Abstract
During the medieval period, hundreds of thousands of Europeans migrated to the Near East to take part in the Crusades, and many of them settled in the newly established Christian states along the Eastern Mediterranean coast. Here, we present a genetic snapshot of these events and their aftermath by sequencing the whole genomes of 13 individuals who lived in what is today known as Lebanon between the 3rd and 13th centuries CE. These include nine individuals from the “Crusaders’ pit” in Sidon, a mass burial in South Lebanon identified from the archaeology as the grave of Crusaders killed during a battle in the 13th century CE. We show that all of the Crusaders’ pit individuals were males; some were Western Europeans from diverse origins, some were locals (genetically indistinguishable from present-day Lebanese), and two individuals were a mixture of European and Near Eastern ancestries, providing direct evidence that the Crusaders admixed with the local population. However, these mixtures appear to have had limited genetic consequences since signals of admixture with Europeans are not significant in any Lebanese group today—in particular, Lebanese Christians are today genetically similar to local people who lived during the Roman period which preceded the Crusades by more than four centuries.
Keywords: aDNA, medieval period, Roman period, whole-genome sequences, Lebanon, Sidon, population history
Main Text
Human migrations, which often accompanied historical battles and invasions, have profoundly reshaped the genetic diversity of local populations in many regions. The Mongols, around Genghis Khan’s time, spread their male lineages throughout Asia from the Pacific to the Caspian Sea,1 while in South America, the arrival of colonial Iberians resulted in the European ancestry becoming the major component in the genomes of many South Americans today.2 But do all mass migrations leave genetic imprints in local populations? For two centuries, starting in 1095 CE, hundreds of thousands of Europeans arrived in the Near East to fight in the Crusades and to settle in the newly established European states along the Eastern Mediterranean coast.3 The historical records from this period report varying episodes of forced displacement of the local people, or co-existence and mixing with them.3, 4 We have previously reported finding European Y chromosome lineages in the present-day population of Lebanon5 and suggested those could have originated from the Crusaders when Lebanon was under their rule during the medieval period. However, more recently, using whole-genome sequences from modern and ancient individuals, we found that present-day Lebanese derive most of their genetic ancestry from the local Bronze Age population and from a Eurasian Steppe-related admixture which occurred around 1,750–170 BCE.6 Thus, the Lebanese autosomal genomes appear not to have been impacted by the Crusades.
Ancient DNA (aDNA) from medieval Crusaders who traveled to the Near East and from local people who were their contemporaries and interacted with them can potentially resolve the discrepancy between the historical records reporting admixed descendants of Crusaders in the Near East and the genetics of modern populations not displaying such a signal. aDNA from this period can also resolve uncertainties in the demographic processes that accompanied the Crusades, providing insight into questions such as what was the provenance of the Crusaders’ armies and the extent to which they were from genetically and geographically diverse groups. How genetically similar are modern Near Easterners to the medieval populations and what genetic changes occurred after the Crusades? These questions can potentially be answered today in great detail using ancient genomics; however, obtaining aDNA from Crusaders is challenging and hindered by two major factors: the hot and humid climate in the Near East negatively impacts the survival of aDNA7 and burials of individuals who can be linked to the Crusaders are rare.8 One of the few known Crusader burial sites is located in the city of Sidon (in the south of present-day Lebanon), an important stronghold in the Crusaders’ Kingdom of Jerusalem and a scene of major battles between the Crusaders and the Arabs from 1110 CE to 1249 CE (Figure S1). A recent archaeological excavation in Sidon near the ruins of a Crusader castle uncovered two mass burials consisting of skeletal remains from a minimum of 25 individuals who had signs of inter-personal violent injuries and dated using radiocarbon to 1025–1283 calCE (calibrated date) (Table S1). The bodies were placed in arbitrary manner into two adjacent simple pits dug in the ground (Figure S2). The location, date, and condition of the burials, together with a Crusader coin issued in Italy in 1245–1250 CE9 and five buckles with designs associated with medieval Europe,8 all suggest that the burials could have been for Crusaders killed in battle during the 13th century CE.
We sampled the petrous portion of the temporal bones from 16 of these individuals and, additionally, sampled five local individuals from an archaeological excavation in Qornet ed-Deir, Jabal Moussa UNESCO Biosphere Reserve in Mount Lebanon (Figure S3),10 who lived during the Roman period (237–632 calCE), and thus these would represent the local ancestry before the time of the Crusades. We extracted DNA, built double-stranded libraries according to published protocols,11, 12, 13 and sequenced the libraries on an Illumina HiSeq 2500 using 2 × 75 bp reads. We processed the sequences using PALEOMIX,14 retained reads ≥30 bp, and collapsed pairs with minimum overlap of 15 bp and a mismatch rate ≤0.1. We mapped the merged sequences to the hs37d5 reference sequence, removed duplicates, removed bases from the end of the reads until the frequency of nucleotide misincorporation dropped to below 5%, and randomly sampled a single sequence with a minimum base quality of ≥20 to represent each SNP. We found that seven samples from the Crusaders’ pit had less than 2% of endogenous DNA from the first exploratory sequencing runs and thus we excluded them from additional sequencing and from further analyses. The remaining nine samples from the Crusaders’ pit and all five samples from the Roman period had 2% to 58% endogenous DNA with post-mortem damage patterns typical of ancient DNA (Figure S4) and produced genomic coverage between 0.4× and 4.4×, plus mitochondrial DNA (mtDNA) genome coverage between 29× and 415× (Table 1). We estimated contamination from the mtDNA genome of all individuals and the X chromosomes of males15, 16 and found that the sequence data were minimally contaminated (Table S2), except for sample QED-9 (from the Roman period) who had 8%–10% estimated mtDNA contamination; thus this individual was not included in subsequent analyses.
Table 1.
Samples Analyzed in This Study
| ENA Number | ID | Excavation Site | Period | Datea(calCE) | Mapped Reads | Mapped Read % | Coverage Genomic | Coverage Mt | Sexb | Mt Haplogroup | Y Haplogroup |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ERS3189349 | SI-38 | Sidon | medieval | – | 33711271 | 4 | 0.5 | 72 | male | J1b4a1 | E-L677 |
| ERS3189350 | SI-39 | Sidon | medieval | 1191–1283 | 35804710 | 3 | 0.6 | 60 | male | H5′36 | R-P312 |
| ERS3189351 | SI-40 | Sidon | medieval | – | 21871263 | 2 | 0.4 | 38 | male | U5a1g | R-P311 |
| ERS3189352 | SI-41 | Sidon | medieval | 1187–1266 | 169086445 | 11 | 3 | 198 | male | HV0a | R-DF27 |
| ERS3189353 | SI-42 | Sidon | medieval | 1154–1281 | 37079898 | 5 | 0.7 | 68 | male | J1b1a1 | T-M70 |
| ERS3189348 | SI-44 | Sidon | medieval | – | 25262493 | 3 | 0.4 | 52 | male | HV1b | J-M304 |
| ERS3189355 | SI-45 | Sidon | medieval | 1219–1278 | 193449685 | 12 | 3.75 | 275 | male | J1d1a1 | Q-M346 |
| ERS3189357 | SI-47 | Sidon | medieval | – | 26722544 | 2 | 0.4 | 44 | male | H2a5 | R-M269 |
| ERS3189343 | SI-53 | Sidon | medieval | 1025–1154 | 266974394 | 19 | 4.4 | 387 | male | T2 | R-CTS300 |
| ERS3189333 | QED-2 | Qornet ed-Deir | Roman | 244–400 | 71266615 | 18 | 1.31 | 127 | male | T1 | T-CTS9882 |
| ERS3189335 | QED-4 | Qornet ed-Deir | Roman | 426–632 | 52457527 | 14 | 0.84 | 78 | female | U3b | N/A |
| ERS3189338 | QED-7 | Qornet ed-Deir | Roman | 237–389 | 95424013 | 22 | 1.55 | 164 | female | HV1b | N/A |
| ERS3189339 | QED-9 | Qornet ed-Deir | Roman | – | 154883416 | 58 | 3.14 | 415 | female | H | N/A |
| ERS3189342 | QED-12 | Qornet ed-Deir | Roman | – | 18591641 | 6 | 0.33 | 29 | female | H2a5 | N/A |
Calibrated radiocarbon date range
Genetically determined
We combined our 13 remaining ancient samples with published ancient and modern data, creating three datasets. The HO set included 2,583 modern humans genotyped on the Human Origins array17, 18, 19 plus 300 ancient individuals6, 18 and consisted of 590,301 SNPs. The SGDP set included 78 modern individuals (West Eurasians and Mbuti) from the Simons Genome Diversity Project20 whole-genome sequenced to high coverage, plus 300 ancient individuals and consisted of 939,107 SNPs. The 1000GP set consisted of 311 modern individuals (CEU, CHB, YRI) from the 1000 Genomes Project phase 321 plus 100 modern Greek (GRK) and 99 modern Lebanese,6 all whole-genome sequenced to ∼8× coverage and consisting of ∼80 million SNPs.
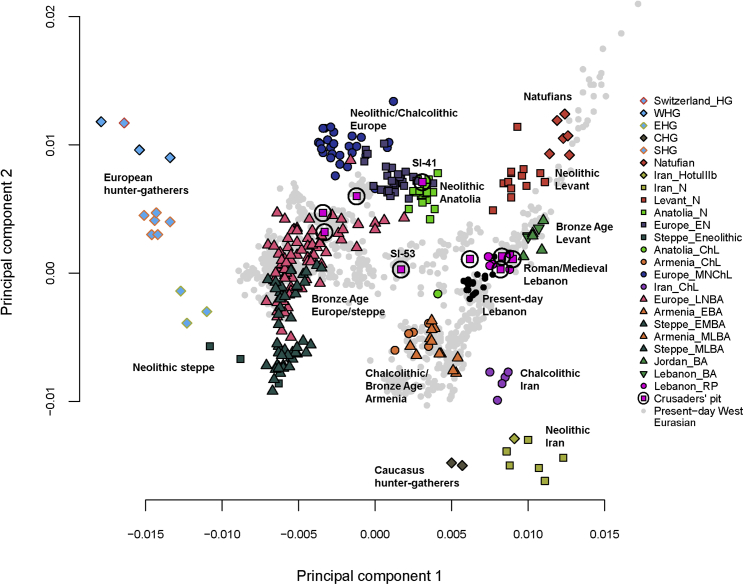
We first identified the biological sex of our samples by determining the ratio of sequences aligning to the X and Y chromosomes22 and found that all individuals in the Crusaders’ pit were genetically males. We then projected the ancient Lebanese samples onto a principal component analysis (PCA)23 plot based on modern West Eurasians of the HO dataset (Figures 1 and S5). The PCA plot revealed a previously described6, 18 structure differentiating between Europeans and Near Easterners on the first principal component with a cline in both regions over the second component. We found that all individuals from Lebanon’s Roman period (Lebanon_RP) clustered with Near Easterners and were close to present-day Lebanese. In contrast, the Crusaders’ pit individuals were more diverse and we classified them into three groups based on their PCA position. First, a group of four individuals appeared to be local Near Easterners since they clustered with the Roman period and present-day Lebanese. Second, three individuals appeared to be Europeans and clustered with different European populations (two clustered with Spaniards and were close to Basque, French, and Northern Italians, and the third clustered with Sardinians). Third, two individuals appeared to have an intermediate position between Europeans and Near Easterners: individual SI-41 overlapped with Neolithic Anatolians on the PCA and was distant from any modern West Eurasian population, and individual SI-53 overlapped with Ashkenazi Jews and South Italians.
Figure 1.
Principal Components Analysis of West Eurasians
Eigenvectors were inferred using present-day populations (gray points) and the ancient samples (colored shapes) were projected onto the plot. The new ancient samples from the Roman period (Lebanon_RP) are represented by solid pink circles and the new ancient samples from the Crusaders period (Crusaders’ pit) are shown in pink squares with black circles. In addition, two samples discussed individually in the text are annotated with their IDs (SI-41 and SI-53) on the plot.
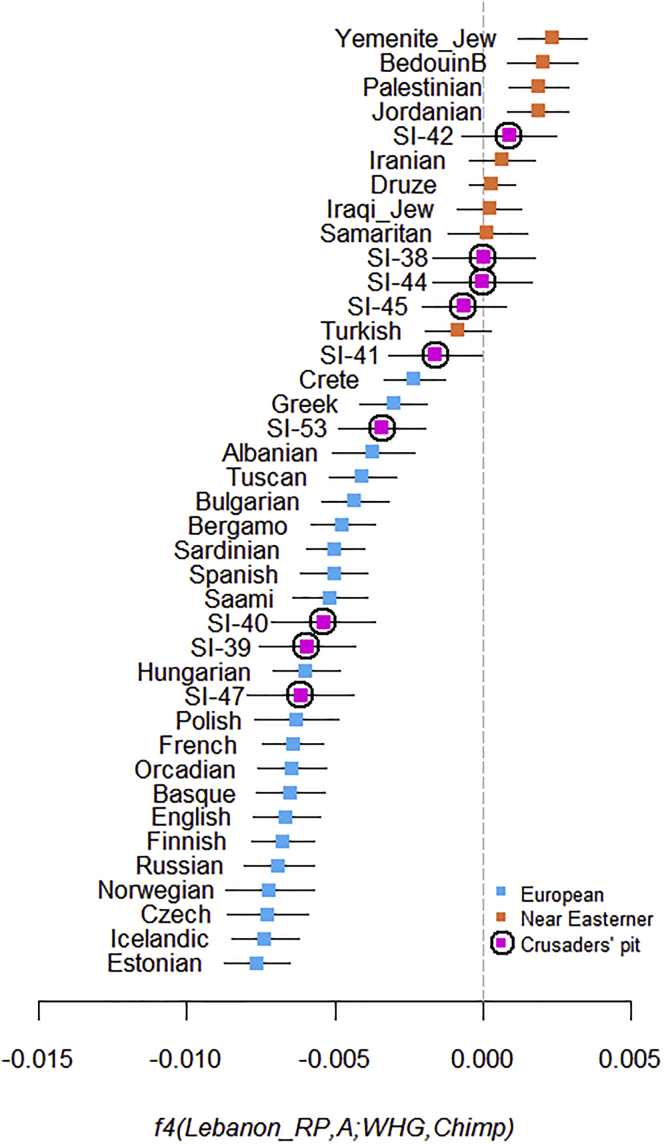
Therefore, the PCA results point to a diverse origin for the individuals buried in the Crusaders’ pit. We sought to confirm these results by testing the genetic affinity of the individuals to West European Hunter-Gatherers (WHG), who contributed ancestry to all Europeans but not to Near Easterners.17 We used the SGDP set to compute19 the statistic f4(Lebanese_RP, A; WHG, Chimpanzee). This is expected to be negative when A is European because of the excess of WHG ancestry in Europeans compared with the Roman period Lebanese, but should not be significantly different from zero when A is a Near Easterner. As expected, we found a significant contrast between Europeans and Near Easterners from their genetic affinity to WHG (Figure 2), confirming the diversity of the Crusaders’ pit individuals: three were Europeans, four were Near Easterners, and the two who had ambiguous ancestry on the PCA appear, based on the f-statistic, to have had European ancestry but with less WHG ancestry than the other Europeans found in the pit.
Figure 2.
Contrast between Europeans and Near Easterners from Their Genetic Affinity to WHG
The statistic f4(Lebanon_RP, A; WHG, Chimpanzee) is significantly negative when A is a European population. The ancient individuals from the Crusaders’ pit are represented by pink squares with black circles. We plot the f4 statistic value and ±3 standard errors.
Finding Europeans in the burial supports the observations from archaeology suggesting that the buried individuals could have been Crusaders. But we also found Near Eastern individuals buried in the same pit along with the Europeans. Two possible scenarios could explain this finding. (1) Following a battle between Crusaders/Europeans and a Muslim army/Near Easterners, the dead from both sides were buried in the same pit. (2) Alternatively, the Crusaders recruited local Near Easterners into their army and thus the burial was for individuals who were all fighting with the Crusaders. The historical records explain that the attacks on the Crusaders were led by Muslim armies mostly recruited from Syria, Iraq, Egypt, Turkey, and Bedouin tribes in the region. But we find the statistic f4(Lebanese_Christian, A; Lebanon_MP, Chimpanzee) is always positive when A is any Near Eastern population (Figure S6), suggesting that the present-day Lebanese Christians are genetically one of the closest populations to the Near Eastern individuals found in the pit. These results support historical records on local Christian integration into the Crusaders’ social structure including the military, with indigenous Christians joining the Crusaders as troops or becoming marshals and knights.4 However, we should note here that we are making an assumption that 800 years ago the medieval Near Easterners were already genetically structured in a way that allows us to differentiate between the different medieval populations in the region, i.e., a medieval Lebanese Christian could be genetically differentiated from a medieval Bedouin. Our tests (see below) suggest that genetic structure between the Lebanese and other Near Easterners could have existed more than 800 years ago but genetic structure between the Lebanese religious groups is probably more recent and therefore our tests might not be able to unambiguously differentiate between the Lebanese Christians and the Lebanese Muslims who lived during the medieval period.
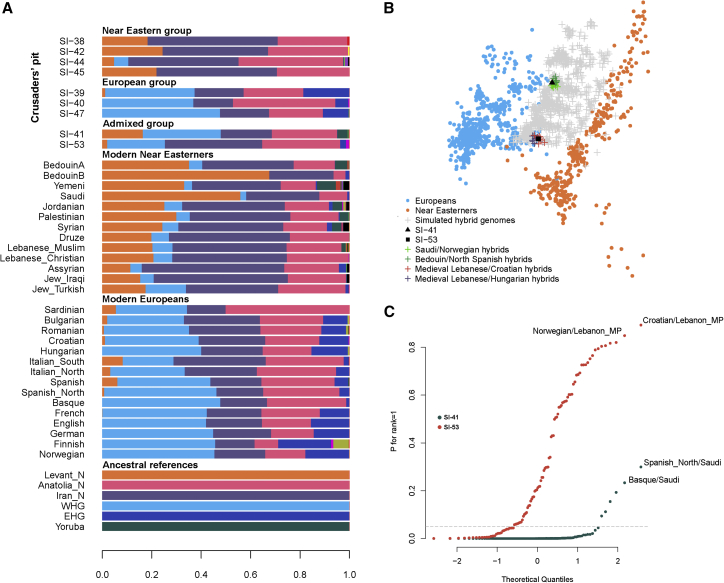
In addition to the three Europeans and four Near Easterners found in the burial, there were two individuals (SI-41 and SI-53) who we could not associate with any specific group. We consequently wanted to explore the possibility that these individuals’ particular pattern on the PCA might be due to their genomes being admixed. Therefore, we first ran an ADMXITURE24 analysis (Figure 3A) which confirmed the presence of three genetically different groups in the Crusaders' pit with SI-41 and SI-53 having intermediate ancestry compositions compared with the other ancient individuals. Next, we simulated hybrid diploid genomes by selecting pairs of individuals, one from the Near East and the second from Europe and we picked a random allele from each individual. We then projected the simulated hybrid genomes onto the PCA plot, looking for hybrid combinations that overlapped with individuals SI-41 and SI-53. We found that a mixture between a medieval Lebanese and a Croatian or a medieval Lebanese and a Hungarian could reproduce the PCA position of SI-53, while a mixture between a Saudi and a Norwegian, or a Bedouin and a Northern Spanish, could reproduce the position of SI-41 (Figure 3B). We assessed these results with formal tests using qpAdm25 and setting the source populations for SI-41 and SI-53 as pairs of populations selected from two groups from the HO set: (1) Medieval Lebanese, Lebanese Christian, Syrian, Palestinian, Assyrian, Iraqi Jew, Turkish Jew, Ashkenazi Jew, Saudi, and Bedouin and (2) French, Italian, Spanish, Basque, English, Norwegian, German, Croatian, Hungarian, Romanian, and Ashkenazi Jew. Then we selected a set of outgroups that are related differently to the source populations: Ust’-Ishim, Kostenki 14, MA1, Onge, Papuans, Chukchi, Karitiana, Eastern hunter-gatherers (EHG), WHG, Natufians, Caucasus hunter-gatherers, Neolithic Iranians, Neolithic Anatolians, and Neolithic Levantines. We found that both SI-41 and SI-53 can be modeled as mixtures of European and Near Eastern ancestries. For individual SI-41, the best-supported model is a descent from a Near Easterner related to a Bedouin or a Saudi and a European related to Northern Spanish or Basques (Figure 3C, Table S3). For individual SI-53, the best-supported models involved a Near Easterner related to medieval Lebanese, Lebanese Christians, or Jews and a European who can be from diverse origins (Figure 3C, Table S4). We investigated the robustness of our qpAdm inferences by testing, similarly to SI-41 and SI-53, the European individuals from the Crusaders’ pit and, as expected, we found that most of their ancestry was derived from Europeans in contrast to SI-41 and SI-53 (Table S5). Individual SI-53 clustered on the PCA with Ashkenazi Jews, Sicilians, and Southern Italians and therefore we wanted to test whether SI-53 could have descended directly from one of these populations who were previously reported to be admixed,26, 27 with the implication that admixture in SI-53 could then have occurred before the Crusaders’ time and in Europe instead of in the Near East. However, our results (Table S6) show that the Europeans have in general a distinct admixture pattern from the one observed in SI-53; among the populations tested, Ashkenazi Jews’ Near Eastern ancestry is mostly related to Near Eastern Jews, Sicilians’ European ancestry is related to Italians, and Southern Italians have Northern Italians as top sources of their European ancestry. We note here that the reference populations tested are not necessarily the precise parental populations; any genetically equivalent populations could have been involved, but the admixture patterns in SI-41 and SI-53 suggest our data (1) provide direct genetic evidence of admixture between Crusaders and Near Easterners, (2) consequently show admixture could have been common (at least in Sidon), as two out of nine sequenced individuals were admixed, (3) show that admixture with the Near Easterners was not limited to only Levantine Christians but also involved people genetically related to Saudis and Bedouins, and (4) provide support for our previous assumption that 800 years ago the Near Easterners were already genetically structured in a way that allows us to differentiate coastal Levantines from inland people such as Bedouins and Saudis.
Figure 3.
Admixed Individuals in the Crusaders’ Pit
(A) ADMIXTURE plot of the HO set using ∼80,000 transversions and showing results from K = 17 in a supervised run with references: Anatolia_N, Iran_N, EHG, WHG, Levant_N, Atayal, Chipewyan, Eskimo, Hadza, Mala, Mbuti, Nganasan, Onge, Papuan, Quechua, Taa_West, and Yoruba. We show results from the ancient individuals from the Crusaders’ pit in addition to the median of the ancestry proportions found in modern populations.
(B) The position on the PCA of two individuals found in the Crusaders’ pit can be explained by admixture. Simulated (first generation) hybrid samples (gray crosses, with some highlighted in color as indicated on the figure) consisting of genomes with positions represented by an allele from a Near Easterner (orange circles) and an allele from a modern European (blue circles) show intermediate positions on the PCA and overlap with the ancient individuals SI-41 and SI-53.
(C) Individuals SI-41 and SI-53 can be successfully modeled as mixture of Europeans and Near Easterners using qpAdm. We show the p values for rank = 1 and annotate the models with the top values.
Next, we analyzed the variations on the Y chromosomes28 of all ancient males and the mtDNA genomes29 of all ancient individuals and determined the respective haplogroups, which can provide information on the place of origin of the paternal and maternal lineages. All ancient individuals assigned in this work as either European or Near Eastern had Y and mtDNA haplogroups that reflected this ancestry, i.e., Europeans had Y-haplogroup R1b and mtDNA-haplogroups H or U5, while the Near Easterners had Y-haplogroups E, T, J, and Q and mtDNA-haplogroups J1 or HV (Table 1). However, the admixed individuals SI-41 and SI-53 had Y-haplogroup R1b-P312 typically found in European males and mtDNA haplogroups HV0 and T2 present in both Europe and the Near East. In particular, SI-41 carried the DF27 lineage, which is highly prevalent in Iberia (up to 70% of males in Basque) and rare elsewhere,30 supporting our previous results from the autosomal variants that this individual descended from a European related to Northern Spanish or Basques. The combined results from the Y and mtDNA haplogroups suggest that SI-41 and SI-53 possibly had a European father and a Near Easterner mother, but a more complex admixture scenario, where the parents themselves are admixed, could also produce this ancestry pattern. Our tests31 on the X chromosome were inconclusive about the ancestry of SI-41 and SI-53, probably because the reduced sequencing coverage and diversity compared with the autosomal genome make them less powerful (Figure S7).
Our data suggest that admixture occurred in Lebanon during the Crusaders’ time; therefore, we sought to investigate the consequences of this admixture for the genomes of the Lebanese by tracking the genetic changes that occurred in Lebanon before and after the Crusades. We started by using the SGDP set to compute the statistic f4(Lebanon_BA, Lebanon_RP; Ancient West Eurasian, Chimpanzee) which quantifies the genetic changes related to West Eurasians that occurred in Lebanon between the Bronze Age and the Roman period. We found there was an increase in Eurasian hunter-gatherer and Steppe population ancestry in Lebanon after the Bronze Age (Figure S8A), which provides direct evidence for our previous inference that this Eurasian ancestry in the Levant predates both the Crusaders and the Roman period.6 Next, we computed f4(Lebanon_RP, Lebanon_MP; Ancient West Eurasian, Chimpanzee) and found that the statistic is not significantly different from zero for any Ancient West Eurasian tested, thus indicating that there were no significant genetic changes between the Roman period and the medieval period in Lebanon (Figure S8B). We then tested f4(Lebanon_MP, Lebanese_Christians; Ancient West Eurasian, Chimpanzee) and f4(Lebanon_MP, Lebanese_Muslims; Ancient West Eurasian, Chimpanzee); there were no significant genetic differences between medieval Lebanese and present-day Lebanese Christians (Figure S8C), but we found that Lebanese Muslims had significantly lower genetic affinity to West Eurasians (Figure S8D). This genetic change in the Lebanese Muslims could potentially be a result of gene flow from a population genetically distant from West Eurasians. We investigated this possibility by computing f3(Lebanese_MP, A; Lebanese_Christians) and f3(Lebanese_MP, A; Lebanese_Muslims), which test whether the Lebanese groups descended from a mixture between medieval Lebanese and another population. We found that Lebanese Christians cannot be modeled in this way (Figure S9A), but Lebanese Muslims had negative f3-statistics when A was an African or a Central/East Asian population (Figure S9B), indicating that they are admixed from these sources. We confirmed these results by analyzing the 1000GP set using rarecoal-tools32 which identifies genetic ancestry using rare variants and thus complements the low sensitivity of the f-statistics to detect admixture when the ancestral mixing fractions are small. We find an enrichment of African and East Asian rare alleles in the Lebanese Muslims compared with the Lebanese Christians, but we found no substantial differences related to their European ancestry (Figure S10). Therefore, our results suggest that the genome-wide genetic legacy of the Crusaders cannot be observed today in the Lebanese even though the Lebanese ancestors have admixed with the Crusaders—as it is evident from the genome of individual SI-53. We propose that admixture during the Crusaders’ time was not widespread enough to leave genetic traces quantifiable in the present-day populations. A shared recent genomic history between Europeans and Near Easterners also contributes to rapidly diluting a specific genetic signal of gene flow from the Crusaders. For example, the Neolithic Anatolians and Bronze Age steppe people, who contributed ancestry to the Europeans,17 were themselves a mixture of populations that included Neolithic Levantines and Chalcolithic Iranians18 who were also major contributors to the genetics of Near Easterners.6 We show by simulating hybrid genomes of decreasing European ancestry that a Lebanese who had a French great grandparent cannot be distinguished—by their genetic relationship to ancient West Eurasians—from a Lebanese who did not have admixed ancestors (Figure S11). Thus our data highlight the power of aDNA in informing on past events that would be otherwise undetectable from modern autosomal DNA alone.
Remarkably, the significant genetic changes in the Lebanese following the Crusades appear to have been not from the Crusaders and were not in the Lebanese Christians, but were mostly restricted to the Lebanese Muslims. We analyzed admixture LD33, 34 in the Lebanese in order to date the time when admixture occurred in their history. We found two signals; the first admixture can be detected with overlapping dates in both Lebanese Christians (850–150 BCE, Z = 6.95) and Lebanese Muslims (900 BCE–500 CE, Z = 3.02) and is consistent with finding this admixture in our ancient Roman period samples 240–400 calCE. However, the second admixture was specific to Lebanese Muslims (Figures S12A and S12B) occurring around 1550–1700 CE (Z = 3.69) and can be detected when Africans and East Asians are used as reference populations. The time of this admixture coincides with the Ottoman Turkish rule over Lebanon and we propose that the Turks, who themselves carry East Asian ancestry (Figure S12C) from their Seljuk ancestors,35 brought this ancestry to the Lebanese Muslims. The African ancestry was introduced into the Lebanese Muslims most likely via the slave trade in the Ottoman Empire and the prohibition of non-Muslims from owning slaves during this period.36
In this report, we have presented new whole-genome sequence data from ancient individuals who lived during times when major historical events were unfolding in the Near East. Our samples from the Crusaders period are evidence of a remarkable genetic diversity that coexisted in this region—Europeans, Near Easterners, and their mixed descendants—but this heterogeneity was transient in the genomic history of the Near East, since, with the exception of some Y chromosome lineages, present-day populations derive most of their ancestry from local people who preceded the Crusades. Ancient DNA from the warm Near East is still problematic to retrieve, and in addition our samples from the Crusaders’ pit were crudely buried and partly burned, but our study shows that recovering such aDNA is feasible and that the historical and genetic insights from it are exceptional and not possible from modern DNA alone.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the Directorate General of Antiquities (DGA) in Lebanon for approving (reference: 3333) transfer and processing of bones from Lebanon. We thank the Association for the Protection of Jabal Moussa (APJM) for supporting the Qornet ed-Deir excavation. M.H., Y.X., and C.T.-S. were supported by The Wellcome Trust (098051). R. Martiniano was supported by an EMBO Long-Term Fellowship (ALTF 133-2017). C.L.S. was supported by the European Regional Development Fund (Project No. 2014-2020.4.01.16-0030) and the Estonian Research Council personal research grant (PRG243).
Published: April 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.015.
Contributor Information
Marc Haber, Email: mh25@sanger.ac.uk.
Chris Tyler-Smith, Email: cts@sanger.ac.uk.
Accession Numbers
Raw sequencing reads for the ancient individuals are available through the European Nucleotide Archive (ENA) under accession number ERP114192. Aligned sequences and genotypes can be obtained from the corresponding authors.
Web Resources
European Nucleotide Archive (ENA), https://www.ebi.ac.uk/ena
Supplemental Data
References
- 1.Zerjal T., Xue Y., Bertorelle G., Wells R.S., Bao W., Zhu S., Qamar R., Ayub Q., Mohyuddin A., Fu S. The genetic legacy of the Mongols. Am. J. Hum. Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortes-Lima C., Bybjerg-Grauholm J., Marin-Padrón L.C., Gomez-Cabezas E.J., Bækvad-Hansen M., Hansen C.S., Le P., Hougaard D.M., Verdu P., Mors O. Exploring Cuba’s population structure and demographic history using genome-wide data. Sci. Rep. 2018;8:11422. doi: 10.1038/s41598-018-29851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maalouf A. Al Saqi; London: 1984. The Crusades through Arab Eyes. [Google Scholar]
- 4.MacEvitt C.H. University of Pennsylvania Press; 2008. The Crusades and the Christian World of the East: Rough Tolerance. [Google Scholar]
- 5.Zalloua P.A., Xue Y., Khalife J., Makhoul N., Debiane L., Platt D.E., Royyuru A.K., Herrera R.J., Hernanz D.F., Blue-Smith J., Genographic Consortium Y-chromosomal diversity in Lebanon is structured by recent historical events. Am. J. Hum. Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber M., Doumet-Serhal C., Scheib C., Xue Y., Danecek P., Mezzavilla M., Youhanna S., Martiniano R., Prado-Martinez J., Szpak M. Continuity and admixture in the last five millennia of Levantine history from ancient Canaanite and present-day Lebanese genome sequences. Am. J. Hum. Genet. 2017;101:274–282. doi: 10.1016/j.ajhg.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haber M., Mezzavilla M., Xue Y., Tyler-Smith C. Ancient DNA and the rewriting of human history: be sparing with Occam’s razor. Genome Biol. 2016;17:1. doi: 10.1186/s13059-015-0866-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins S. Sidon in the time of St. Louis: archaeological discoveries of the 13th century AD. Archaeology & History in the Lebanon. 2012;35:415–432. [Google Scholar]
- 9.Moorhead S., Cook B. The coins from the excavation at Sidon. Archaeology & History in the Lebanon. 2012;35:399–405. [Google Scholar]
- 10.Fischer-Genz B., Genz H., Elias N., Doumet-Serhal C. Report on the 2017 archaeological soundings at Qornet ed-Deir, Jabal Moussa Biosphere Reserve. Bulletin d’Archéologie et d’Architecture Libanaises. 2018;18:245–262. [Google Scholar]
- 11.Gamba C., Jones E.R., Teasdale M.D., McLaughlin R.L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dabney J., Knapp M., Glocke I., Gansauge M.T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.L., Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010:t5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 14.Schubert M., Ermini L., Der Sarkissian C., Jónsson H., Ginolhac A., Schaefer R., Martin M.D., Fernández R., Kircher M., McCue M. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 15.Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen M., Guo X., Wang Y., Lohmueller K.E., Rasmussen S., Albrechtsen A., Skotte L., Lindgreen S., Metspalu M., Jombart T. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazaridis I., Nadel D., Rollefson G., Merrett D.C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skoglund P., Stora J., Gotherstrom A., Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. [Google Scholar]
- 23.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray S.M., Mulle J.G., Dodd A.F., Pulver A.E., Wooding S., Warren S.T. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc. Natl. Acad. Sci. USA. 2010;107:16222–16227. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarno S., Boattini A., Pagani L., Sazzini M., De Fanti S., Quagliariello A., Gnecchi Ruscone G.A., Guichard E., Ciani G., Bortolini E. Ancient and recent admixture layers in Sicily and Southern Italy trace multiple migration routes along the Mediterranean. Sci. Rep. 2017;7:1984. doi: 10.1038/s41598-017-01802-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poznik G.D. Identifying Y-chromosome haplogroups in arbitrarily large samples of sequenced or genotyped men. bioRxiv. 2016 [Google Scholar]
- 29.Weissensteiner H., Forer L., Fuchsberger C., Schöpf B., Kloss-Brandstätter A., Specht G., Kronenberg F., Schönherr S. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw247. W64-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solé-Morata N., Villaescusa P., García-Fernández C., Font-Porterias N., Illescas M.J., Valverde L., Tassi F., Ghirotto S., Férec C., Rouault K. Analysis of the R1b-DF27 haplogroup shows that a large fraction of Iberian Y-chromosome lineages originated recently in situ. Sci. Rep. 2017;7:7341. doi: 10.1038/s41598-017-07710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meisner J., Albrechtsen A. Inferring population structure and admixture proportions in low-depth NGS data. Genetics. 2018;210:719–731. doi: 10.1534/genetics.118.301336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiffels S., Haak W., Paajanen P., Llamas B., Popescu E., Loe L., Clarke R., Lyons A., Mortimer R., Sayer D. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 2016;7:10408. doi: 10.1038/ncomms10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh P.R., Lipson M., Patterson N., Moorjani P., Pickrell J.K., Reich D., Berger B. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickrell J.K., Patterson N., Loh P.R., Lipson M., Berger B., Stoneking M., Pakendorf B., Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haber M., Mezzavilla M., Xue Y., Comas D., Gasparini P., Zalloua P., Tyler-Smith C. Genetic evidence for an origin of the Armenians from Bronze Age mixing of multiple populations. Eur. J. Hum. Genet. 2016;24:931–936. doi: 10.1038/ejhg.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haber M., Platt D.E., Khoury S., Badro D.A., Abboud M., Tyler-Smith C., Zalloua P.A. Y-chromosome R-M343 African lineages and sickle cell disease reveal structured assimilation in Lebanon. J. Hum. Genet. 2011;56:29–33. doi: 10.1038/jhg.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.