Abstract
Holoprosencephaly is the incomplete separation of the forebrain during embryogenesis. Both genetic and environmental etiologies have been determined for holoprosencephaly; however, a genetic etiology is not found in most cases. In this report, we present two unrelated individuals with semilobar holoprosencephaly who have the identical de novo missense variant in the gene CCR4-NOT transcription complex, subunit 1 (CNOT1). The variant (c.1603C>T [p.Arg535Cys]) is predicted to be deleterious and is not present in public databases. CNOT1 has not been previously associated with holoprosencephaly or other brain malformations. In situ hybridization analyses of mouse embryos show that Cnot1 is expressed in the prosencephalic neural folds at gestational day 8.25 during the critical period for subsequent forebrain division. Combining human and mouse data, we show that CNOT1 is associated with incomplete forebrain division.
Keywords: holoprosencephaly, CNOT1, neonatal diabetes mellitus
Main Text
Holoprosencephaly (HPE) is defined by varying degrees of separation of the embryonic forebrain. While occurring in approximately 1 in 10,000 live births, HPE is estimated to occur in 1 in 250 embryos, making it one of the most common human developmental abnormalities.1 The etiology of HPE is complex and most likely involves the interaction of genetic and environmental factors. The most common cause is trisomy 13, but in cases not associated with aneuploidy, only a fraction of affected subjects have a known genetic etiology.2, 3 In this report, we describe the identical de novo missense variant in two unrelated families in the gene CCR4-NOT transcription complex, subunit 1 (CNOT1 [MIM: 604917]) and show that this gene is expressed during early neurulation in the mouse embryo.
CNOT1 is one of at least nine components of the CCR4-NOT complex, which has an important role in posttranscriptional regulation and is conserved from yeast to mammals.4 The CCR4-NOT complex is the main enzyme responsible for mRNA deadenylation, which shortens the poly(A) tail in mRNA, thus leading to mRNA degradation.5 Cnot1 is expressed in the embryonic brain of the mouse and has drastically decreased expression after gestational day 13.4 Whether Cnot1 is expressed during the critical period for induction of HPE, between gestational day 7.0 and 8.5, has not been studied previously.6, 7
To expand the genetic etiology of HPE and uncover novel regulators of forebrain development, we have applied whole-exome sequencing (WES) to 134 trios (proband and parents) with holoprosencephaly in an ongoing HPE research protocol (Table S1). The individuals and families with HPE in this study are recruited from multiple clinical genetics centers from the United States. Within the participating institutions, the phenotype was evaluated by clinical exam and brain imaging (MRI or CT) or autopsy. The study was approved by National Human Genome Research Institute (NHGRI) Institutional Review Board (protocol 98-HG-0249); procedures followed were in accordance with the ethical standards of NHGRI for human experimentation, and proper consent was obtained.
DNA samples from study participants underwent WES at the National Intramural Sequencing Center (NISC) (Supplemental Material and Methods). The mean read depth for each sample was 79.8. Variant calling, annotation, and filtering is described in the Supplemental Material and Methods. Copy-number variation (CNV) prediction from exome data was done using the eXome-Hidden Markov Model (XHMM) caller (Supplemental Material and Methods).8
All probands were first searched for four common genes known to cause HPE—SHH (MIM: 600725) on 7q36, ZIC2 (MIM: 603073) on 13q32, SIX3 (MIM: 603714) on 2p21, and TGIF1 (MIM: 602630)—on 18p11.3 using Sanger sequencing as recommended.3 20% (27 probands) of the discovery cohort had damaging variants in these genes. With the goal of gene discovery, minimizing false positives, and sacrificing sensitivity, the discovery cohort was filtered with stringent criteria including de novo inheritance in genes intolerant of variation,9 variant absence in the Exome Aggregation Consortium (ExAC) database,9 and Combined Annotation-Dependent Depletion (CADD) scores above 20.10 Variants that met these criteria were considered deleterious. An identical de novo missense variant (c.1603C>T [p.Arg535Cys]) in CNOT1 (GenBank: NM_001265612.1) was found in two unrelated families by WES and verified by Sanger sequencing (Supplemental Material and Methods). In proband 1, the WES alternate allele frequency for the c.1603C>T (p.Arg535Cys) variant was 39% (read depth 57), and for proband 2 the alternate allele frequency was 54% (read depth 54). Proband 1 (Figure 1) is a male born at 33 weeks’ gestation after a pregnancy complicated by intrauterine growth restriction (IUGR). Semilobar holoprosencephaly was confirmed by brain MRI. Medical problems (Table 1) included bilateral microtia, hearing loss, diabetes insipidus, neonatal diabetes mellitus requiring insulin, pancreatic exocrine insufficiency requiring enzyme therapy, and global developmental delay. Facial characteristics (Figure 1) include epicanthal folds, depressed nasal bridge, hypotelorism, and long philtrum. Proband 1 died at age 16 months. Proband 2 is female and was a term uncomplicated pregnancy without IUGR. Semilobar holoprosencephaly was confirmed by brain MRI postnatally and other medical problems included severe bilateral sensorineural hearing loss, global developmental delay, and hypertonia. Proband 2 was last seen in clinic at age 6.5 years, and her exam (Table 1) was significant for microcephaly, epicanthal folds, and long philtrum (photo unavailable). With the exception of insulin-requiring diabetes in proband 1, both probands have similar phenotypes including semilobar HPE, similar facial features, hearing loss, and global developmental delay.
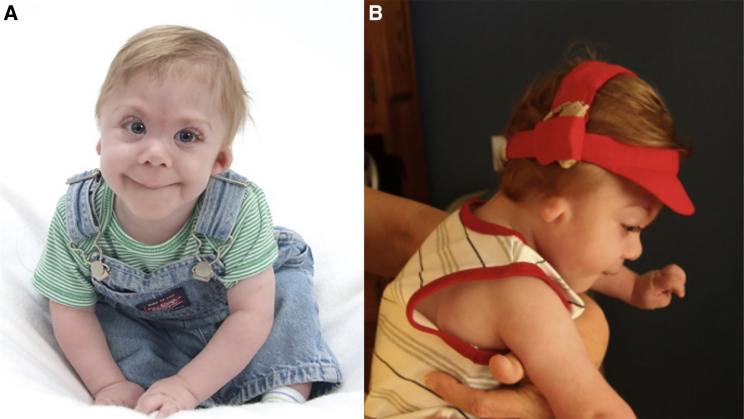
Figure 1.
Patient Images
(A) Proband 1 at age 12 months; facial characteristics include hypotelorism, epicanthal folds, depressed nasal bridge, and long philtrum.
(B) Proband 1 at 15 months, note right ear microtia.
Photo not available for proband 2.
Table 1.
Summary of Clinical Characteristics
| Proband 1 | Proband 2 | |
|---|---|---|
| Age at last exam | 16 months | 6.5 years |
| Gender | male | female |
| Prenatal history | IUGR | increased risk for Down syndrome on prenatal quad screen; amniocentesis not done |
| Birth history | Cesarean section at 35 weeks for IUGR | term vaginal delivery |
| Brain MRI | semilobar holoprosencephaly | semilobar holoprosencephaly |
| Craniofacial exam | microcephaly, epicanthal folds, long philtrum | microcephaly, epicanthal folds, long philtrum |
| Ears/hearing | bilateral microtia, bilateral conductive and sensorineural hearing loss with right ear worse than left, CT scan showed ossicle anomalies | severe bilateral sensorineural hearing loss |
| Seizure history | isolated seizure associated with fentanyl administration, normal EEG | none |
| Diabetes insipidus | present, treated with desmopressin | none |
| Neurologic history | global developmental delay, low muscle tone, non-ambulatory | global developmental delay, muscle spasticity, non-ambulatory |
| Other anomalies | pancreatic insufficiency: neonatal diabetes mellitus requiring insulin therapy and pancreatic exocrine deficiency treated with enzyme therapy | none |
Abbreviations: IUGR, intrauterine growth restriction; MRI, magnetic resonance imaging; CT, computed tomography; EEG, electroencephalogram.
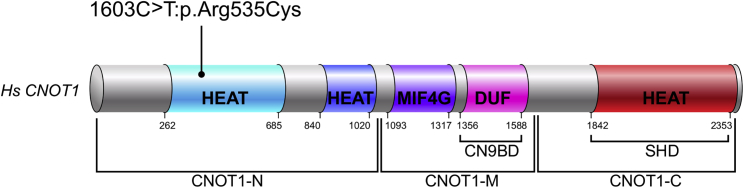
The c.1603C>T (p.Arg535Cys) variant is located in the conserved HEAT domain in the N-terminal (Figure 2). The N-terminal of CNOT1 associates with another complex protein, CNOT11.11 The CADD score for the CNOT1 c.1603C>T (p.Arg535Cys) variant is 35, and it is not present in the ExAC database (accessed January 18, 2019). XHMM analysis of exomes showed no copy number variations in either proband. Two identical variants in unrelated probands with holoprosencephaly is unlikely by chance in a relatively small cohort of 134 trios with HPE; especially, given that CNOT1 is intolerant of both missense change (z = 7.44) and loss of function (pLi = 1.00) (constraint metrics accessed from ExAC database on January 18, 2019).9 There were two other de novo variants in CNOT1 in two other unrelated individuals in our HPE cohort, but neither met the above criteria for a deleterious variant. One variant was a synonymous change (c.6057C>T [p.(=)]) in CNOT1 (GenBank: NM_001265612.1) and the other variant was a missense change (c.1394A>C [p.Gln465Pro]) in CNOT1 (GenBank: NM_001265612.1) with an ExAC database allele frequency of 8.25E−06 and a CADD score of 17.9. Additionally, Table S2 lists all de novo variants found in proband 1 and proband 2. The CNOT1 c.1603C>T (p.Arg535Cys) was the only de novo variant found for proband 2. Proband 1 had three additional de novo variants; two were synonymous (Table S2) and a missense variant occurred in the gene RNF150 (MIM: not listed) (GenBank: NM_020724.2; c.510G>A [p.Met170Ile]). The variant in RNF150 is a variant of unknown significance. RNF150 is not intolerant of variation based on ExAC constraint metrics: z = 2.15 for missense and pLI = 0.01 (accessed from ExAC database on January 18, 2019).9
Figure 2.
CNOT1 Protein Domains
CNOT1 c.1603C>T (p.Arg535Cys) (GenBank: NM_001265612.1) variant is located in the conserved HEAT domain
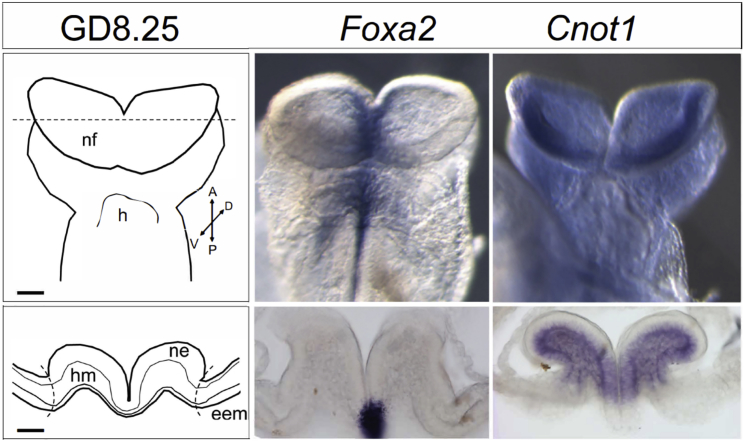
Genes that regulate forebrain patterning and play a role in HPE pathogenesis are expected to be expressed in the prosencephalic neural folds that give rise to the forebrain during primary neurulation.12 We therefore conducted in situ hybridization on mouse embryos at GD8.25, a stage representing early neurulation and within the critical period for HPE genesis.7 Mouse in situ hybridization studies were conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the University of Wisconsin-Madison School of Veterinary Medicine Institutional Animal Care and Use Committee (protocol number G005396). CD-1 mice (Mus musculus) were purchased from Charles River and C57BL/6J mice from The Jackson Laboratory. Timed pregnancies were established as previously described.13 Embryos were dissected at GD8.25 and fixed overnight in 4% PFA. In situ hybridization (ISH) was conducted on whole C57BL/6J embryos or 50 μm sections cut from CD-1 embryos with a vibrating microtome in the transverse plane along the anterior-posterior axis. ISH was conducted as previously described and analysis was limited to the prosencephalic regions of the neural fold from which the forebrain will develop.14 As seen in Figure 3, Cnot1 expression is detectable in both the neuroectoderm and the mesenchyme of the neural folds but not in extraembryonic membranes (Figure 3). Specificity of staining is additionally shown by staining for Foxa2 (Hnf-3β), which is expressed in the ventral neuroectoderm.15
Figure 3.
Gestational Day (GD) 8.25 Mouse Embryos
A ventral view (top) is shown for whole mounts. Transverse sections (bottom) through the prosencephalic neural folds (at the level of the dashed line in schematic) were stained to visualize gene expression in specific cellular compartments. Abbreviations: nf, neural folds; h, heart; ne, neuroectoderm; hm, head mesenchyme; eem, extra-embryonic membranes. Scale bar = 100 μm.
Disruption of the sonic hedgehog signaling pathway is known to result in holoprosencephaly.16 Using human osteosarcoma cells, Cheng et al. showed that knockdown of CNOT1 using short hairpin (sh) RNA inhibited the sonic hedgehog signaling pathway based on decreased expression of genes downstream of SHH including GLI1 and PTCH1.17 These experiments in osteosarcoma cells have established a link between CNOT1 knockdown and decreased sonic hedgehog signaling and raise the possibility that the CNOT1 p.Arg535Cys variant may inhibit Sonic hedgehog signaling by a loss-of-function or a dominant-negative mechanism. This report allows for further research into the molecular mechanisms involved in CNOT1 and Hedgehog signaling.
In summary, we report identical de novo missense variants in CNOT1 in two unrelated individuals with semilobar holoprosencephaly and show in the mouse model that Cnot1 is expressed during the critical period for holoprosencephaly.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Human Genome Research Institute Intramural Research program. Work in the R.J.L. lab was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under award numbers R01ES026819 and T32ES007015.
Published: April 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.017.
Accession Numbers
The accession number for the CNOT1 c.1603C>T (p.Arg535Cys) variant is ClinVar: SUB5130764.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Matsunaga E., Shiota K. Holoprosencephaly in human embryos: epidemiologic studies of 150 cases. Teratology. 1977;16:261–272. doi: 10.1002/tera.1420160304. [DOI] [PubMed] [Google Scholar]
- 2.Kruszka P., Muenke M. Syndromes associated with holoprosencephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 2018;178:229–237. doi: 10.1002/ajmg.c.31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruszka P., Martinez A.F., Muenke M. Molecular testing in holoprosencephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 2018;178:187–193. doi: 10.1002/ajmg.c.31617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C., Ito K., Takahashi A., Wang G., Suzuki T., Nakazawa T., Yamamoto T., Yokoyama K. Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 2011;411:360–364. doi: 10.1016/j.bbrc.2011.06.148. [DOI] [PubMed] [Google Scholar]
- 5.Bartlam M., Yamamoto T. The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell. 2010;1:443–452. doi: 10.1007/s13238-010-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyne G.W., Everson J.L., Ansen-Wilson L.J., Melberg C.G., Fink D.M., Parins K.F., Doroodchi P., Ulschmid C.M., Lipinski R.J. Gli2 gene-environment interactions contribute to the etiological complexity of holoprosencephaly: evidence from a mouse model. Dis. Model. Mech. 2016;9:1307–1315. doi: 10.1242/dmm.026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyne G.W., Melberg C.G., Doroodchi P., Parins K.F., Kietzman H.W., Everson J.L., Ansen-Wilson L.J., Lipinski R.J. Definition of critical periods for Hedgehog pathway antagonist-induced holoprosencephaly, cleft lip, and cleft palate. PLoS ONE. 2015;10:e0120517. doi: 10.1371/journal.pone.0120517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromer M., Moran J.L., Chambert K., Banks E., Bergen S.E., Ruderfer D.M., Handsaker R.E., McCarroll S.A., O’Donovan M.C., Owen M.J. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bawankar P., Loh B., Wohlbold L., Schmidt S., Izaurralde E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 2013;10:228–244. doi: 10.4161/rna.23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng X., Oliver G. Pathogenesis of holoprosencephaly. J. Clin. Invest. 2009;119:1403–1413. doi: 10.1172/JCI38937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyne G.W., Plisch E.H., Melberg C.G., Sandgren E.P., Peter J.A., Lipinski R.J. A simple and reliable method for early pregnancy detection in inbred mice. J. Am. Assoc. Lab. Anim. Sci. 2015;54:368–371. [PMC free article] [PubMed] [Google Scholar]
- 14.Everson J.L., Fink D.M., Yoon J.W., Leslie E.J., Kietzman H.W., Ansen-Wilson L.J., Chung H.M., Walterhouse D.O., Marazita M.L., Lipinski R.J. Sonic hedgehog regulation of Foxf2 promotes cranial neural crest mesenchyme proliferation and is disrupted in cleft lip morphogenesis. Development. 2017;144:2082–2091. doi: 10.1242/dev.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki H., Hogan B.L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Roessler E., Muenke M. The molecular genetics of holoprosencephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 2010;154C:52–61. doi: 10.1002/ajmg.c.30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng D.D., Li J., Li S.J., Yang Q.C., Fan C.Y. CNOT1 cooperates with LMNA to aggravate osteosarcoma tumorigenesis through the Hedgehog signaling pathway. Mol. Oncol. 2017;11:388–404. doi: 10.1002/1878-0261.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.