Abstract
The receptor for hyaluronic acid-mediated motility (RHAMM) is upregulated in various cancers. We previously screened genes upregulated in human hepatocellular carcinomas for their metastatic function in a mouse model of pancreatic neuroendocrine tumor (PNET) and identified that human RHAMMB promoted liver metastasis. It was unknown whether RHAMMB is upregulated in pancreatic cancer or contributes to its progression. In this study, we found that RHAMM protein was frequently upregulated in human PNETs. We investigated alternative splicing isoforms, RHAMMA and RHAMMB, by RNA-Seq analysis of primary PNETs and liver metastases. RHAMMB, but not RHAMMA, was significantly upregulated in liver metastases. RHAMMB was crucial for in vivo metastatic capacity of mouse and human PNETs. RHAMMA, carrying an extra 15-amino acid-stretch, did not promote metastasis in spontaneous and experimental metastasis mouse models. Moreover, RHAMMB was substantially higher than RHAMMA in pancreatic ductal adenocarcinoma (PDAC). RHAMMB, but not RHAMMA, correlated with both higher EGFR expression and poorer survival of PDAC patients. Knockdown of EGFR abolished RHAMMB-driven PNET metastasis. Altogether, our findings suggest a clinically relevant function of RHAMMB, but not RHAMMA, in promoting PNET metastasis in part through EGFR signaling. RHAMMB can thus serve as a prognostic factor for pancreatic cancer.
Electronic supplementary material
The online version of this article (10.1186/s12943-019-1018-y) contains supplementary material, which is available to authorized users.
Keywords: RHAMM, Isoforms, Pancreatic cancer, PNETs, PDAC, Metastasis
Main text
Metastasis accounts for 90% of cancer deaths. We developed a mouse model of well-defined multistage tumorigenesis: RIP-Tag; RIP-tva to identify metastatic factors [1]. We identified that the receptor for hyaluronic acid (HA)-mediated motility, isoform B (RHAMMB), significantly promotes liver metastasis of pancreatic neuroendocrine tumors (PNET) in RIP-Tag; RIP-tva mouse models [2]. Expression of RHAMM is restricted in normal adult tissues, but is upregulated in cancers [3, 4]. Increased production of glycosaminoglycan, HA, is correlated with increased migration and invasion in aggressive cancers [5]. CD44 and RHAMM are two major HA receptors. The roles of CD44 isoforms in cancer have been studied extensively, but the functions of RHAMM isoforms in tumorigenesis are less clear. RHAMM encodes 18 exons and alternative splicing generates different isoforms. RHAMMA includes all 18 exons and RHAMMB lacks exon 4 (Fig. 1a). Here we aimed to determine the clinical relevance of RHAMMA and RHAMMB isoforms and their functions in pancreatic cancer.
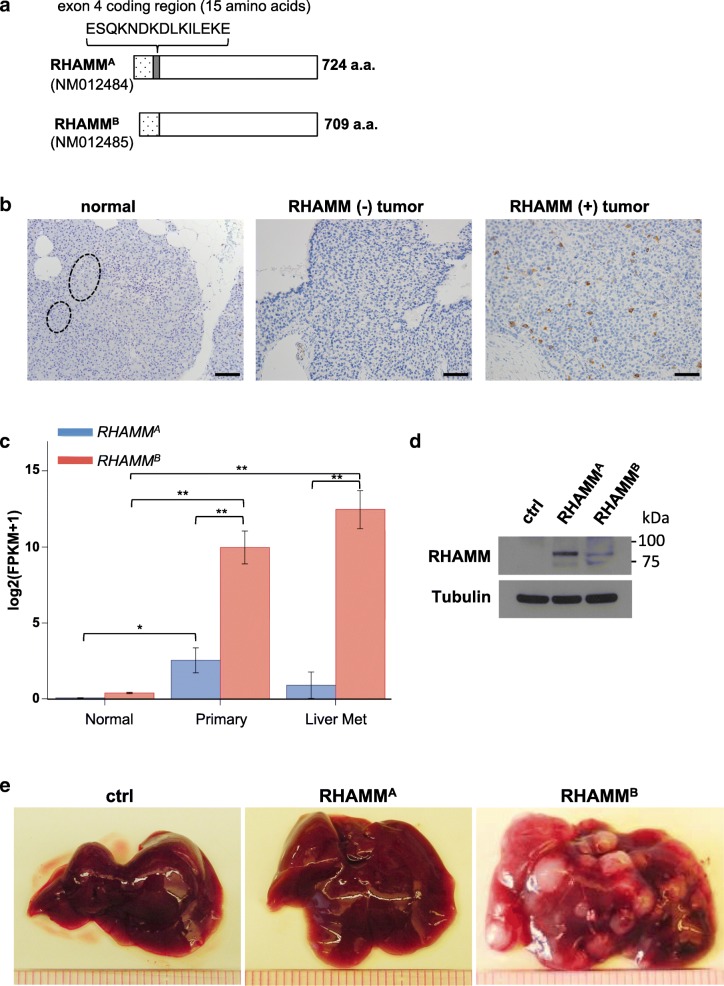
Fig. 1.
RHAMMB, but not RHAMMA, is upregulated in human PNETs and promotes liver metastasis of mouse PNET cells. a Diagram of RHAMMA and RHAMMB proteins. b RHAMM is upregulated in 54 of 83 cases (65%) of human PNETs in immunohistochemical staining. Left: Normal pancreas with islets in dashed circle. Middle: RHAMM negative PNET. Right: RHAMM positive PNET. Original magnification: 20X. Scale bar, 50 μm. c RNA-seq analysis showed that RHAMMB is significantly upregulated compared to RHAMMA in primary human PNETs and liver metastases. The p value was calculated using two-way ANOVA followed by Tukey’s test. **: p < 0.0001, *: p < 0.05. Error bars represent standard error of mean. d Western blot analysis of human RHAMM in mouse N134 cell line (control), N134-RHAMMA cells, and N134-RHAMMB cells. e A total of 1 million N134 cells, N134-RHAMMA cells, or N134-RHAMMB cells were injected into the tail vein of NSG mice (n = 5 for each group). Five weeks later, the recipient mice were euthanized to survey for metastatic sites and incidence. Representative liver photos were shown
RHAMMB, but not RHAMMA, is upregulated in human PNET liver metastases
To investigate RHAMM expression in human PNETs, a tissue microarray consisting of 83 PNETs was immunostained for RHAMM. RHAMM was not detectable in the normal pancreas, while 54 of 83 (65%) PNETs exhibited cytoplasmic staining using an antibody that recognizes common region in RHAMM isoforms (Fig. 1b). Because isoform-specific RHAMM antibodies were not available, we investigated the mRNA levels of RHAMMA and RHAMMB by performing RNA-Seq analysis on 27 primary PNETs and 12 liver metastases, using 89 human islets from NCBI Gene Expression Omnibus database for comparison. Consistent with our immunohistochemical data, normal islets had very low RHAMMA and RHAMMB mRNA (Fig. 1c). RHAMMB was significantly higher than RHAMMA in both primary and metastatic PNETs, suggesting that RHAMMB is the predominant isoform naturally expressed in PNETs. Although RHAMMA levels in primary tumors were significantly higher than those in normal islets (p = 0.0002), RHAMMA levels in metastatic tumors were not significantly higher than those in normal islets (p = 0.8928). The mRNAs of RHAMMA and RHAMMB were readily detectable in additional primary PNETs and metastases by RT-qPCR using isoform-specific primers (Additional file 1: Figure S1A-B).
We compared metastatic potential of RHAMMA to RHAMMB in RIP-Tag; RIP-tva models of spontaneous metastasis and tail vein assays [1, 2]. In contrast to RHAMMB, RHAMMA did not promote spontaneous metastasis (Additional file 2: Table S1). Then, we generated N134 cells overexpressing RHAMMA (N134-RHAMMA). N134 is a cell line derived from a PNET of RIP-Tag; RIP-tva mouse [1]. Although there were more RHAMMA than RHAMMB for unknown reasons (Fig. 1d), only one visible tumor was found in 5 immunodeficient NOD/scid-IL2Rgc knockout (NSG) mice receiving N134-RHAMMA cells after 5 weeks, while all 5 mice receiving N134-RHAMMB cells developed large liver metastases within 5 weeks (Fig. 1e and Additional file 1: Figure S2A). To detect micrometastases, we performed immunostaining for synaptophysin, a neuroendocrine marker. Mice receiving N134 cells and N134-RHAMMA cells had an average of 1.8 and 0.6 liver micrometastases, respectively (Additional file 1: Figure S2B). These data suggested that the unique 15-amino acid-stretch, ESQKNDKDLKILEKE, which is present in RHAMMA but not in RHAMMB, inhibits the metastatic function of RHAMM.
RHAMMB is crucial for the metastatic potential of human PNET cell line BON1-TGL
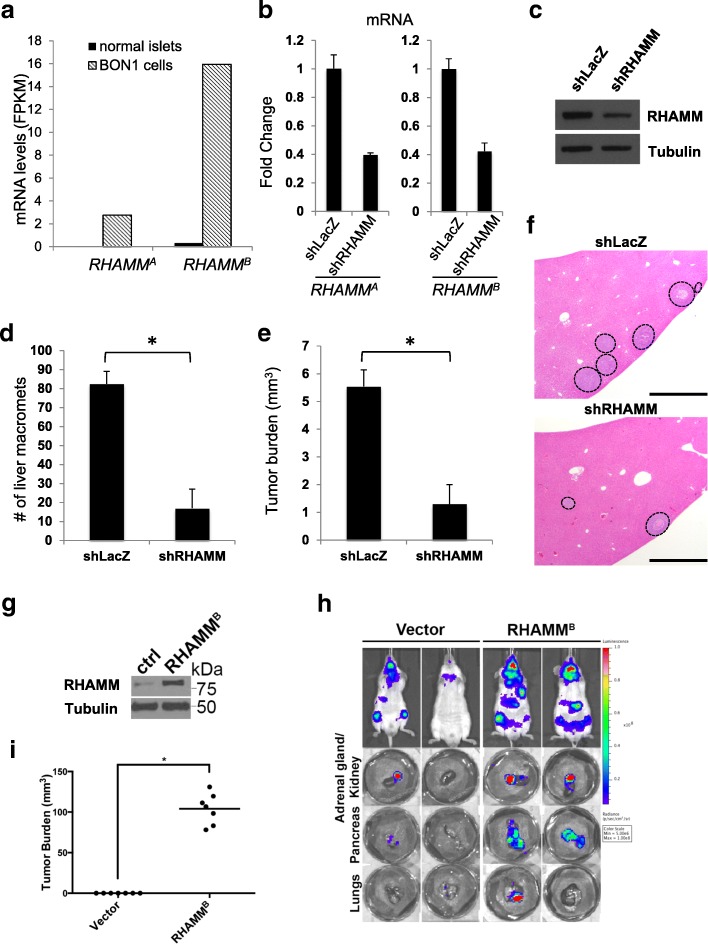
BON1 is the most utilized human PNET cell line and was established from a peri-pancreatic lymph node in a patient with metastatic PNET. We found that BON1 had much higher expression of RHAMMB than RHAMMA as determined by RNA-Seq (Fig. 2a). We performed shRNA-mediated knockdown of total RHAMM to investigate whether this reduces metastasis of BON1-TGL cells, which carry the thymidine kinase/green fluorescent protein /luciferase fusion reporter (TGL). Knockdown of RHAMM by shRNA was confirmed (Fig. 2b, c). We used an orthotopic model of PNET liver metastases by injecting cells into the spleen of NSG mice [6]. Mice receiving control cells developed an average of 82.5 liver metastases after 3 weeks, while mice receiving BON1-TGL-shRHAMM cells developed an average of 17 liver metastases with significantly lowered tumor burden (Fig. 2d-f).
Fig. 2.
RHAMMB is crucial for metastatic potential of human PNET cell line, BON1-TGL. a RHAMMA and RHAMMB expression in BON1 cell line compared to those in normal islets. b RHAMMA and RHAMMB knockdown in BON1-TGL cell line by shRHAMM as determined by RT-qPCR analysis. c Western blot analysis of RHAMM and tubulin (as a loading control) in BON1-TGL-shLacZ cell line and BON1-TGL-shRHAMM cell line. d-e RHAMM knockdown greatly inhibited liver metastasis of BON1-TGL cells. A total of 0.5 million each BON1-TGL-shLacZ (control) or BON1-TGL-shRHAMM were injected into the spleen of NSG mice (n = 4 for each group). After 3 weeks, the recipient mice were euthanized to survey for metastatic sites and incidence. The number of liver macrometastases (d) and the tumor burden of liver macrometastases (e) were recorded. *: statistically significantly different (p < 0.05, one-tailed Mann-Whitney U test). Error bars represent standard deviation. f Liver sections with hematoxylin and eosin stain. Dashed circles indicate metastases. Original magnification: 10X. Scale bar, 1 mm. g RHAMMB overexpression in BON1-TGL-RHAMMB cell line. Western blot analysis of RHAMM and tubulin (as a loading control) are shown. h Representative bioluminescent images of NSG mice 4 weeks after injection (upper panel) and their organs (lower panel). A total of 1 million cells was injected into NSG mice via intracardiac injection (n = 7). i The tumor burden of macrometastases at adrenal glands was documented. *: statistically significantly different, p < 0.05, t-test
To determine whether even higher RHAMMB levels would further enhance metastasis of BON1-TGL cells, we generated BON1-TGL cells overexpressing RHAMMB (Fig. 2g). We injected the cells into NSG mice via intracardiac injection. The increased levels of RHAMMB enhanced metastasis of BON1-TGL in mice throughout the mouse body as visualized by bioluminescence imaging and signals from multiple organs were higher in mice receiving BON1-TGL-RHAMMB than those in mice receiving BON1-TGL overexpressing a control vector (Fig. 2h). Notably, we observed macrometastases at the adrenal glands of mice receiving BON1-TGL-RHAMMB, but not in control mice (Fig. 2i). Taken together, RHAMMB is crucial for PNET metastasis.
RHAMMB is upregulated in human pancreatic ductal adenocarcinoma (PDAC) and correlates with poor survival
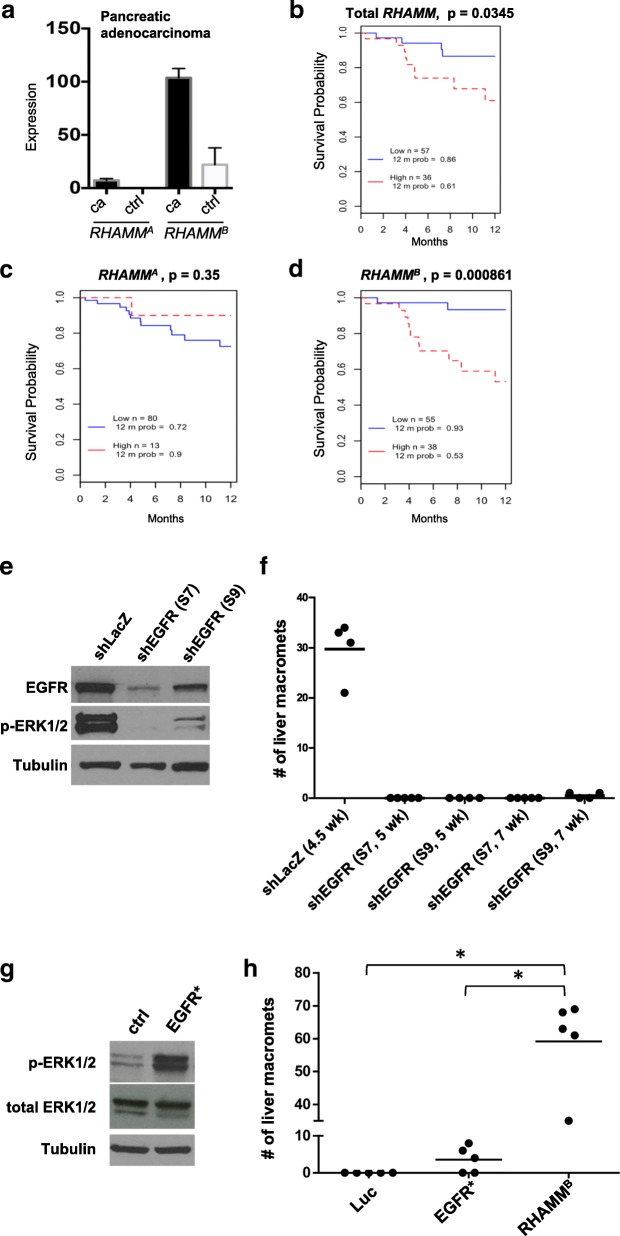
PDAC is the most common pancreatic cancer type. It was shown that total RHAMM is upregulated in primary PDAC by RT-qPCR of 14 matched tumors and adjacent normal tissues [7]. To compare RHAMMA and RHAMMB levels in PDAC, we analyzed publicly available The Cancer Genome Atlas (TCGA) datasets. Both RHAMMA and RHAMMB were expressed at significantly higher levels in PDAC than in normal pancreatic tissues, and RHAMMB was substantially higher than RHAMMA (Fig. 3a). Survival analysis showed that high RHAMM levels were correlated with a worse outcome (Fig. 3b). Furthermore, patients with high RHAMMB had inferior survival compared to those with high RHAMMA (Fig. 3c, d), suggesting that RHAMMB, but not RHAMMA, is a prognostic factor for survival of PDAC patients. Due to limited information available, we could not perform survival analysis for PNETs.
Fig. 3.
RHAMMB is upregulated in human pancreatic ductal adenocarcinoma (PDAC) and correlates with poor survival. a RHAMMA and RHAMMB expression values from TCGA PDAC dataset. RHAMMA: uc003lzf or NM_012484. RHAMMB: uc003lzg or NM_012485. cancer (ca): n = 124; control (ctrl): n = 4. Bars and error bars represent means and standard errors. b-d Kaplan-Meier survival analysis of TCGA cohort with 93 PDAC cases. High and low represent the status of the RHAMM mRNA expression levels compared to average values. e Knockdown efficiency of two EGFR shRNAs. S7 and S9 reduced EGFR protein expression and p-Erk1/2 levels in N134 cells overexpressing RHAMMB by Western blot analysis. α-tubulin was used a loading control. f EGFR knockdown greatly inhibited the liver metastasis of N134 cells overexpressing RHAMMB. A total of 1 million N134_RHAMMB_shLacZ, N134_RHAMMB_shEGFR(S7), or N134_RHAMMB_shEGFR(S9) cells were injected into the tail vein of NSG mice (n = 5 for each group). At the indicated time points, the recipient mice were euthanized to survey for metastatic sites and incidence. The number of liver macrometastases was recorded. g Western blot analysis of p-Erk1/2 and total Erk from N134 overexpressing luciferase (control), and N134_EGFR*. h N134 cells overexpressing luciferase (Luc), N134_EGFR* (EGFR*), N134_RHAMMB (RHAMMB) were injected into the tail vein of NSG mice (n = 5, each group). Five weeks later (for Luc and EGFR* groups) or when mice were lethargic (for RHAMMB), mice were euthanized to survey for metastatic sites and incidence. *: p < 0.0001, One-way ANOVA and pairwise comparison with Tukey’s adjustment
EGFR signaling is required for RHAMMB-induced metastasis
EGFR activation is associated with worse survival in many malignancies. Analysis of TCGA PDAC dataset showed a good correlation between EGFR and RHAMMB expression, but not between EGFR and RHAMMA expression (Additional file 1: Figure S3A-B). We previously showed that EGFR signaling is activated in N134-RHAMMB cells and an EGFR inhibitor, gefitinib, induces apoptosis of N134-RHAMMB cells [2]. These findings led us to hypothesize that enhanced EGFR signaling involves RHAMMB-induced metastasis. To test this hypothesis, we used two different shRNAs targeting mouse EGFR (S7 and S9) as well as a control shRNA targeting LacZ (shLacZ). shEGFR(S7 and S9) decreased the levels of both EGFR and p-ERK1/2, a downstream target of EGFR signaling, with S7 exhibiting a better knockdown efficiency than S9 (Fig. 3e).
In tail vein metastasis assays, mice receiving N134-RHAMMB-shLacZ became lethargic, showing a mean of 29.8 liver macrometastases per mouse within 5 weeks, but mice injected with N134-RHAMMB-shEGFR(S7) and N134-RHAMMB-shEGFR(S9) cells did not develop metastases 5 weeks post-injection (Fig. 3f). An additional cohort of mice injected with N134-RHAMMB-shEGFR(S7) still did not develop metastases 7 weeks post-injection, and only a mean of 0.5 liver macrometastases appeared in mice injected with N134-RHAMMB-shEGFR(S9) at this time point (Fig. 3f).
We investigated whether a constitutively active form of EGFR, EGFR*, is sufficient to recapitulate RHAMMB activity in metastasis. Five of the 8 RIP-Tag; RIP-tva mice receiving RCASBP-EGFR* developed pancreatic lymph node metastases (62.5%) and 2 developed liver metastases (25%) (Additional file 2: Table S1). We generated N134 cells overexpressing EGFR* (N134-EGFR*) for experimental metastasis (Fig. 3g). While no metastasis was found in mice receiving control N134-Luciferase after 6 weeks, an average of 6 liver macrometastases was detected in mice receiving N134-EGFR* (Fig. 3h). Although EGFR* promotes liver metastasis of PNETs in both spontaneous and experimental metastasis mouse models, the degree of metastasis is less than that of RHAMMB (Additional file 2: Table S1 and Fig. 3h). Therefore, these EGFR knockdown and EGFR* overexpression data suggest that activation of EGFR contributes to RHAMMB-induced PNET metastasis, but EGFR* cannot fully recapitulate the metastatic phenotype of RHAMMB. Further studies are required to identify signals other than EGFR provided by RHAMMB and to understand whether endogenous levels of RHAMMA activate EGFR signaling.
Conclusion
We provide evidence that upregulation of RHAMMB is a valuable prognostic marker for pancreatic cancer. We demonstrated that only the shorter isoform RHAMMB, but not RHAMMA with 15 extra amino acids encoded by exon 4, is significantly upregulated in PNET and PDAC. RHAMMB, but not RHAMMA, promotes metastasis in spontaneous and experimental metastasis mouse models of PNET. EGFR signaling is required for RHAMMB-induced liver metastasis, but is not sufficient to promote PNET metastasis. RHAMMB, but not RHAMMA, is correlated with inferior survival in PDAC patients.
Additional files
Figure S1. RT-qPCR analysis of RHAMMA (A) and RHAMMB (B) in 9 primary human PNETs and 3 metastatic PNETs from the livers. Figure S2. Unlike RHAMMB, RHAMMA did not promote liver metastasis of mouse PNET N134 cell line in a tail vein experimental metastasis assay. (A) The number of liver macrometastases was recorded. (B) Immunohistochemical staining of synaptophysin in the liver sections to reveal the presence of metastatic PNETs. Arrows indicate micrometastases. Original magnification: 10X. Scale bar, 200 μm. Figure S3. Correlation between EGFR expression and RHAMMA (A) or RHAMMB (B) expression in TCGA PDAC dataset (RNA-Seq V2). (PPTX 3371 kb)
Table S1. Impact of genes on lymph node and liver metastasis of PNETs in RIP-Tag; RIP-tva mice. (DOCX 37 kb)
Supplementary materials and methods. (DOCX 37 kb)
Acknowledgments
The authors thank Danny Huang for mouse database design; Harold Varmus, Yi Li, Jihye Paik, Todd R. Evans, David Foster, Bi-Sen Ding, Katie Politi, Jeffrey Yongchun Zhao, Romel Somwar, Selina Chen-Kiang, Stephanie Azzopardi, Samantha Li, Megan Wong, Joseph Na, and Robin Zhang for their valuable input and excellent assistance; Diane L. Reidy and David S. Klimstra for contributing the approval of the tumor materials.
Funding
This work is partially supported by NIH grants 2U01DK072473 (to Y.-C.N.D.), 1R21CA173348-01A1 (to Y.-C.N.D.), 1 R01CA204916-01A1 (to Z.C., G.Z., Y.-C.N.D.), UL1TR000457 (to Z.C.), DOD grants W81XWH-13-1-0331 (to S.C., Z.C., Y.-C.N.D.) W81XWH-16-1-0619 (to Y.-C.N.D.), and Goldhirsh Foundation (to A.V., T.J. F.III, O.E., Y.-C.N.D.).
Availability of data and materials
The RNA-Seq and TCGA datasets that support the findings of this study are available in Gene Expression Omnibus (GEO accession GSE50398, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50398) [8–10], and http://cancergenome.nih.gov/cancersselected, and the other data are available from the authors upon reasonable request.
Abbreviations
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
Fetal bovine serum
- FPKM
Fragments per kilobase of transcripts per million reads
- HA
Hyaluronic acid
- PDAC
Pancreatic ductal adenocarcinoma
- PNET
Pancreatic neuroendocrine tumor
- RHAMM
Receptor for hyaluronic acid-mediated motility
- RT-qPCR
Quantitative real-time reverse transcription PCR
- TCGA
The Cancer Genome Atlas (TCGA)
Authors’ contributions
SC, XC, BJK, GZ, SP, TH, and LS performed the experiments and analyzed the data. DW conducted the analysis on the TCGA dataset. DW, GZ, SP, TJFIII, and KR edited the manuscript. LHT contributed immunohistochemical interpretation. AV, CSC, and OE contributed to bioinformatics analyses. ZC performed statistical analysis. LHT, KR, and TJFIII contributed to samples. Y-CND designed and performed the experiments, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures involving mice were approved by the institutional animal care and use committee. Retrospective and prospective review of PNETs was performed using the pathology files and pancreatic cancer database at the authors’ institutions with IRB approval. Experimental details are provided in Additional file 3: Supplementary materials and methods.
Consent for publication
All authors agreed on the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Du YC, Lewis BC, Hanahan D, Varmus H. Assessing tumor progression factors by somatic gene transfer into a mouse model: Bcl-xL promotes islet tumor cell invasion. PLoS Biol. 2007;5:e276. doi: 10.1371/journal.pbio.0050276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du YC, Chou CK, Klimstra DS, Varmus H. Receptor for hyaluronan-mediated motility isoform B promotes liver metastasis in a mouse model of multistep tumorigenesis and a tail vein assay for metastasis. Proc Natl Acad Sci U S A. 2011;108:16753–16758. doi: 10.1073/pnas.1114022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greiner J, Ringhoffer M, Taniguchi M, Schmitt A, Kirchner D, Krahn G, Heilmann V, Gschwend J, Bergmann L, Dohner H, Schmitt M. Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol. 2002;30:1029–1035. doi: 10.1016/S0301-472X(02)00874-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen YT, Chen Z, Du YN. Immunohistochemical analysis of RHAMM expression in normal and neoplastic human tissues: a cell cycle protein with distinctive expression in mitotic cells and testicular germ cells. Oncotarget. 2018;9:20941–20952. doi: 10.18632/oncotarget.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sironen RK, Tammi M, Tammi R, Auvinen PK, Anttila M, Kosma VM. Hyaluronan in human malignancies. Exp Cell Res. 2011;317:383–391. doi: 10.1016/j.yexcr.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Fraedrich K, Schrader J, Ittrich H, Keller G, Gontarewicz A, Matzat V, Kromminga A, Pace A, Moll J, Blaker M, et al. Targeting aurora kinases with danusertib (PHA-739358) inhibits growth of liver metastases from gastroenteropancreatic neuroendocrine tumors in an orthotopic xenograft model. Clin Cancer Res. 2012;18:4621–4632. doi: 10.1158/1078-0432.CCR-11-2968. [DOI] [PubMed] [Google Scholar]
- 7.Cheng XB, Sato N, Kohi S, Koga A, Hirata K. Receptor for hyaluronic acid-mediated motility is associated with poor survival in pancreatic ductal adenocarcinoma. J Cancer. 2015;6:1093–1098. doi: 10.7150/jca.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y, Park SY, Su J, Bailey K, Ottosson-Laakso E, Shcherbina L, Oskolkov N, Zhang E, Thevenin T, Fadista J, et al. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet. 2014;23:6419–6431. doi: 10.1093/hmg/ddu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, Storm P, Osmark P, Ladenvall C, Prasad RB, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taneera J, Fadista J, Ahlqvist E, Atac D, Ottosson-Laakso E, Wollheim CB, Groop L. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum Mol Genet. 2015;24:1945–1955. doi: 10.1093/hmg/ddu610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RT-qPCR analysis of RHAMMA (A) and RHAMMB (B) in 9 primary human PNETs and 3 metastatic PNETs from the livers. Figure S2. Unlike RHAMMB, RHAMMA did not promote liver metastasis of mouse PNET N134 cell line in a tail vein experimental metastasis assay. (A) The number of liver macrometastases was recorded. (B) Immunohistochemical staining of synaptophysin in the liver sections to reveal the presence of metastatic PNETs. Arrows indicate micrometastases. Original magnification: 10X. Scale bar, 200 μm. Figure S3. Correlation between EGFR expression and RHAMMA (A) or RHAMMB (B) expression in TCGA PDAC dataset (RNA-Seq V2). (PPTX 3371 kb)
Table S1. Impact of genes on lymph node and liver metastasis of PNETs in RIP-Tag; RIP-tva mice. (DOCX 37 kb)
Supplementary materials and methods. (DOCX 37 kb)
Data Availability Statement
The RNA-Seq and TCGA datasets that support the findings of this study are available in Gene Expression Omnibus (GEO accession GSE50398, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50398) [8–10], and http://cancergenome.nih.gov/cancersselected, and the other data are available from the authors upon reasonable request.