 P12,6 and P138 and four unreported individuals, P8–11) were included in the current open-label study and supplemented with 0.5 to 1.5 g of oral galactose/kg/day for 18 weeks, and they remained on long-term treatment (for 12–31 months per Table S1) at 1 g of galactose/kg/day (maximum dose of 50 g daily) as described.
Biochemical data were collected from five individuals (P8–12) for the 18-week trial period, and long-term data were collected from three individuals (P8, P11, and P12) treated for 12–31 months.
The clinical progression scores of all eight individuals were evaluated per the TPCRS (Tulane PGM1-CDG Rating Scale) scoring system,8 which consists of three sections: TPCRS section I, history and current function; section II, system-specific involvement since birth; and section III, current clinical assessment. Higher scores indicate a more severe phenotype, and the total TPCRS score infers the overall severity of the disease. These clinical progression scores were collected a maximum of 4 weeks prior to the start of galactose treatment and again during the latest, most current clinical follow up (12–31 months after treatment). The same investigator evaluated the individuals at each study site.
Fibroblast Cell Lines
Control and PGM1-CDG fibroblasts (from P1–P7) at less than passage 15 were maintained in low-glucose DMEM (Thermo Fisher Scientific) with 5.5 mM glucose (the closest approximation to the physiological glucose concentration in plasma) and 2 mM glutamine supplemented with 10% fetal calf serum (FCS) at 37°C with 5% CO2 in a humidified incubator. As described, fibroblasts were additionally cultured with 2 mM galactose, chosen because 2 mM galactose reflects the concentration in serum in individuals supplemented with 1 g galactose/kg/day (see description in Clinical Data).7 We used an amount of 5 mM galactose for measuring ICAM1 (GenBank: NM_000201.3) expression with qPCR.
Reagents
All reagents were from Sigma unless otherwise specified. All stable isotopes (tracers) were purchased from Cambridge Isotope Laboratories.
Stable Isotope Tracer Studies
Fibroblasts were seeded at 1,700 cells/cm2/0.15 mL of DMEM (GIBCO) with 5.5 mM glucose and 2 mM glutamine in 6-well plates. At day 1, the cells and empty wells (for background noise control), were washed once in PBS, then replenished with DMEM containing 2 mM glutamine and either A) 5.5 mM 13C6 glucose and 2 mM 12C6 galactose; B) 5.5 mM 12C6 glucose and 2 mM 13C6 galactose; C) 5.5 mM 12C6 glucose and 2 mM 12C6 galactose; D) 5.5 mM 13C6 glucose; or E) 5.5 mM 12C6 glucose. At day 3, the tracer media were refreshed as described above. At day 5, we harvested the cells on ice by removing the media and washing them once in ice-cold saline solution (9 g/L NaCl). We then immersed the cells in 250 μL of ice-cold extraction buffer A (80% methanol [Merck] and 2 μM d27 myristic acid) or B (50% methanol, 30% acetonitrile [Thermo Fisher Scientific], 10 mM ammonium acetate [pH 9.3], and 2 μM d27 myristic acid), allowed them to incubate for 2 min, and finally scraped and transferred the cells to a 1.5 mL tube and stored them overnight at −80°C. The cellular debris was then pelleted (for 15 min at 20,000 g and 4°C), and the supernatant was transferred to a new vial. The cell pellet was dissolved in 100 μL of 200 mM NaOH (for 20 min at 95°C), and the protein content was determined by the bicinchoninic acid (BCA) method (Thermo Fisher Scientific). Cells harvested in extraction buffer B were additionally dried by vacuum centrifugation and suspended in 250 μL ddH2O. 10 μL of sample (buffer A or B) was then separated on an ion-pairing liquid-chromatography column, and the metabolites were resolved on a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer in negative ion mode with the following ESI settings: 50 sheet gas flow rate, auxiliary gas flow rate 10, spray voltage of 4 kV, S-lens RF level of 60, and the capillary temperature at 350°C. A full scan (resolution set at 140,000 at 200 m/z, AGC at 3 e6, 512 ms ion fill time, and 70–1,050 m/z scan range) was applied.
Finally, we quantified the metabolites according to their elution time and m/z ratios by using the Thermo Fisher Scientific XCalibur Quan Browser. We performed a correction of naturally occurring carbon isotopes.10 We normalized relative metabolite abundances to protein content.
UDP-Glucose and UDP-Galactose Separation by UGDH Assay
We used UDP-glucose-6-dehydrogenase (UGDH; EC: 1.1.1.22) to convert UDP-glucose to UDP-glucuronic acid, thus enabling the separation of the UDP-hexoses UDP-glucose and UDP-galactose. In brief, 60 μL of metabolic cell extract from the tracer-based metabolomics experiments (above) and standards (1 mM of each metabolite comprising UDP-glucose, UDP-galactose, and UDP-glucose + UDP-galactose) were dried (vacuum centrifugation at 6°C, overnight) and re-suspended in 90 μL of buffer (12.5 mM glycine and 160 μM NAD+ with or without >0.002 U UGDH from S. pyrogenes), shaken (overnight at 750 rpm and 37°C), and dried. The samples were re-suspended in 150 μL 80% methanol (Merck) containing 2 mM d27 myristic acid, then vortexed and pelleted (for 15 min at 20,000 g and 4°C), and the supernatant was transferred to a new vial. We detected UDP-hexose and UDP-glucuronic acid by using liquid chromatography-tandem mass spectrometry (LC-MS) methods, as reported for the stable-isotope tracer experiments (see above). The UDP-hexose to UDP-glucuronic acid ratio was calculated in standards to evaluate the efficiency of the enzymatic reaction, resulting in >95 % conversion of UDP-glucose to UDP-glucuronic acid. The relative metabolite abundances were normalized to protein content, and finally we calculated the abundances of UDP-galactose (UDP-hexose minus [UDP-hexose with UGDH]) and UDP-glucose (UDP-hexose minus UDP-galactose).
Fragmentation Analysis of UDP-Hexose and UDP-HexNAc
Parallel reaction monitoring (PRM) was performed on a Q Exactive Focus Mass Spectrometer operating in negative ion mode, and the fragmentation of metabolites was performed via high collision dissociation (HCD). In brief, 10 μL of tracer-based metabolomics experiment samples (described above) were loaded onto a Hilicon iHILIC-Fusion column (Achrom). A linear gradient (flow rate 200 μL/min at 25°C) was carried out under the following conditions: from 0–2 min, 90% solvent A (LC-MS-grade acetonitrile) and 10% solvent B (10 mM ammonium acetate [pH 9.3]); from 2–20 min, increase to 80% solvent B; from 21–23 min, 80% solvent B; from 23–25 min, decrease to 40% solvent B; from 26–27 min, decrease to 10% solvent B; and from 28–35 min, hold at 10% solvent B. The heated electrospray ionization (HESI) probe settings were: sheath gas flow, 40; auxiliary gas flow rate, 10 at 260°C; spray voltage, 4.8 kV; capillary temperature, 300°C; and S-lens RF level, 50. The targeted PRM method focused on the ions C15H23N2O17P2 (m/z 565.047) and C17H26N3O17P2 (m/z 606.074) from UDP-hexose and UDP-hexNAc, respectively. The ions were fragmented via HCD (with helium gas) at normalized voltages of 10, 30, and 60 eV. The resolution was set at 35,000, AGC target at 2 e5, the automatic injection time was 128 ms, the isolation window was set at 20 m/z, and the isolation offset was set at 8 m/z (ensuring a fragmentation of the full isotopologue profiles of the target metabolites). We manually verified the fragments by using the Thermo Fisher Scientific Xcalibur Quan Browser, and we matched them to the METLIN MS/MS metabolite database on the basis of their fragment structures and m/z ratios (https://metlin.scripps.edu).
N-Glycan Analysis by Gas Chromatography-Mass Spectrometry
Cells were prepared as described in the Stable Isotope Tracer Studies section. The mature N-glycans were isolated and processed according to previously reported methods,11 and the incorporation of 13C6 glucose and 13C6 galactose into specific, mature N-glycans monosaccharides was evaluated by gas chromatography-mass spectrometry (GC-MS). In brief, the media was removed from the cells, the cells were washed once in PBS, then the cells were scraped in fresh PBS and pelleted by centrifugation (for 15 min at 800 g and 4°C). The supernatant was removed, and the cell pellet was dissolved in 250 μL of a 10 mM ammonium bicarbonate (AB) buffer. The cells were then lysed through 3 successive cycles of freeze-thawing on dry ice.
The membrane fraction was then pelleted (for 15 min at 20,000 g and 4°C) and the supernatant discarded. The pellet was washed in 1 mL of the AB buffer.
Next, the membranes were incubated in 40 μL of a Peptide: N-glycosidase F (PNGase F) buffer (Bioke protocol, New England Biolabs) (overnight at 750 rpm and 37°C). Debris was then pelleted (for 15 min at 20,000 g and 4°C), and the supernatant containing the N-glycans was transferred to a fresh 1.5 mL tube and dried with a vacuum centrifuge. For the hydrolysis of the N-glycans into individual monosaccharides, the samples were incubated with 160 μL of 2 M trifluoroacetic acid (TFA) (for 2 h at 120°C) and then dried.
For the acetylation of the monosaccharides, samples were then rehydrated in 200 μL of a fresh 10 mg/mL Na-borohydride solution dissolved (for 1 hr at 25°C) in 1 M ammonium hydroxide buffer. Next, 50 μL of a 100% methanol solution was added, and then the samples were dried. The samples were rehydrated in 100 μL of a 9:1 methanol:acetate solution and subsequently dried again. Then, 18 μL of ddH2O was added, the samples were vortexed (for 1 min) and sonicated (for 2 min), and then incubated with 250 μL of concentrated acetic anhydride (for 10 min at 50°C). After, the samples were incubated with 230 μL of concentrated TFA (for 10 min at 50°C). The reaction mix was then transferred to a 15 mL tube, and 1.8 mL of dichloromethane was added, then 2 mL of ddH2O. The samples were vortexed, and after they were incubated (for 10 min at 25°C), the upper aqueous phase was discarded, and the samples were washed again with ddH2O. Then, 1.4 mL of the remaining lower dichloromethane phase was transferred to a fresh 1.5 mL tube, dried, and solubilized in 100 μL concentrated ethyl acetate. Finally, the samples were vortexed (for 1 min) and transferred to MS-vials.
GC-MS analysis of the acetylated monosaccharides was performed with an Agilent 7890A GC equipped with an HP-5ms 5% phenyl methyl silox capillary column (30 m, 0.25 mm, 0.25 μm; Agilent Technologies), interfaced with a triple-quadruple tandem mass spectrometer (Agilent 7000B, Agilent Technologies) and operating under ionization by electron impact at 70 eV. The injection port, interface, and ion source temperatures were kept at 230°C. The temperature of the quadrupoles was maintained at 150°C. The injection volume was 1 μL, and the samples were injected at a 1:10 split ratio. Helium flow was kept constant at 1 mL/min.
The temperature of the column started at 80°C for 1 min, was increased to 220°C at Δ40°C/min, and then remained at 220°C for 1 min. Next, a 5°C/min gradient was carried out until the temperature reached 270°C and remained at 270°C for 1 min. Finally, the temperature was increased to 300°C at a Δ60°C/min ramp. After the gradient was carried out, the column was heated for another 2 min at 325°C. The GC-MS analyses were performed with single ion monitoring (SIM) scanning for the isotopic pattern of acetylated galactose, mannose (m/z = 289, 290, 291, 292, 293, and 294), fucose (m/z = 217, 218, 219, 220, and 221), and glcNAc (m/z = 318, 319, 320, 321, 322, 323, 324, and 326).
GC-MS Analysis of Hexose-Phosphate Sugars
Analysis of the subspecies of hexose-phosphates (glucose-6-P, glucose-1-P, fructose-6-P, mannose-6-P, and galactose-1-P) was performed as described by Chen et al., 2002.12 In brief, metabolite extracts from fibroblast cultures were dried via vacuum centrifugation. Next, 60 μL of pyridine and 60 μL of TMS (N,O-bis[Trimethylsilyl] trifluoroacetamide with trimethylchlorosilane [BSTFA + TMCS]) were added, and the mixture was heated (for 30 min at 90°C). The samples were centrifuged (for 15 min at 20,000 g and 4°C), and the supernatant was transferred to an MS vial. The GC-MS analyses were performed on an Agilent 7890A GC equipped with an HP-5ms 5% phenyl methyl silox capillary column (30 m, 0.25 mm, 0.25 μm; Agilent Technologies), interfaced with a triple-quadruple tandem mass spectrometer (Agilent 7000B, Agilent Technologies) and operating under ionization by electron impact at 70 eV. The injection port, interface, and ion source temperatures were kept at 230°C. The temperature of the quadrupoles was maintained at 150°C. The injection volume was 1 μl, and the samples were in splitless mode. Helium flow was kept constant at 1 mL/min.
The column gradient temperature was started at 100°C for 2 min, increased to 175°C at Δ20°C/min, increased to 230°C at Δ4°C/min, held at 230°C for 3 min, increased to 300°C at Δ40°C/min, and finally held at 300°C for 5 min. After the gradient, the column was baked out for another 3 min at 325°C. The GC-MS analyses were performed by SIM scanning for fragments with 299 m/z. Data were processed on the MassHunter Workstation software with the Quantitative Analysis Version B.06.00/Build 6.0.388.0.
Statistics
Error bars describe ± standard error of the mean (SEM). We generated the p values in GraphPad Prism (version 7.0) from biological replicates by using either paired or un-paired (as described) parametric Student’s-t tests. The p values are reported to three decimal places for consistency and to allow the reader to make their own interpretation about statistical “significance.” We use the word “significance” here only to convey the perceived importance.
Results
Galactose Treatment Biochemically and Clinically Improves the Outcome in PGM1-CDG
Here, we report on additional biochemical and clinical progression data in eight individuals with PGM1 deficiency. Although limited biochemical data have been previously reported, this study additionally reports on clinical improvement in fully compliant PGM1-CDG-affected individuals on long-term galactose treatment.
Five individuals (P8–12) underwent an 18-week, open-label, escalating-dose dietary galactose supplementation trial (Figure 2). All five individuals were evaluated prior to galactose treatment and after 18 weeks of the trial. P8–P11 were newly identified (see Table S1 and Figure S2 for genetic and pedigree details, respectively), whereas P12, who has been previously reported on, was re-trialed with galactose after a washout period. The biochemical data (Figures 2A and 2C) collected from the five affected individuals during the 18-week clinical trial period showed accruing improvement in liver function (alanine aminotransferase [ALT] and aspartate aminotransferase [AST]), coagulation (antithrombin III), and muscle function (creatine kinase).
Figure 2.
Clinical and Biochemical Improvements After Galactose Supplementation in PGM1-CDG-Affected Individuals and Cells
(A) TPCRS (Tulane PGM1-CDG Rating Scale) scoring for individuals before galactose treatment and at the most recent follow up (12–31 months; see Table S1). The scale describes TPCRS scores: higher scores represent a more severe phenotype (see Material and Methods).
(B) TPCRS scores from individuals prior to oral galactose treatment and at long term follow-up (LT; 12–31 months) for TPCRS sub-categories I, II, and III and the combined scores.
(C) The effect of oral galactose supplementation on liver function (ALT, AST), coagulation systems (antithrombin III), and muscle function (creatine kinase) in affected individuals prior to treatment, during an 18-week trial period, and at long-term follow-up (LT; 12–31 months). The control reference ranges are shown in green, and numerical values are displayed on the right.
(D) ICAM1 expression in PGM1-CDG fibroblasts (technical n ≥ 2) relative to untreated controls (biological n = 3, technical n = 3) in the absence and presence of 5 mM galactose in the medium.
The statistics are from paired (panels B and C) and unpaired (panel D) Student’s t tests, and error bars are ± SEM. Abbreviations are as follows: ALT = alanine aminotransferase and AST = aspartate aminotransferase.
More importantly, these five individuals with PGM1-CDG and three previously reported individuals (P2, P7, and P13) had an extension of galactose supplementation and additional long-term follow-up observations. Long-term laboratory data were also collected (from P8, P11, and P12; Figure 2C). The clinical progression scores (TPCRS scores) of the eight individuals with PGM1-CDG were collected prior to galactose supplementation and again at the most recent clinical follow up for all individuals (Figures 2A and 2B). None of them reported any adverse effects. TPCRS scores improved during the 18-week trial and also in long-term (12–31 months) follow up (Figures 2A and 2B). The galactose therapy improved liver impairment, transaminase levels, and coagulation (TPCRS-II), as previously reported (Figure 2B). All individuals who were trialed on galactose had hypoglycemia before starting the treatment. P2 and P11 had recurrent hypoglycemia that had to be managed by tube feedings. Both individuals’ hypoglycemia resolved after treatment. P7–P10 had hypoglycemic episodes several times a year before the treatment. P12 also had monthly hypoglycemic episodes, which resulted in an inability to work. Though this individual still presented with asymptomatic episodes of fasting hypoglycemia even after being treated with galactose, she regained full functionality.
Long-term therapy resulted in the improvement of muscle impairment, as well (TPCRS-III; Figures 2A and 2C). Notably, two of the individuals (P8 and P12) in our study presented with at least one rhabdomyolysis episode, and two other individuals (P10 and P11) suffered from muscle weakness. All individuals improved on these measures when on galactose. Myopathy is, however, very slowly progressive in PGM1-CDG-affected individuals, though the progression is quicker in individuals who have presented with rhabdomyolysis.6, 13, 14 In addition, the natural history of PGM1-CDG is not well described, and most of the individuals were started on galactose treatment soon after diagnosis. Therefore, predicting the expected frequency of rhabdomyolysis, as well as hypoglycemia and seizures, without therapy in these individuals is difficult, thus limiting our ability to fully assess the functional relevance of these measures on galactose.
Strengthening the functional significance of the scores, P11, who had steatofibrosis of the liver at the beginning of the galactose trial, was subsequently removed from the liver transplantation list. Critically, no hypoglycemic episodes, rhabdomyolysis, or seizures (common presentations in PGM1-CDG as described above) were observed in any of the individuals with PGM1-CDG during the treatment trial, further strengthening the efficacy of the treatment.
In parallel, we also show that ICAM1 expression (ICAM-1 expression is downregulated when it is hypoglycosylated)15 is restored in individuals’ fibroblasts that are treated with galactose, further indicating that the in vitro system mimics key aspects of the in vivo biology, and that PGM1-CDG fibroblasts are a relevant model in which to study the disease and treatment mechanisms (Figure 2D).7
Galactose Treatment Does Not Alter PGM1 Activity
Despite the success of galactose therapy in PGM1-CDG, the biochemical mechanisms, notably the mixed type-I and type-II CDG profile,5 that underlie galactose therapy have remained elusive.
To understand the mode(s) of action of galactose therapy in PGM1-CDG, we first investigated whether galactose pharmacologically regulates PGM1 at the transcript or protein level, potentially compensating for the enzymatic defect. We found no evidence for either phenomenon in vitro (Figure S1).
The expression of PGM1, relative to its expression in untreated control fibroblasts, was equivalent with or without galactose treatment (Figure S1A). At the protein level, we found that neither galactose nor its derivative (galactose-1-P) at an excess of ligand have any significant binding affinity change for the recombinant PGM1 enzyme (there was a less than 1° change in the thermal-shift bindings assays [TSAs]). In comparison, the stable analog (Xylose-1-P) of PGM1’s natural substrate (glucose-1-P) increased melting temperature (TM) by 4.7°C, thus confirming the specificity of PGM1 binding (Figure S1B).
Similarly, the enzymatic activity of recombinant PGM1 in a time-course assay was equivalent in the presence or absence of galactose or its derivative, galactose-1-P (Figure S1C).
Such activity data are also consistent with the modeling of galactose in the active site of the reported crystal structure of PGM1; the modeling shows that galactose would have unfavorable binding because of a lack of critical enzyme interactions with the O4 hydroxyl residue of galactose, as well as possible steric conflicts (Figure S1D).16, 17
These results clearly demonstrate that galactose does not modulate PGM1 expression or activity and that galactose therapy affects glycosylation in PGM1-CDG by (an)other mechanism(s).
Galactose Treatment Does Not Modulate Central Carbon Metabolism Linked through Glucose-6-P, nor Mitochondrial Function
We then undertook to map the effect of galactose therapy in the metabolic context of disrupted PGM1 activity (Figure 1A), which interfaces between central carbon metabolism (through glucose-6-P) and glycogen and UDP-nucleotide monosaccharide metabolism (through glucose-1-P).
To address this question, we turned to the technique of tracer metabolomics.18, 19 This technique allows for the study of systemic effects, as opposed to the study of just a single enzyme pathway. By using this technique, we aimed to understand the metabolic adaptions in our PGM1-CDG cells and thereby highlight opportunities for dietary therapies in PGM1-CDG and other CDG.
Focusing first on central carbon metabolism linked through glucose-6-P, we analyzed the fate of both 13C6 glucose and 13C6 galactose (Figures S3A–C; statistics provided in Figure S4) in different culturing conditions (see Material and Methods). Although PGM1-CDG fibroblasts cultured in the presence of both glucose and galactose appear to have altered activity (notably increased abundance of the pentose phosphate pathway metabolites [Figure S3A]) through a number of compartments of central carbon metabolism linked through glucose-6-P, galactose does not directly fuel this system (as seen by the lack of labeling by 13C6 galactose). Similarly, galactose treatment does not consistently alter the metabolic profile of the central carbon metabolic network linked through glucose-6-P (Figures S3A–C).
We further monitored both mitochondrial activity in PGM1-CDG fibroblasts and the effect of galactose treatment. We found that the energy charge ratio in PGM1-CDG fibroblasts that were cultured in the presence of either glucose only or both glucose and galactose is equivalent to that in untreated control fibroblasts (Figure S3D), suggesting that there was no mitochondrial impairment.
To further evaluate mitochondrial activity on glucose media, we examined the respiratory capacity of untreated PGM1-CDG fibroblasts through oxygraphy (Figure S3E). We observed no consistent change for coupled (complex V [CV]-linked), uncoupled (maximal rate), or isolated CIV-specific rates of respiration, although some individuals did appear to have reproducibly elevated rates (Figure S3E).
To evaluate the effect of galactose on respiratory function, we monitored the activity of the mitochondrial respiratory chain: CI–IV in PGM1-deficient fibroblasts cultured in the presence or absence of galactose. With the exception of CIII, none of the complexes in untreated or galactose-treated PGM1-CDG fibroblasts had altered rates of activity (Figure S3F), as normalized to either citrate synthase (CS) activity (a marker for mitochondrial volume) or total protein (a marker for cell volume). For complex III, rates of activity appeared elevated in both untreated and galactose-treated PGM1-CDG fibroblasts relative to those in untreated controls (Figure S3F), matching the tendency for increased respiration in these cells (Figure S3E). The basis of this increased activity is unknown, but it could stem from altered levels of hexose-phosphates (see below), which are reported to regulate CIII activity.20
This is critical, as fibroblasts cultured with galactose alone as a carbon source are more susceptible to mitochondrial stress.21 By comparison, our fibroblasts cultured here with glucose and galactose together show no such susceptibility, further supporting the idea that galactose treatment in PGM1-CDG in the presence of physiologic glucose intake (as it is currently advised, and including frequent feeding and corn starch therapy) is safe.
Galactose Treatment Replenishes Depleted Levels of Galactose-1-P and the Nucleotide Monosaccharides UDP-Glucose and UDP-Galactose
Accordingly, our focus shifted to mapping the metabolic alterations that occur in PGM1-CDG fibroblasts in the Leloir pathway that directly connects galactose and glucose metabolism.
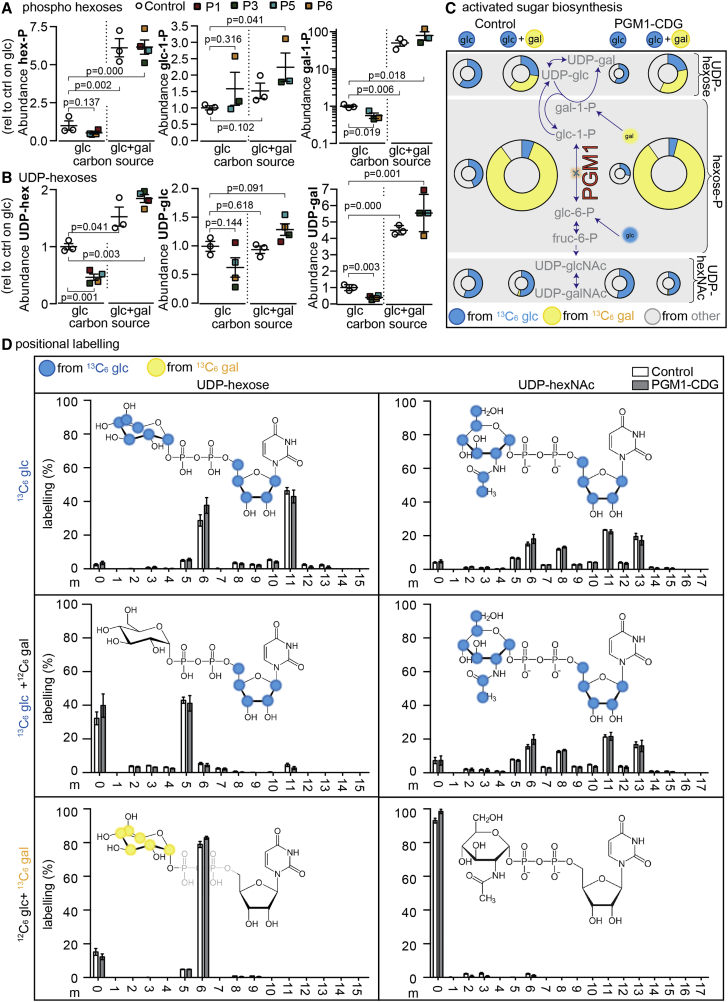
We observed via LC-MS that the total levels of hexose-P species (glucose-P, galactose-P, fructose-P, and mannose-P; Figure 1A) were depleted in PGM1-CDG fibroblasts cultured on glucose only, and they were massively boosted by the addition of galactose (Figure 3A; PGM1-CDG fibroblasts untreated at 0.5-fold and galactose treated at 6.1-fold, relative to untreated control fibroblasts, respectively). Analysis of these sub-populations in PGM1-CDG fibroblasts by GC-MS strikingly reveals that galactose-1-P is the only hexose-P isomer which is A) depleted and B) significantly boosted by galactose therapy (Figures 3A and S5; galactose-1-P in PGM1-CDG fibroblasts untreated at 0.6-fold and galactose treated at 79.4-fold, relative to untreated control fibroblasts, respectively). That glucose-1-P was not also depleted and only slightly increased in abundance after galactose treatment in PGM1-CDG cells is surprising. This could stem from the fact that these individuals have a secondary defect in the enzyme GALT (UDP-glucose + galactose 1-phosphate ⇌ glucose 1-phosphate + UDP-galactose) as previously reported.6 This secondary defect in GALT may then cause a secondary block at glucose-1-P. Because it is directly connected to galactose-1-P metabolism, we also observed via LC-MS that UDP-hexoses (UDP-glucose and UDP-galactose) were depleted in untreated PGM1-CDG fibroblasts and were boosted above untreated control fibroblast levels by galactose treatment (Figure 3B; PGM1-CDG fibroblasts untreated at 0.5-fold and galactose treated at 1.5-fold, relative to untreated controls, respectively).
Figure 3.
Depleted Hexose-P and UDP-Hexoses in PGM1-CDG Fibroblasts are Rescued with Galactose Supplementation
(A) The relative abundances of total hexose-P (as resolved by LC-MS) and galactose-1-P and glucose-1-P (as resolved by GC-MS) in control (biological n = 3, technical n = 1–4 for all panels) and PGM1-CDG fibroblasts (technical n = 2–4 in panels A and B) cultured in glucose (glc)-only medium or in glucose and galactose (glc+gal).
(B) The relative abundances (as resolved by LC-MS) of total UDP-hexoses and of UDP-glucose and UDP-galactose (as resolved by the selective enzymatic conversion of UDP-glucose to UDP-glucuronic acid by UDP-glucose-6-dehydrogenase [UGDH]) in control and PGM1-CDG fibroblasts cultured in glucose-only medium or in glucose and galactose medium.
(C) Tracer-based metabolomics in control and PGM1-CDG fibroblasts (from P1, P3, P5, and P6; technical n = 2–4) cultured with or without 2 mM galactose and with 13C6 glucose or 13C6 galactose. The metabolomics describe nucleotide-monosaccharide (UDP-hexose and UDP-hexNAc) biosynthesis from hexose-Ps (galactose-1-P, glucose-1-P, glucose-6-P, fructose-6-P, mannose-6-P, and mannose-1-P). The different areas of the pie charts represent abundance relative to the untreated controls. Blue, yellow, and gray shading represents fractional contributions from 13C6 glucose, 13C6 galactose, or other sources, respectively.
(D) Isotopologue profiling (m = the total number of carbons labeled from the described source) in UDP-hexose and UDP-hexNAc in control and PGM1-CDG fibroblasts (from P1, P3, P5, and P6; technical n = 1) treated with or without 2 mM galactose and with 13C6 glucose or 13C6 galactose. Blue and yellow shading over target molecules depicts a significant fractional contribution from 13C6 glucose or 13C6 galactose tracers, respectively, as identified by fragmentation analysis (see Figure S6). The statistics are from unpaired Student’s t tests, and the error bars are ± SEM.
To help resolve the identity and abundance of the UDP-hexoses, we enzymatically converted UDP-glucose to UDP-glucuronic acid with UDP-glucose-6-dehydrogenase (UGDH; EC: 1.1.1.22). By using this approach, we observed that both UDP-glucose and UDP-galactose levels mirrored changes in glucose-1-P and galactose-P levels. Specifically, UDP-glucose and UDP-galactose appear depleted in untreated PGM1-CDG fibroblasts, and levels of both metabolites were boosted with galactose treatment, although UDP-galactose was boosted to much higher levels than UDP-glucose (Figure 3B; UDP-glucose, 0.5- and 1.5-fold and UDP-galactose, 0.4- and 7.8-fold in untreated and galactose-treated PGM1-CDG fibroblasts, relative to untreated controls, respectively). The depletion of both UDP-glucose, necessary for ER-related glycosylation, and UDP-galactose, necessary for GA-related glycosylation, critically provides a possible explanation for the mixed CDG type I-II phenotype seen in affected individuals and for its correction upon galactose treatment.
Tracer-based metabolomics provided another tool for understanding how metabolic activity in these cells was altered (Figure 3C). Beyond observing that hexose-P and UDP-hexose pools were depleted in untreated PGM1-CDG fibroblasts, we also found that the fractional contribution (relative contribution of labeled carbons derived from the tracer) from 13C6 glucose into hexose-P is much lower in PGM1-CDG fibroblasts than in controls (control = 44%, PGM1-CDG = 28%; p value of a two-tailed, un-paired t test = 0.032).
We further observed that 13C6 galactose is specifically linked to the production of the nucleotide monosaccharides UDP-glucose and UDP-galactose in treated fibroblasts (Figure 3C), but it is completely absent in the formation of other key nucleotide monosaccharides such as UDP-N-acetylhexosamines (UDP-hexNAc: UDP-glcNAc and UDP-galNAc; Figure 3C)
Together, these data again suggest that galactose therapy in PGM1-CDG is ultimately effective through the modulation of UDP-monosaccharide levels.
To more specifically define the contribution of glucose and galactose to the composite molecules the UDP-hexoses and UDP-hexNAcs, we investigated their isotopologue profiles (Figures 3D). We found that not only is the contribution of glucose massively reduced in UDP-hexoses in the presence of galactose, but also that UDP-hexoses are metabolically re-wired, as evidenced by distinct changes in their isotopologue profiles (Figure 3D).
Specifically, in culture conditions only containing glucose, both the ribose (m5 isotopologue) and hexose (m6 isotopologue) moieties (combined m5 + m6 = m11) of UDP-hexoses appear to be substantially derived from 13C6 glucose (Figure 3D). In contrast, in culture conditions with glucose and galactose, 13C6 glucose appears to solely contribute to the ribose moiety of UDP-hexose (unique m5 isotopologue), and 13C6 galactose appears to contribute selectively to the hexose moiety (unique m6 isotopologue; Figure 3D). By comparison, the formation of UDP-hexNAcs is invariant in the presence or absence of galactose (Figure 3D).
To validate our interpretation of the isotopologue data, we performed a fragmentation analysis of UDP-hexoses and UDP-hexNAcs (Figure S6). This approach generates fragments that contain solely the ribose, uracil, or hexose moieties (or combinations thereof) of the parent molecules. The aforementioned metabolic re-wiring of the cell by galactose treatment was validated by this approach.
Together, these data strongly suggest that galactose therapy functions through the restoration of UDP-hexoses (Figure 1A) by refueling the hexose moiety of these nucleotide monosaccharides. In turn, impaired ER glycosylation requiring UDP-glucose for N-glycan maturation (Figure 1B), and GA glycosylation requiring UDP-galactose (Figure 1C), may be restarted.
PGM1-CDG Fibroblasts Incorporate 13C6 Galactose in De Novo N-Glycans
Finally, we followed the molecular fate of the nucleotide monosaccharides (as described above) into the construction of mature N-glycans in the presence and absence of galactose. This was achieved by hydrolyzing membrane-associated N-glycans to yield their individual monosaccharides, notably fucose, mannose, glcNAc, and galactose. Using 13C6 tracer metabolomics, we re-traced their origins from 13C6 galactose or 13C6 glucose in the culture media, to nucleotide-monosaccharides including those described above, and finally, as described here, into the mature N-glycans. Fucose, mannose, glcNAc, and galactose were the focus of our analysis because they are present in mature membrane N-glycans, whereas glucose only exists in the ER-associated, immature N-glycans (Figures 1B and 1C).
The result was a switch in the origin of galactose in the mature N-glycans (Figure 4). Specifically, fibroblasts cultured in media containing glucose only (untreated) sourced the galactose found in mature N-glycans largely from the supplied 13C6 glucose (Figure 4A; fractional contribution from 13C6 glucose in the control = 51%; in PGM1-CDG = 68%). In comparison, fibroblasts cultured in media containing both glucose and galactose derived the galactose found in N-glycans from the 13C6 galactose in the culture media (Figure 4B; fractional contribution from 13C6 galactose in the control = 63%; in PGM1-CDG = 75%).
Figure 4.
13C6 Galactose is Incorporated into Mature N-Glycans
(A and B) Membrane N-glycans from control (biological n = 2, technical n = 1–2) and PGM1-CDG (from P3, P5, and P6; technical n = 1–2) fibroblasts cultured in the presence of either (A) 13C6 glucose (glc) only or (B) glucose + galactose (glc+gal) with 13C6 glucose or 13C6 galactose were hydrolyzed, and the fractional contribution (% of 13C6-labeled carbons in the molecule) from 13C6 glucose (blue), 13C6 galactose (yellow), or other sources (gray) in fucose (F), mannose (man), galactose (gal), and glcNAc derived from the mature N-glycans was resolved. The statistics are from unpaired Student’s t tests, and the error bars are ± SEM. The bottom panels display the route of nucleotide-monosaccharide construction for glucose and galactose monosaccharides under the different culture conditions in PGM1-CDG fibroblasts, and they also display the resulting N-glycans that are produced through the ER and GA. Other abbreviations are as follows: SA = sialic acid.
This contrasts with the origins of the other monosaccharides (fucose, mannose, and glcNAc) that we monitored and that were derived from the N-glycans; these were invariably derived from 13C6 glucose, irrespective of the culture conditions tested (Figure 4).
These data are significant, as they concretely link galactose therapy to glycosylation and reiterate that the mechanism is through replenishing depleted levels of galactose-1-P and subsequently the nucleotide monosaccharides UDP-glucose and UDP-galactose (Figure 4B). The restoration of both metabolites can then restore both ER-linked glycosylation (Figure 1B), requiring UDP-glucose, and GA-linked glycosylation (Figure 1C), requiring UDP-galactose.
Discussion
Treatment of IEMs, such as CDG, is inherently extremely challenging and has so far met with limited success.22, 23 PGM1-CDG is one of the few such CDG for which a successful nutritional treatment exists in the form of oral galactose therapy.7, 24 Here, we show in eight individuals with PGM1-CDG that long-term galactose supplementation improves the clinical progression of PGM1 deficiency, and not just in their laboratory data. However, why PGM1 deficiency leads to its specific biochemical abnormality (detectable as a mixed type-I and type-II CDG profile characterized by the loss of glycans and glycan chains with decreased galactose content) remained unclear. Additionally, despite galactose’s apparent success with respect to the clinical improvement of laboratory and glycosylation abnormalities, the biochemical mechanisms that underlie its positive effect—notably the reversal of deficient ER-related glycan synthesis and GA-localized galactosylation—had remained elusive.
To address this question, we deployed tracer metabolomics to understand the metabolic consequences of the mutated PGM1 and its role in galactose therapy. Tracer metabolomics is still an underexploited tool in the field of IEMs,25, 26 least of all in CDG, but it is powerful because it allows for the study of systemic effects and not just the single-enzyme analysis of old. By taking such a view, we propose that we might take into account other, previously overlooked metabolic adaptions in IEMs, and in doing so, highlight opportunities for dietary therapies.
To this end, we report here that the mechanism by which galactose therapy is effective in PGM1-CDG is through bypassing metabolic defects in nucleotide-sugar production by boosting depleted levels of galactose-1-P and subsequently the nucleotide sugars UDP-glucose and UDP-galactose (Figure 1A). Replenishing both metabolite levels restores both ER-linked glycosylation that requires UDP-glucose and GA-linked glycosylation that requires UDP-galactose. In this study, we demonstrated the functional significance of this labeling by showing that exogenous galactose replaces glucose for labeling galactose sugars in mature glycans.
These observations are further relevant beyond just PGM1-CDG because they offer significant clues on how to approach the treatment of other CDG wherein it has been observed that disrupted nucleotide-sugar activation (e.g., in PMM2-CDG), transfer and transport (e.g., in SLC35C1-, SLC35A1-, B4GALT1-, and SLC35A2-CDG), or transport regulation (e.g., inSLC38A9-, and TMEM165-CDG) is at play.
Declaration of Interests
Bart Ghesquière, Tamas Kozicz, and Eva Morava declare that they have filed a provisional patent relating to this work for the treatment of CDG at application number: 07039-1808P01 / 2018-132.
Acknowledgments
B.G., D.C., and P.V. were generously supported by a KU Leuven internal C1 funding grant (EFF-D2860-C14/17/110), B.G. by the Nutricia Metabolic Research Fund, T.H. by the General University Hospital in Prague in the Czech Republic (RVO-VFN 64165) and the Ministry of Health of the Czech Republic (MZ CR AZV 16-31932A). D.C. acknowledges generous support from the Shire Research Chair. We also thank Dr. Gert Matthijs (UZ Leuven) and Dr. Kucukcongar A. (Gazi University Medical School) for kindly providing us with PGM1-CDG fibroblast cultures. Finally, we are very grateful for the help related to the GC-MS analyses of hexose-P sugars by Dr. Frank David (Research Institute Chromatography, Kortrijk, Belgium).
Published: April 11, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.003.
Contributor Information
Eva Morava, Email: morava-kozicz.eva@mayo.edu.
Bart Ghesquière, Email: bart.ghesquiere@kuleuven.vib.be.
Accession Numbers
The accession numbers for the genes, PGM1 and ICAM1, that are reported in this paper are GenBank: NM_002633.2 and GenBank: NM_000201.3, respectively. Patient variants have been submitted to the Leiden Open Variation Database (LOVD) available to the general public (PGM1_000005, PGM1_000004).
Web Resources
Online Mendelian Inheritance in Man, http://www.omim.org
Supplemental Data
References
- 1.Jaeken J., van Eijk H.G., van der Heul C., Corbeel L., Eeckels R., Eggermont E. Sialic acid-deficient serum and cerebrospinal fluid transferrin in a newly recognized genetic syndrome. Clin. Chim. Acta. 1984;144:245–247. doi: 10.1016/0009-8981(84)90059-7. [DOI] [PubMed] [Google Scholar]
- 2.Roth Z., Yehezkel G., Khalaila I. Identification and quantification of protein glycosylation. Int. J. Carbohydr. Chem. 2012;2012:1–10. [Google Scholar]
- 3.Jaeken J., Péanne R. What is new in CDG? J. Inherit. Metab. Dis. 2017;40:569–586. doi: 10.1007/s10545-017-0050-6. [DOI] [PubMed] [Google Scholar]
- 4.Witters P., Cassiman D., Morava E. Nutritional therapies in congenital disorders of glycosylation (CDG) Nutrients. 2017;9:e1222. doi: 10.3390/nu9111222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timal S., Hoischen A., Lehle L., Adamowicz M., Huijben K., Sykut-Cegielska J., Paprocka J., Jamroz E., van Spronsen F.J., Körner C. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum. Mol. Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- 6.Tegtmeyer L.C., Rust S., van Scherpenzeel M., Ng B.G., Losfeld M.E., Timal S., Raymond K., He P., Ichikawa M., Veltman J. Multiple phenotypes in phosphoglucomutase 1 deficiency. N. Engl. J. Med. 2014;370:533–542. doi: 10.1056/NEJMoa1206605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong S.Y., Gadomski T., van Scherpenzeel M., Honzik T., Hansikova H., Holmefjord K.S.B., Mork M., Bowling F., Sykut-Cegielska J., Koch D. Oral D-galactose supplementation in PGM1-CDG. Genet. Med. 2017;19:1226–1235. doi: 10.1038/gim.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong S.Y., Beamer L.J., Gadomski T., Honzik T., Mohamed M., Wortmann S.B., Brocke Holmefjord K.S., Mork M., Bowling F., Sykut-Cegielska J. Defining the phenotype and assessing severity in phosphoglucomutase-1 deficiency. J Pediatr. 2016;175:130–136e8. doi: 10.1016/j.jpeds.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Ondruskova N., Honzik T., Vondrackova A., Tesarova M., Zeman J., Hansikova H. Glycogen storage disease-like phenotype with central nervous system involvement in a PGM1-CDG patient. Neuroendocrinol. Lett. 2014;35:137–141. [PubMed] [Google Scholar]
- 10.Fernandez C.A., Des Rosiers C., Previs S.F., David F., Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y., Borges C.R. A spin column-free approach to sodium hydroxide-based glycan permethylation. Analyst (Lond.) 2017;142:2748–2759. doi: 10.1039/c7an00396j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Yager C., Reynolds R., Palmieri M., Segal S. Erythrocyte galactose 1-phosphate quantified by isotope-dilution gas chromatography-mass spectrometry. Clin. Chem. 2002;48:604–612. [PubMed] [Google Scholar]
- 13.Stojkovic T., Vissing J., Petit F., Piraud M., Orngreen M.C., Andersen G., Claeys K.G., Wary C., Hogrel J.Y., Laforêt P. Muscle glycogenosis due to phosphoglucomutase 1 deficiency. N. Engl. J. Med. 2009;361:425–427. doi: 10.1056/NEJMc0901158. [DOI] [PubMed] [Google Scholar]
- 14.Voermans N.C., Preisler N., Madsen K.L., Janssen M.C., Kusters B., Abu Bakar N., Conte F., Lamberti V.M., Nusman F., van Engelen B.G. PGM1 deficiency: Substrate use during exercise and effect of treatment with galactose. Neuromuscul. Disord. 2017;27:370–376. doi: 10.1016/j.nmd.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 15.He P., Ng B.G., Losfeld M.E., Zhu W., Freeze H.H. Identification of intercellular cell adhesion molecule 1 (ICAM-1) as a hypoglycosylation marker in congenital disorders of glycosylation cells. J. Biol. Chem. 2012;287:18210–18217. doi: 10.1074/jbc.M112.355677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beamer L.J. Mutations in hereditary phosphoglucomutase 1 deficiency map to key regions of enzyme structure and function. J. Inherit. Metab. Dis. 2015;38:243–256. doi: 10.1007/s10545-014-9757-9. [DOI] [PubMed] [Google Scholar]
- 17.Regni C., Naught L., Tipton P.A., Beamer L.J. Structural basis of diverse substrate recognition by the enzyme PMM/PGM from P. aeruginosa. Structure. 2004;12:55–63. doi: 10.1016/j.str.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Nowak-Sliwinska P., Alitalo K., Allen E., Anisimov A., Aplin A.C., Auerbach R., Augustin H.G., Bates D.O., van Beijnum J.R., Bender R.H.F. Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis. 2018;21:425–532. doi: 10.1007/s10456-018-9613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buescher J.M., Antoniewicz M.R., Boros L.G., Burgess S.C., Brunengraber H., Clish C.B., DeBerardinis R.J., Feron O., Frezza C., Ghesquiere B. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Díaz-Ruiz R., Avéret N., Araiza D., Pinson B., Uribe-Carvajal S., Devin A., Rigoulet M. Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J. Biol. Chem. 2008;283:26948–26955. doi: 10.1074/jbc.M800408200. [DOI] [PubMed] [Google Scholar]
- 21.Robinson B.H., Petrova-Benedict R., Buncic J.R., Wallace D.C. Nonviability of cells with oxidative defects in galactose medium: A screening test for affected patient fibroblasts. Biochem. Med. Metab. Biol. 1992;48:122–126. doi: 10.1016/0885-4505(92)90056-5. [DOI] [PubMed] [Google Scholar]
- 22.Parikh S., Saneto R., Falk M.J., Anselm I., Cohen B.H., Haas R., Medicine Society T.M. A modern approach to the treatment of mitochondrial disease. Curr. Treat. Options Neurol. 2009;11:414–430. doi: 10.1007/s11940-009-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campeau P.M., Scriver C.R., Mitchell J.J. A 25-year longitudinal analysis of treatment efficacy in inborn errors of metabolism. Mol. Genet. Metab. 2008;95:11–16. doi: 10.1016/j.ymgme.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Morava E. Galactose supplementation in phosphoglucomutase-1 deficiency; review and outlook for a novel treatable CDG. Mol. Genet. Metab. 2014;112:275–279. doi: 10.1016/j.ymgme.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reijngoud D.J. Flux analysis of inborn errors of metabolism. J. Inherit. Metab. Dis. 2018;41:309–328. doi: 10.1007/s10545-017-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaude E., Schmidt C., Gammage P.A., Dugourd A., Blacker T., Chew S.P., Saez-Rodriguez J., O’Neill J.S., Szabadkai G., Minczuk M. NADH shuttling couples cytosolic reductive carboxylation of glutamine with glycolysis in cells with mitochondrial dysfunction. Mol Cell. 2018;69:581–593e7. doi: 10.1016/j.molcel.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.