Abstract
We previously characterized major components of mitotic chromosomes assembled in Xenopus laevis egg extracts and collectively referred to them as Xenopus chromosome–associated polypeptides (XCAPs). They included five subunits of the condensin complex essential for chromosome condensation. In an effort to identify novel proteins involved in this process, we have isolated XCAP-F and found it to be the Xenopus ortholog of ISWI, a chromatin remodeling ATPase. ISWI exists in two major complexes in Xenopus egg extracts. The first complex contains ACF1 and two low-molecular-weight subunits, most likely corresponding to Xenopus CHRAC. The second complex is a novel one that contains the Xenopus ortholog of the human Williams syndrome transcription factor (WSTF). In the absence of the ISWI complexes, the deposition of histones onto DNA is apparently normal, but the spacing of nucleosomes is greatly disturbed. Despite the poor spacing of nucleosomes, ISWI depletion has little effect on DNA replication, chromosome condensation or sister chromatid cohesion in the cell-free extracts. The association of ISWI with chromatin is cell cycle regulated and is under the control of the INCENP-aurora B kinase complex that phosphorylates histone H3 during mitosis. Apparently contradictory to the generally accepted model, we find that neither chromosome condensation nor chromosomal targeting of condensin is compromised when H3 phosphorylation is drastically reduced by depletion of INCENP-aurora B.
INTRODUCTION
The faithful rearrangements of chromatin structure are essential for a variety of nuclear functions in eukaryotic cells, including DNA replication, repair, gene expression, and chromosome segregation. Malfunctions in any of these processes cause DNA damage, aneuploidy, and chromosome breakage and translocations, potentially leading to cancers or birth defects (reviewed by Lengauer et al., 1998; Hoeijmakers, 2001).
DNA is packaged into the nucleosome, the basic unit of chromatin that is made up of a histone octamer and ∼200 base pairs of DNA. Emerging lines of evidence suggest that nucleosome function is tightly regulated by two different mechanisms. The first involves covalent modifications of histones (reviewed by Cheung et al., 2000). For example, the N-terminal tail of histone H3 is phosphorylated at serine 10 during mitosis and meiosis in a wide range of eukaryotes, and this phosphorylation is believed to be essential for proper chromosome condensation and segregation (Wei et al., 1999). Recent studies have shown that a protein kinase known as aurora B is responsible for this modification (Hsu et al., 2000). Perturbation of this kinase function leads to defects in a number of mitotic events, such as chromosome segregation and cytokinesis (Biggins et al., 1999; Speliotes et al., 2000; Adams et al., 2001; Giet and Glover, 2001). The second mechanism involves energy-dependent mobilization of nucleosomes that is mediated by chromatin remodeling complexes (reviewed by Aalfs and Kingston, 2000). Although such local “remodeling” of nucleosomes has been shown to be important for proper gene expression in numerous model systems, its potential impact on global chromatin dynamics is just beginning to be elucidated (Deuring et al., 2000).
Nucleosome fibers are further folded and packaged within the interphase nucleus. At the onset of mitosis, the nuclear envelope disassembles and the chromatin is converted into an even more organized structure, the mitotic chromosome (reviewed by Koshland and Strunnikov, 1996; Hirano, 2000). An understanding of the molecular mechanisms underlying higher-order chromosome dynamics and its cell cycle regulation is among the biggest challenges in modern cell biology.
Cell-free extracts derived from Xenopus laevis eggs provide a powerful biochemical system for studying the global rearrangements of chromatin structure. The addition of the physiological substrate, sperm chromatin, to such extracts allows reconstitution of key nuclear events in vitro, including DNA replication and chromosome condensation and segregation (Blow and Laskey, 1986; Newport and Spann, 1987; Shamu and Murray, 1992). The development of a simple method for isolating mitotic chromosomes assembled in vitro made it possible to characterize their structural components systematically (Hirano and Mitchison, 1994). The major protein components identified in this way were collectively referred to as Xenopus chromosome–associated polypeptides (XCAPs). It was found that XCAP-B and -D are topoisomerase II and the chromokinesin Xklp1 (Vernos et al., 1995), respectively. XCAP-C and -E were initially identified as members of the structural maintenance of chromosomes (SMC) family of ATPases (Hirano and Mitchison, 1994) and subsequently were shown to be part of a chromosome condensation complex (termed condensin) that contains three additional non-SMC subunits, XCAP-D2, -G and -H (Hirano et al., 1997; Kimura et al., 1998). The five-subunit condensin complex is highly conserved from yeast to humans. Genetic studies in different model organisms have shown that each of the condensin subunits is required for proper condensation and segregation of mitotic chromosomes (reviewed by Hirano, 2000).
In an attempt to identify novel protein components involved in the regulation of higher-order chromosome dynamics, we sought to characterize the remaining XCAPs. In this study, we focus on XCAP-F and find it to be the Xenopus ortholog of ISWI, the ATPase subunit of a subclass of chromatin remodeling complexes. We show that this polypeptide is present in two major complexes in Xenopus egg extracts and is required for regular spacing of nucleosomes but not for DNA replication or chromosome condensation. Immunodepletion of the INCENP-aurora B kinase complex reduces the level of histone H3 phosphorylation and affects the association of ISWI with chromosomes in mitosis. However, we find little, if any, disturbance in chromosome condensation or chromosomal targeting of condensin under this condition.
MATERIALS AND METHODS
Preparation and Fractionation of Xenopus Egg Extracts
Mitotic low-speed supernatants (LSSs) were prepared in XBE2 buffer (10 mM potassium-HEPES, pH 7.7, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA) according to Murray (1991). To prepare interphase LSS, cyclohexamide (0.1 mg/ml) and CaCl2 (0.4 mM) were added to mitotic LSS, and then the extract was incubated at 22°C for 30 min. LSSs were centrifuged at 50,000 rpm for 2 h using a TLS 55 rotor (Beckman, Palo Alto, CA) to yield high-speed supernatants (HSSs; Hirano and Mitchison, 1991). Sucrose gradient centrifugation of HSS was performed as described previously (Hirano et al., 1997).
Microsequencing and Cloning of XISWI, XACF1, and XWSTF
Mitotic chromosomes were assembled from sperm chromatin in a mitotic HSS, isolated, fractionated by 6% SDS-PAGE, and stained with Coomassie blue G (Hirano et al., 1997). A gel fragment corresponding to XCAP-F was excised and digested in situ with lysylendopeptidase. The resulting peptides were fractionated by reverse-phase chromatography and sequenced by Edman degradation as described previously (Bell et al., 1993). The following sequences were obtained: TVRVFRFITDNTVEERIVEXA; FNXRYLVIDEAHRI; WGRDDIENIAREVEGK; and KLLTQGFTNWNK (X represents an ambiguous residue). A database search revealed that these sequences were highly homologous to hSNF2H/hISWI (BAA25173), a human ortholog of Drosophila ISWI. Taking advantage of this information, we amplified a human cDNA fragment by PCR using a λgt10 library as a template. Oligonucleotides used for PCR were as follows: snf2h 551A, 5′-TACGGATCCGTTTTCATGTTAAGCACGCG-3′ (BamHI-tag sequence is underlined); and snf2h 651B, 5′-CTCAAGCTTAAGCATTTCATCTTTCCCAA-3′ (HindIII-tag sequence is underlined). The two primers amplified a ∼300-base pair fragment encoding a highly conserved region of the hSNF2H/hISWI cDNA (amino acids 551–651), which was then used as a hybridization probe to screen a Xenopus λZAP oocyte cDNA library (Stratagene, La Jolla, CA). Three overlapping cDNAs of varying lengths were isolated. All clones contained sequences encoding the C-terminal coding region of XISWI and were found to be part of the recently reported full-length sequence of XISWI (AF292095; Guschin et al., 2000).
XISWI-containing protein complexes were immunoprecipitated from mitotic HSS with anti-XISWI antibody and fractionated by 6% SDS-PAGE. Two polypeptides of 190 and 180 kDa were excised and processed as described above. The following three sequences were obtained for p190: QVPDYFDIIQRPIALNLIRE; AIYLQSFFVTEAQN; and YVPEGDD. The following four sequences were obtained for p180: KQVAEMTEEQREXYMIR; SLDLLER; KLETSEFFESTTEE; and KVISFVPVDSLYR. The peptide sequences of p190 were homologous to the Drosophila ATP-dependent chromatin assembly factor 1 (ACF1; Ito et al., 1999) and its human ortholog (WCRF180/hACF1/BAZ1A; Bochar et al., 2000; Jones et al., 2000; LeRoy et al., 2000; Poot et al., 2000). The sequences of p180 displayed a high degree of similarity to the human Williams syndrome transcription factor (hWSTF, Lu et al., 1998; also called BAZ1B, Jones et al., 2000). To clone cDNAs encoding p190 and p180 (termed XACF1 and XWSTF, respectively), a Xenopus oocyte cDNA library was screened using DNA probes derived from the corresponding human sequences (a generous gift of Dr. M. H. Jones). Three overlapping clones of XACF1 and a single clone of XWSTF were isolated and sequenced. Partial sequences of XACF1 and XWSTF are available from GenBank/EMBL/DDBJ under accession numbers AF412332 and AF412333, respectively.
Preparation of Antibodies
Rabbit antisera were raised against synthetic peptides corresponding to the C-terminal sequences of hISWI/hSNF2H (DGAPDGRGRKKKLKL; the underlined alanine is replaced by threonine in XISWI), XACF1 (RAPAKTPPAKRSRF), XWSTF (PETANPGRGRKQKK), XINCENP (SNRHHLAVGYGLKY; Adams et al., 2000), and Xaurora B/AIRK2 (RRVLPPVYQSTQSK; Adams et al., 2000). Immunization and purification of antibodies were carried out as described previously (Hirano et al., 1997). Antibodies were also raised against a hexa-histidine–tagged recombinant fragment of XISWI that contains its C-terminal 619-amino acid sequence. An antibody that recognizes a phosphorylated form of histone H3 at serine 10 was described previously (Kimura and Hirano, 2000).
Immunodepletion and Immunoprecipitation
For complete depletion, 25 μl of protein A agarose beads (GIBCO-BRL, Gaithersburg, MD) were coated with a mixture containing 25 μg of anti-XISWI, 10 μg of anti-XWSTF, and 15 μg of anti-XACF1 antibodies. For depletion of individual subunits, the same volume of beads was coated with either 35 μg of anti-XISWI, 12.5 μg of anti-XWSTF, or 17.5 μg of anti-XACF1 antibody. As a control, 50 μg of preimmune IgG was used. The antibody-coupled beads were washed twice with TBS and then twice with XBE2 containing protease inhibitors (10 μg/ml each of leupeptin, chymostatin, and pepstatin at a final concentration). After removing excess buffer, the beads were mixed with 50 μl of mitotic or interphase HSS that had been supplemented with energy mix (1 mM MgATP, 10 mM creatine phosphate, and 50 μg/ml creatine kinase at final concentrations). After incubating at 4°C for 1 h, supernatants were recovered by two rounds of brief spins and used as depleted HSSs. For immunodepletion of LSS, protein A agarose beads were replaced by Affi-Prep protein A support (Bio-Rad, Hercules, CA). Complete depletion of XISWI complexes was achieved with a mixture of 40 μg of anti-XISWI, 15 μg of anti-XWSTF, and 20 μg of anti-XACF1 antibodies. For depletion of XWSTF or XACF1 alone, 15 μg of anti-XWSTF or 20 μg of anti-XACF1 antibody, respectively, were used. Immunoprecipitation and 32P-labeling of extracts were carried out according to the methods described by Hirano et al. (1997).
Chromatin Assembly
To assemble mitotic chromosomes, mitotic HSS was supplemented with energy mix and diluted twofold with XBE6 (XBE2 containing 6 mM MgCl2 rather than 2 mM), and sperm chromatin was added at a final concentration of 2.5 × 103 nuclei/μl (7.5 ng DNA/μl). After incubating at 22°C for 2 h, samples were placed on ice for 10 min, and chromatin was isolated by centrifugation through a 30%-sucrose cushion in XBE2 at 10,000 rpm for 15 min (Sorval HB-4 rotor; DuPont, Wilmington, DE; Hirano and Mitchison, 1994). Interphase chromatin was assembled in the same way after incubation of sperm chromatin with interphase HSS. When required, half a volume of mitotic HSS was added to the interphase assembly mixture to convert the cell cycle state into mitosis and incubated at 22°C for another 90 min (e.g., Figure 8B). To assemble interphase nuclei competent for DNA replication, sperm chromatin was added to interphase LSS at a final concentration of 500 nuclei/μl (1.5 ng DNA/μl). To visualize the efficiency of DNA replication, biotin-14-dATP (GIBCO-BRL) was added at a final concentration of 4 μM. To convert the cell cycle state of the extract into mitosis, half a volume of mitotic LSS or recombinant cyclin B Δ90 was added (Losada et al., 1998).
Figure 8.
Effects of immunodepletion of INCENP (XINC) and aurora B (XAUB) from Xenopus egg extracts. (A) Mitotic HSS was depleted either with control IgG, (lane 1) or with a mixture of antibodies against XINC and XAUB (lane 2) and analyzed by immunoblotting using anti-XINC (top panel) or anti-XAUB antibody (bottom panel). (B) Chromatin was assembled in either interphase (lanes 1 and 2) or mitotic HSS (lanes 5 and 6). Alternatively, chromatin was first assembled in interphase HSS, which was then driven into mitosis by addition of half a volume of mitotic HSS (lanes 3 and 4). In each case, the HSSs had been mock-depleted (lanes 1, 3, and 5) or depleted of XINC and XAUB (lanes 2, 4, and 6). After incubation, chromatin was isolated and associated polypeptides were analyzed by immunoblotting using the antibodies indicated. (C) Mitotic HSS was mock-depleted (lanes 1 and 3) or depleted of XINC and XAUB (lanes 2 and 4) and labeled by addition of [γ-32P]ATP. The XISWI complexes were immunoprecipitated with anti-XACF1 and anti-XWSTF antibodies, fractionated by SDS-PAGE, and analyzed by Coomassie blue staining (lanes 1 and 2) or autoradiography (lane 3 and 4). (D) Sperm chromatin was incubated with mitotic HSS that had been mock-depleted (a and b) or depleted of XINC and XAUB (c and d). The assembled chromosomes were stained with DAPI (a and c) or anti–XCAP-E antibody (b and d). Bar, 10 μm.
Immunofluorescence Staining
Immunofluorescence staining of chromatin and chromosomes assembled in the cell-free extracts was performed as described previously (Losada et al., 1998, 2000) with minor modifications. For fixation, 4% paraformaldehyde rather than 2% was used. Affinity-purified antibodies were used at 1 μg/ml, and antisera were used at 1:500 dilution. For detection of incorporated biotin-14-dATP, FITC-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA) was used at 1:100 dilution. Images were taken using a Zeiss Axophot microscope equipped with a cooled charge-coupled device camera, processed with Oncor Image v2.0.5 (Oncor Inc., Gaithersburg, MD), and assembled with Photoshop v5.5 (Adobe, San Jose, CA). XL177 interphase cells were stained according to the method of Losada et al. (1998) except that the cells were fixed with 4% paraformaldehyde in PBS. Staining of unfixed metaphase spreads was carried out according to McDowell et al. (1999). Briefly, XL177 cells were treated with colcemid at 0.1 μg/ml for 1 h to increase the number of metaphase cells before harvest. The cells were swollen in 75 mM KCl and spun at 2000 rpm for 10 min onto a coverslip using a cytospin (Shandon, Pittsburgh, PA). Coverslips were immediately immersed in KCM buffer (10 mM Tris-HCl, pH 7.7, 120 mM KCl, 20 mM NaCl, 0.1% Triton X-100) and were incubated with anti-XISWI serum followed by FITC-conjugated secondary antibody. Chromosomes were then fixed with 4% paraformaldehyde in KCM buffer and counterstained with DAPI.
Supercoiling Assay
The supercoiling assay to monitor histone deposition was carried out according to Hirano and Mitchison (1991) with the following modifications. Supercoiled pRSETA plasmid DNA was incubated at a final concentration of 12.5 ng/μl in interphase HSS that had been mock-depleted or depleted of XISWI, XACF1, or XWSTF. At intervals, aliquots were removed and reactions were terminated by adding 10 volumes of stop solution (20 mM Tris-HCl, pH 8.0, 20 mM EDTA, 0.5% SDS, and 500 μg/ml proteinase K) and incubated at 37°C for 1 h. Fractions were extracted with phenol:chloroform, precipitated with ethanol, resuspended in TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) containing 50 μg/ml RNase A, and incubated at 37°C for 30 min. The resulting DNA was separated by gel electrophoresis on a 1.25% agarose gel in 0.5× TBE and visualized with ethidium bromide.
Micrococcal Nuclease Assay
Micrococcal nuclease treatment was carried out as described previously (Sandaltzopoulos and Becker, 1999) except that sperm chromatin instead of immobilized DNA was used as a substrate for nucleosome assembly. Briefly, 500 ng DNA equivalent of sperm chromatin (1.6 × 105 nuclei) were incubated for 90 min in interphase HSS that had been mock-depleted or depleted of XISWI, XACF1, or XWSTF. Samples were divided into three and supplemented with 10 mM CaCl2. Micrococcal nuclease was then added to a final concentration of 0.36, 1.2, and 3.6 U/ml. After incubation at 22°C for 10 min, reactions were stopped by addition of 25 mM EDTA and deproteinated in digestion buffer (20 mM Tris-HCl, pH 8.0, 20 mM EDTA, 0.5% SDS, and 500 μg/ml proteinase K) at 37°C for 45 min. Samples were extracted with phenol:chloroform, ethanol-precipitated, separated on a 1.25% agarose gel, and visualized with ethidium bromide.
DNA Replication Assay
DNA replication assays were carried out according to Chong et al. (1997). Briefly, interphase LSS was depleted using control IgG, anti-XISWI, anti-XACF1, or anti-XWSTF antibody and supplemented with [α-32P]dATP (specific radioactivity of 3000 Ci/mmol) at a final concentration of 0.05 mCi/ml. Sperm chromatin was then added at 500 nuclei/μl (1.5 ng DNA/μl). At regular intervals, aliquots were taken, and the reactions were stopped and deproteinated. The DNA was precipitated with 10% trichloroacetic acid (TCA) and placed on a filter for scintillation counting of incorporated radioactive nucleotides.
RESULTS
Identification of XISWI as a Major Component of Mitotic Chromosomes Assembled in Xenopus Egg Extracts
To identify novel chromosome-associated polypeptides, mitotic chromosomes were assembled from sperm chromatin in mitotic high-speed supernatant (HSS) prepared from Xenopus eggs. Chromosomes were purified from the assembly mixture by centrifugation through a sucrose cushion, and polypeptides associated with the chromosomes were analyzed by SDS-PAGE (Figure 1A). In addition to core histones and the histone H1-like protein B4, topoisomerase II and four of the five subunits of the condensin complex (XCAP-C, -D2, -E and -G) were readily detectable as we have described previously (Hirano and Mitchison, 1994). Among the remaining uncharacterized XCAPs, we focused on XCAP-F, a 135-kDa polypeptide. To determine the identity of XCAP-F, a large-scale preparation of mitotic chromosomes was separated by SDS-PAGE, and the 135-kDa band was excised from the gel and microsequenced. Analysis of the peptide sequences obtained allowed us to identify XCAP-F as the Xenopus ortholog of the Drosophila imitation switch protein (ISWI; see MATERIALS AND METHODS). We therefore renamed this polypeptide Xenopus ISWI (XISWI). ISWI is a member of the SNF2 family of ATPases (reviewed by Längst and Becker, 2001) and is the common subunit of several different chromatin remodeling complexes including NURF (Tsukiyama et al., 1995), ACF (Ito et al., 1997), and CHRAC (Varga-Weisz et al., 1997).
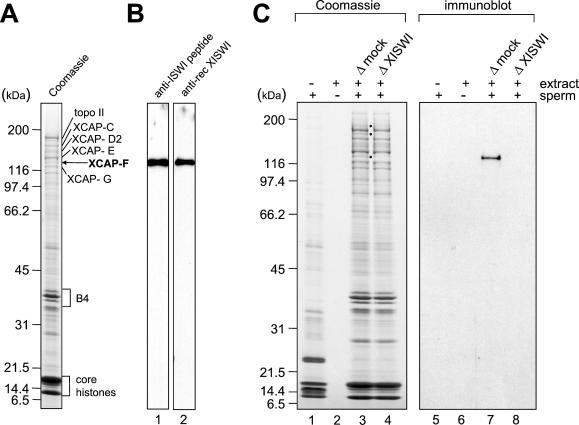
Figure 1.
Identification of XCAP-F as the Xenopus ortholog of ISWI. (A) Mitotic chromosomes were assembled from sperm chromatin in Xenopus egg mitotic HSS and purified through a sucrose cushion. Chromosomal proteins were separated by 7.5–15% SDS-PAGE and stained with Coomassie blue. (B) Mitotic HSS was fractionated by 7.5–15% SDS-PAGE and analyzed by immunoblotting with anti-XISWI antibodies against a synthetic peptide (lane 1) or a recombinant fragment (lane 2). (C) Mitotic chromosomes were assembled from sperm chromatin in mock-depleted (lanes 3 and 7) or XISWI-depleted mitotic HSS (lanes 4 and 8), and chromosome-associated polypeptides were analyzed by Coomassie blue staining (lanes 1–4) or immunoblotting with anti-XISWI antibody (lanes 5–8). As negative controls, reactions using sperm chromatin alone (lanes 1 and 5) or mitotic HSS alone (lanes 2 and 6) were processed in parallel.
To biochemically characterize XISWI, we raised antibodies against a synthetic peptide corresponding to the C-terminal sequence of human ISWI (hISWI/hSNF2H). In parallel, we isolated cDNAs for XISWI by screening a Xenopus oocyte library using an hISWI cDNA as a probe and prepared antisera against a recombinant fragment of XISWI (see MATERIALS AND METHODS). Both antibodies recognized a single 135-kDa polypeptide on an immunoblot of Xenopus egg mitotic HSS (Figure 1B, lanes 1 and 2). To confirm that XISWI is indeed identical to XCAP-F, mitotic chromosomes were assembled from sperm chromatin in mitotic HSS that had been mock-depleted or depleted of XISWI. The efficiency of immunodepletion was >95% as judged by quantitative immunoblotting (see Figure 5A). We found that the 135-kDa band corresponding to XCAP-F was present on chromosomes assembled in the mock-depleted HSS (Figure 1C, lane 3) but was missing on those assembled in the XISWI-depleted HSS (Figure 1C, lane 4). This result was further confirmed by immunoblotting using anti-XISWI antibody (Figure 1C, lanes 7 and 8). No XISWI signal was detected in assembly mixtures containing sperm chromatin alone (Figure 1C, lanes 1 and 5) or HSS alone (Figure 1C, lanes 2 and 6). These results demonstrated convincingly that the abundant 135-kDa component of mitotic chromosomes is the Xenopus ortholog of ISWI. It should be noted that, in addition to XISWI, two polypeptides of 180 and 190 kDa were specifically depleted from chromosomes assembled in the XISWI-depleted HSS (Figure 1C, lane 4). As shown below, we found that the two polypeptides associate with XISWI to form different complexes in Xenopus egg extracts.
Figure 5.
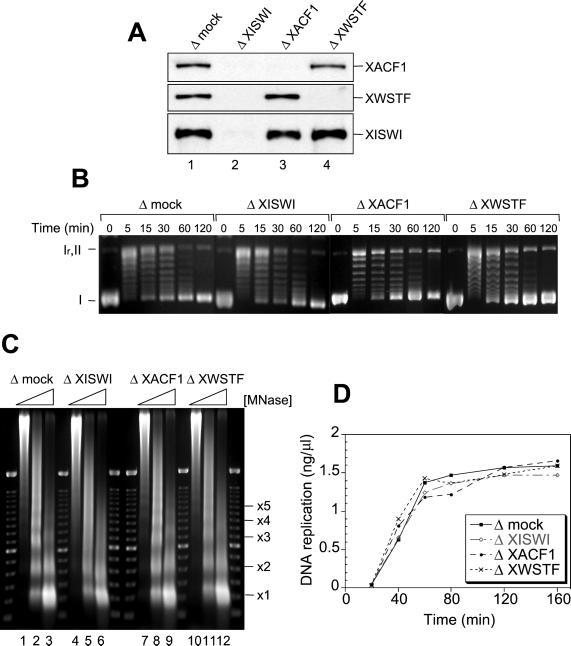
Effect of XISWI-, XACF1-, or XWSTF-depletion on interphase nuclear functions. (A) Interphase HSS was depleted with control IgG (lane 1), anti-XISWI (lane 2), anti-XACF1 (lane 3), or anti-XWSTF antibody (lane 4). Equal volumes of each HSS were analyzed by immunoblotting with the antibodies indicated. (B) Histone deposition assay. Supercoiled plasmid DNA was incubated with the control HSS (Δmock) or the depleted HSSs (ΔXISWI, ΔXACF1, and ΔXWSTF). Aliquots were taken at the time points indicated, deproteinated, separated on a 1.25% agarose gel, and stained with ethidium bromide. I, supercoiled DNA; Ir, relaxed DNA; II, nicked circular DNA. (C) Nucleosome spacing assay. Sperm chromatin was incubated with control or depleted HSS for 90 min. Each sample was then divided into three aliquots and treated with increasing concentrations of micrococcal nuclease (MNase: 0.36 U/ml, lanes 1, 4, 7, and 10; 1.2 U/ml, lanes 2, 5, 8, and 11; and 3.6 U/ml, lanes 3, 6, 9, and 12). After deproteination, the DNA was separated on a 1.25% agarose gel and stained with ethidium bromide. A 100-base pair ladder was used as a molecular weight marker. The positions of nucleosomal oligomers are indicated. (D) DNA replication assay. Sperm chromatin was incubated with control or depleted LSS in the presence of [α-32P]dATP. Aliquots were taken at the time points indicated, deproteinated, precipitated with TCA, and placed on a filter for scintillation counting. The extent of replication is expressed as nanograms of DNA replicated per microliter of extract.
XISWI Is Present in Two Major Protein Complexes in Xenopus Egg Extracts
It has been shown in Drosophila and humans that ISWI associates with distinct sets of subunits to form several different chromatin remodeling complexes (reviewed by Längst and Becker, 2001). To test whether this is also the case in Xenopus egg extracts, XISWI was immunoprecipitated from mitotic HSS. We found that anti-XISWI antibody immunoprecipitated not only XISWI, but also two major polypeptides with apparent molecular weights of 180 and 190 kDa (tentatively designated p180 and p190, respectively; Figure 2A, lane 2). Neither p180 nor p190 was detectable when a control antibody was used (Figure 2A, lane 1) or when a competing peptide was added into the immunoprecipitation reaction (Figure 2A, lane 3). We also noticed that the anti-XISWI antibody precipitated three additional polypeptides of low abundance with apparent molecular weights of 215, 90, and 55 kDa (Figure 2A, lane 2).
Figure 2.
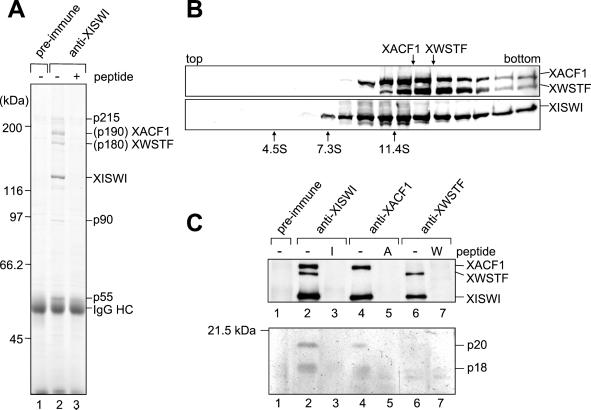
Biochemical characterization of two ISWI complexes in Xenopus egg extracts. (A) Mitotic HSSs were incubated with preimmune (lane 1) or anti-XISWI antibody (lanes 2 and 3) in the absence (lanes 1 and 2) or presence (lane 3) of ISWI peptide. Immunoprecipitates were recovered on protein A agarose beads, washed, separated by 7.5% SDS-PAGE and stained with Coomassie blue. IgG heavy chain is indicated by IgG HC. Microsequencing of p190 and p180 identified them as XACF1 and XWSTF, respectively. The identity of three minor polypeptides (indicated by p215, p90, and p55) remains to be determined. (B) Mitotic HSS was fractionated by centrifugation through a 5–20% sucrose gradient in XBE2. Fractions were TCA-precipitated and analyzed by immunoblotting using anti-XISWI, -XACF1, and -XWSTF antibodies. The positions of three protein standards (BSA [4.6S], aldolase [7.3S] and catalase [11.4S]) are indicated. (C) Mitotic HSSs were incubated with either preimmune (lane 1), anti-XISWI (lanes 2 and 3), anti-XACF1 (lanes 4 and 5), or anti-XWSTF antibody (lanes 6 and 7) in the absence (lanes 1, 2, 4, and 6) or presence (lanes 3, 5, and 7) of competing peptides (I, ISWI peptide; A, XACF1 peptide; W, XWSTF peptide). Immunoprecipitates were recovered on protein A agarose beads, and analyzed by immunoblotting using a mixture of anti-XISWI, -XACF1, and -XWSTF antibodies (top panel). Alternatively, the samples were separated by 15% SDS-PAGE and stained with silver (bottom panel).
The identity of p180 and p190 was determined by microsequencing of the corresponding bands excised from an SDS-polyacrylamide gel. Database searches with the resulting peptide sequences allowed us to conclude that p190 is the Xenopus ortholog of the Drosophila ATP-utilizing chromatin accessibility factor protein 1 (ACF1; Ito et al., 1999), and p180 is the Xenopus ortholog of the human Williams syndrome transcription factor (WSTF; Lu et al., 1998). We screened a Xenopus oocyte library to isolate cDNAs encoding these polypeptides (named XACF1 and XWSTF, respectively) and raised antibodies against their C-terminal peptide sequences (see MATERIALS AND METHODS). Each of the antibodies recognized a single polypeptide of the expected size by immunoblotting against mitotic HSS. When mitotic HSS was fractionated by sucrose gradient centrifugation, XACF1 and XWSTF sedimented at different peaks of 12.5S and 13.6S, respectively, whereas XISWI formed a very broad peak spreading from 11S to 14S (Figure 2B). This result suggests that XISWI associates with XACF1 or XWSTF to form two separate complexes. To test this further, immunoprecipitations were performed with antibodies against each of the three polypeptides, and coprecipitated polypeptides were analyzed by immunoblotting. Consistent with the data shown in Figure 2A, anti-XISWI antibody immunoprecipitated XACF1 and XWSTF along with XISWI (Figure 2C, top, lane 2). Anti-XACF1 antibody immunoprecipitated XACF1 and XISWI, but not XWSTF (Figure 2C, top, lane 4), whereas anti-XWSTF antibody precipitated XWSTF and XISWI, but not XACF1 (Figure 2C, top, lane 6). None of these polypeptides were precipitated with preimmune IgG (Figure 2C, top, lane 1) or in the presence of competing peptides (Figure 2C, top, lanes 3, 5, and 7). To determine whether XISWI associates with low-molecular-weight proteins, as has been found in one of the ISWI-containing complexes termed CHRAC (Corona et al., 2000; Poot et al., 2000), the same set of immunoprecipitates was fractionated on a 15% SDS-polyacrylamide gel and stained with silver. Anti-XISWI antibody specifically immunoprecipitated two polypeptides of 20 and 18 kDa (Figure 2C, bottom, lane 2; indicated by p20 and p18, respectively). A similar set of polypeptides was precipitated with anti-XACF1 antibody (Figure 2B, bottom, lane 4) but not with anti-XWSTF antibody (Figure 2B, bottom, lane 6). Taken all the results together, we conclude that XISWI is present in at least two different protein complexes in Xenopus egg extracts: one contains XACF1 and two low-molecular-weight subunits and the other contains XWSTF.
Cell Cycle Regulation of the XISWI Complexes in Xenopus Egg Extracts
To investigate the cell cycle regulation of the two XISWI complexes, their biochemical properties were analyzed in interphase and mitotic HSSs prepared from Xenopus eggs. XISWI, XACF1, and XWSTF were present at the same level in the two extracts, as judged by immunoblotting. When anti-XISWI antibody was used for immunoprecipitation from the two HSSs, a similar amount of XACF1 and XWSTF was recovered (Figure 3A), suggesting that there is no subunit rearrangement between interphase and mitosis. The same antibody was then used to immunoprecipitate the complexes from 32P-labeled HSSs. We found that all three polypeptides were weakly phosphorylated in interphase (Figure 3B, lane 1), whereas XISWI and XACF1, but not XWSTF, were hyperphosphorylated in mitosis (Figure 3B, lane 3).
Figure 3.
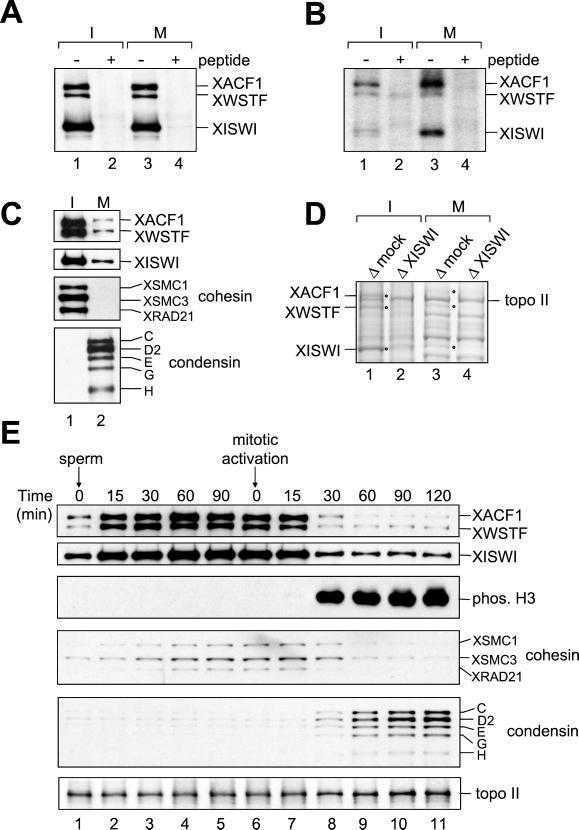
Cell cycle regulation of two ISWI complexes in Xenopus egg extracts. (A) XISWI was immunoprecipitated from interphase (lanes 1 and 2) or mitotic HSS (lanes 3 and 4) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of ISWI peptide and analyzed by immunoblotting with antibodies against XISWI, XACF1, and XWSTF. (B) XISWI was immunoprecipitated from 32P-labeled HSSs as in A, separated by SDS-PAGE and analyzed by autoradiography. (C) Chromatin was assembled in interphase (lane 1) or mitotic HSS (lane 2) and analyzed by immunoblotting with the antibodies indicated. (D) Chromatin was assembled in interphase (lanes 1 and 2) or mitotic HSS (lanes 3 and 4) that had been mock-depleted (lanes 1 and 3) or depleted of XISWI (lanes 2 and 4). Chromatin-associated polypeptides were separated by 7.5% SDS-PAGE and stained with Coomassie blue. The positions of XACF1, XWSTF, and XISWI are indicated by dots. (E) Sperm chromatin was incubated with interphase LSS. After 120 min, recombinant sea urchin cyclin BΔ90 was added to convert the extract into a mitotic state. At the indicated time points, aliquots were taken and chromatin-bound polypeptides were analyzed by immunoblotting using the antibodies indicated.
The cell cycle–specific interaction of the XISWI complexes with chromatin was first characterized in HSS. Sperm chromatin was incubated in interphase or mitotic HSS, and the protein components bound to the chromatin were analyzed by immunoblotting (Figure 3C). We found that XISWI, XACF1,and XWSTF were approximately four times more abundant on interphase chromatin compared with mitotic chromosomes. A consistent result was obtained when the chromatin and chromosome fractions were analyzed by Coomassie blue staining (Figure 3D). It was estimated by quantitative immunoblotting that XISWI is present on every ∼5 kb of DNA in interphase chromatin and every ∼20 kb in mitotic chromosomes. Cohesin and condensin were also used as specific markers for chromosomal components (Figure 3C; Hirano et al., 1997; Losada et al., 1998). Condensin was very abundant on mitotic chromosomes (one XCAP-E per ∼5 kb of DNA), whereas virtually no cohesin was detectable on the same structures (one XSMC3 per >400 kb). Thus, the cell cycle–dependent association of the XISWI complexes with chromatin is reminiscent of that of cohesin, but is quantitatively different.
We then examined the interaction of the XISWI complexes with chromatin in Xenopus egg LSSs, in which additional key events including nuclear envelope formation and DNA replication can be reconstituted (e.g., Blow and Laskey, 1986). In interphase LSS, the association of XISWI, XACF1, and XWSTF with chromatin was rapid and preceded the accumulation of cohesin on chromatin (Figure 3E, lanes 1–5). When the cell cycle state of the extract was converted into mitosis, ∼75% of XISWI, XACF1, and XWSTF dissociated from chromatin, coincidentally with phosphorylation of histone H3 at serine 10 (Figure 3E, lanes 6–11). The kinetics of dissociation was again slightly faster than that of cohesin. Consistent with our previous data (Losada et al., 1998), condensin was targeted to chromosomes in a mitosis-specific manner, whereas the chromosomal level of topoisomerase II (topo II) remained constant throughout the cell cycle.
Immunolocalization of the XISWI Complexes In Vitro and In Vivo
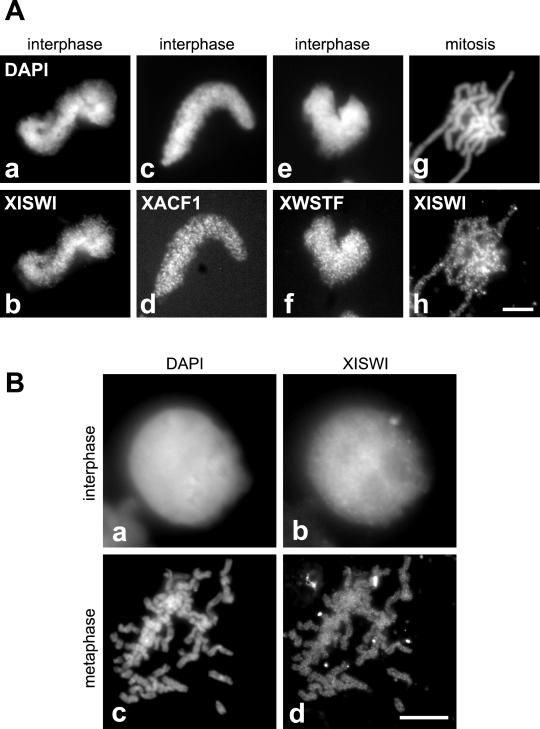
To determine the localization of the XISWI complexes on chromosomes assembled in vitro, sperm chromatin was incubated with interphase or mitotic HSS, fixed, and stained with antibodies against the individual subunits. We found that XISWI, XACF1, and XWSTF were distributed on interphase chromatin in a similar punctate pattern (Figure 4A, a–f). XISWI was weakly detected throughout mitotic chromosome arms, again, in a punctate pattern (Figure 4A, g and h). No specific enrichment of the signals on subchromosomal structures (e.g., centromeres) was observed. Consistent with the in vitro result, XISWI was localized to the interphase nucleus in Xenopus tissue culture cells (Figure 4B, a and b). At metaphase, XISWI was detected weakly throughout mitotic chromosomes (Figure 4B, c and d). These results are in agreement with our immunoblotting data showing a reduction in the association of XISWI with chromosomes during mitosis.
Figure 4.
Immunolocalization of XISWI, XACF1, and XWSTF. (A) Interphase chromatin (a–f) or mitotic chromosomes (g and h) were assembled from sperm chromatin in interphase or mitotic HSS, respectively, fixed, and stained with antibodies against XISWI (b and h), XACF1 (d), or XWSTF (f). DNA was counterstained with DAPI (a, c, e, and g). Bar, 10 μm. (B) XISWI localization in Xenopus tissue culture cells. Interphase (a and b) or mitotic cells (c and d) were stained with an antibody against recombinant XISWI (b and d) and counterstained with DAPI (a and c). Bar, 10 μm.
XISWI Complexes Are Required for Nucleosome Spacing, but not for Histone Deposition or DNA Replication
To determine the functions of the XISWI complexes in Xenopus egg extracts, we immunodepleted XISWI, XACF1, or XWSTF using the corresponding antibodies. The efficiency of immunodepletion was >95% in each case, as judged by quantitative immunoblotting (Figure 5A). We used a DNA supercoiling assay to test whether these polypeptides are required for the deposition of histones onto protein-free DNA (Hirano and Mitchison, 1991). On incubation with interphase HSS, supercoiled plasmid DNA was rapidly relaxed by the action of endogenous topoisomerases (Figure 5B, 0–5 min). Progressive introduction of negative supercoils over time reflected the efficiency of histone deposition (Figure 5B, 15–120 min). No significant difference was detected either in the kinetics or extent of supercoiling between the mock-depleted and the depleted interphase HSSs. Next, sperm chromatin was incubated with the mock-depleted or depleted interphase HSSs, and the reaction mixtures were digested with micrococcal nuclease. In the mock-depleted HSS, a discrete ladder of nucleosomes with an interval of ∼200 base pairs was observed (Figure 5C, lanes 1–3). In contrast, the nucleosomes were poorly spaced in the XISWI-depleted HSS (Figure 5C, lanes 4–6). Immunodepletion of XACF1 caused a subtle disturbance in nucleosome spacing (Figure 5C, lanes 7–9), whereas depletion of XWSTF resulted in a more severe defect (Figure 5C, lanes 10–12). Thus, the XISWI complexes are not required for histone deposition onto DNA but do contribute to the formation of regularly spaced nucleosomes in Xenopus egg cell-free extracts.
To determine the effect of poorly spaced nucleosomes on DNA replication, interphase nuclei were assembled in LSS that had been mock-depleted or depleted of XISWI, XACF1, or XWSTF, and the incorporation of [α-32P]dATP into newly synthesized DNA was measured (Figure 5D). Depletion of XISWI, XACF1, or XWSTF had little effect on the extent or kinetics of DNA replication. We also found no difference in the kinetics of nuclear envelope assembly (our unpublished results), which is known to be a prerequisite for the initiation of DNA replication in Xenopus egg extracts.
XISWI Complexes Are Not Required for Chromosome Condensation
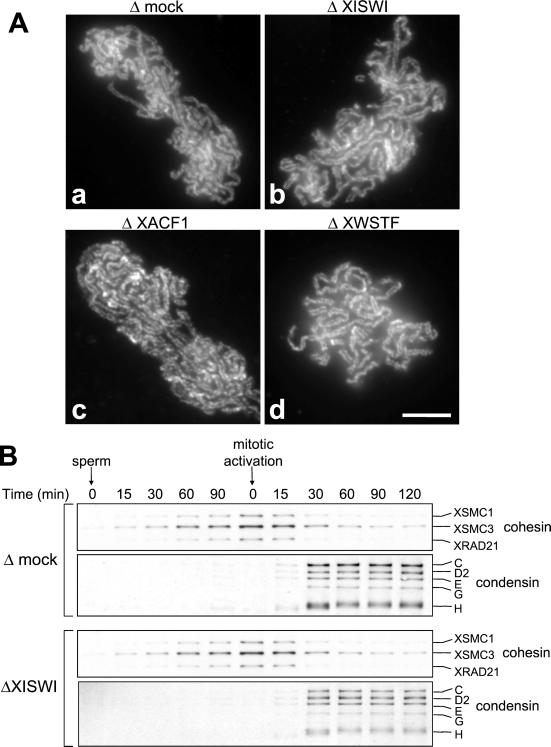
The identification of the XISWI complexes as major components of mitotic chromosomes suggested that they could play a role in chromosome condensation. We tested the possibility that local nucleosome mobilization mediated by XISWI might contribute to large-scale reorganization of chromatin fibers during mitosis. Sperm chromatin was incubated with mitotic HSS that had been mock-depleted or depleted of XISWI, XACF1, or XWSTF. The assembled structures were fixed and stained with DAPI and an antibody against the condensin subunit XCAP-E. We found little difference in the morphology of the chromosomes assembled in the control (Figure 6A, a and b) and the depleted extracts (Figure 6A, c–h). In all cases, DAPI staining showed uniformly condensed chromosomes, and XCAP-E was localized to an internal filamentous network as reported previously (Hirano and Mitchison, 1994). Immunoblotting of isolated chromosome fractions showed that depletion of XISWI had no detectable effect on the timing of chromosomal binding of condensin or on phosphorylation of histone H3 at serine 10 (Figure 6B).
Figure 6.
Effect of XISWI-, XACF1-, or XWSTF-depletion on chromosome condensation. (A) Sperm chromatin was incubated in mitotic HSS that had been mock-depleted (a and b) or depleted of XISWI (c and d), XACF1 (e and f) or XWSTF (g and h) for 2 h. Chromosomes were fixed and stained with DAPI (a, c, e, and g) and anti–XCAP-E antibody (b, d, f, and h). Bar, 10 μm. (B) Sperm chromatin was incubated with mock-depleted (Δmock) or XISWI-depleted (ΔXISWI) mitotic HSS. Aliquots were taken at the times indicated, and chromatin-bound polypeptides were analyzed by immunoblotting.
XISWI Complexes Are Not Required for Sister Chromatid Cohesion
Next we tested whether the XISWI complexes might play a role in the formation of metaphase chromosomes with duplicated sister chromatids, a reaction that can be reconstituted only in LSS but not in HSS (Losada et al., 1998). Nuclei were assembled in interphase LSS that had been mock-depleted or depleted of XISWI, XACF1, or XWSTF. After DNA replication, half a volume of mock-depleted or the corresponding depleted mitotic LSS was added to drive the extract into mitosis. The resulting metaphase chromosomes were fixed and stained with anti–XCAP-E antibody (Figure 7A). We found that sister chromatids were tightly paired along their entire length in all cases, and no apparent defect in chromosome cohesion or condensation was observed. Efficient DNA replication in each chromosome was confirmed by incorporation of biotin-dATP (our unpublished results). We also analyzed the time course of the interactions of cohesin and condensin with chromatin and found that XISWI depletion had little effect on the behavior of these protein complexes (Figure 7B).
Figure 7.
Effect of XISWI-, XACF1-, or XWSTF-depletion on sister chromatid cohesion. (A) Sperm chromatin was incubated for 2 h with interphase LSS that had been mock-depleted (a) or depleted of XISWI (b), XACF1 (c) or XWSTF (d), and half a volume of the corresponding mitotic LSS was added to drive the extract into mitosis. After incubation for 90 min, chromosomes were fixed and stained with anti–XCAP-E antibody. Bar, 10 μm. (B) Sperm chromatin was incubated with either mock-depleted (Δmock) or XISWI-depleted (ΔXISWI) LSS. After 120 min, recombinant sea urchin cyclin B Δ90 was added to drive the extract into mitosis. Aliquots were taken at the indicated time points, and chromatin was purified and analyzed by immunoblotting with antibodies against cohesin and condensin subunits.
Depletion of INCENP-aurora B Reduces Histone H3 Phosphorylation and Suppresses Dissociation of XISWI, but Has Little Effect on Chromosome Condensation
Emerging lines of evidence, from different organisms, suggest that aurora B is one of the major protein kinases that directly phosphorylate serine 10 of histone H3 in mitosis (Hsu et al., 2000; Speliotes et al., 2000; Adams et al., 2001; Giet and Glover, 2001). We tested whether there is any functional relationship between H3 phosphorylation and bulk dissociation of XISWI from chromatin, because the two events are tightly coupled at the onset of mitosis (e.g., Figure 3E). To this end, we raised antibodies specific to Xenopus aurora B (XAUB) and its binding partner Xenopus INCENP (XINC). Consistent with a previous report (Adams et al., 2000), we found that the two polypeptides tightly associate with each other in Xenopus egg extracts (our unpublished results). We then used a mixture of anti-XINC and anti-XAUB antibodies to deplete >95% of the XINC-XAUB complex from interphase HSS or mitotic HSS (Figure 8A). In interphase HSS, immunodepletion of the complex had little effect on the association of XISWI, XACF1, or XWSTF with chromatin (Figure 8B, lanes 1 and 2). XAUB was found in the chromatin assembled in the control HSS but not in that assembled in the depleted HSS. The chromosomal level of histone H3 phosphorylation at serine 10 was negligible in both cases. When the mock-depleted interphase HSS was converted into mitosis, condensin bound to chromosomes and histone H3 became phosphorylated (Figure 8B, lane 3). The chromosomal level of the two XISWI complexes decreased under this condition as was shown in LSS (Figure 3E). We found that, in the absence of XINC and XAUB, mitotic phosphorylation of histone H3 was greatly diminished (Figure 8B, lane 4). Interestingly, the dissociation of the XISWI complexes from chromatin was partially suppressed under this condition. In contrast, very little, if any, difference was observed in the chromosomal association of condensin and topoisomerase II in the presence or absence of XINC-XAUB. Similar results were obtained when sperm chromatin was incubated directly with mitotic HSS (Figure 8B, lanes 5 and 6).
To determine whether any one of XISWI, XACF1, and XWSTF are directly phosphorylated by the aurora B kinase, we immunoprecipitated these polypeptides from 32P-labeled mitotic HSS that had been mock-depleted or depleted of XINC-XAUB (Figure 8C, lanes 1 and 2). Virtually no difference was detected in phosphorylation of XISWI, XWSTF, or XACF1 (Figure 8C, lanes 3 and 4), suggesting that none of these polypeptides are likely to be a direct target of aurora B kinase in Xenopus egg extracts.
To examine the effect of XINC-XAUB depletion on chromosome condensation, sperm chromatin was incubated with the mock-depleted or depleted mitotic HSS. Surprisingly, we found little difference in the morphology of chromosomes assembled in the presence or absence of XINC-XAUB, as judged by DAPI staining (Figure 8D, a and c). The chromosomal localization of XCAP-E was also indistinguishable between the two conditions (Figure 8D, b and d). Thus, in Xenopus egg HSS, chromosome condensation is apparently normal when histone H3 phosphorylation is drastically reduced by depletion of XINC-XAUB.
DISCUSSION
In this study, we have identified ISWI ATPase as a major component of mitotic chromosomes assembled in Xenopus egg extracts. This finding has prompted us to take advantage of this powerful cell-free system to explore the functional interfaces between nucleosome remodeling, histone H3 phosphorylation, and higher-order chromosome dynamics.
ISWI Protein Complexes in Xenopus Egg Extracts
Our results show that ISWI is present in at least two different protein complexes in Xenopus egg extracts. The first complex contains XACF1 and two low-molecular-weight polypeptides, p20 and p18. Previous studies identified a heterodimer of ISWI and ACF1 (called ACF) from Drosophila (Ito et al., 1997, 1999) and humans (Bochar et al., 2000; LeRoy et al., 2000). A similar ISWI-ACF1 complex with two additional histone-fold polypeptides (called CHRAC) has also been found in the two organisms (Corona et al., 2000; Poot et al., 2000). Given the presence of the two small subunits, it is most likely that the XACF1-containing complex we report here is the Xenopus version of CHRAC. The second ISWI complex in Xenopus egg extracts contains XWSTF, the Xenopus ortholog of human WSTF. It should be emphasized that this is a novel protein complex that has never been reported from any organisms. WSTF is one of several genes deleted in Williams syndrome (WS), a developmental disorder with multisystemic defects including congenital heart disease and mental retardation (Lu et al., 1998). Although it remains to be determined whether WSTF is indeed responsible for some of the defects in WS, mutations in another SNF2 family member have also been implicated in severe mental retardation (Picketts et al., 1996), further emphasizing the clinical importance of this class of chromatin remodeling ATPases.
The primary sequences of ACF1 and WSTF share several structural motifs including a PHD finger motif and a bromodomain. Recent sequence analysis indicates that ACF1 and WSTF, together with two additional members, constitute a new protein family called BAZ (bromodomain adjacent to zinc finger; Jones et al., 2000) or WAL (WSTF-, ACF1-like; Poot et al., 2000). It is unknown whether the remaining members of the BAZ/WAL family might associate with ISWI or other remodeling ATPases. Neither is it known whether they are present in Xenopus egg extracts. Double immunodepletion using anti-XACF1 and anti-WSTF antibodies removed 60–70% of XISWI from the extracts (our unpublished results), suggesting the occurrence of other ISWI complexes in Xenopus eggs. The relative abundance of the ISWI-ACF1 and ISWI-WSTF complexes is ∼3:1 as judged by Coomassie blue staining of the anti-XISWI immunoprecipitates. Our current results provide complementary information to the recent work by Guschin et al. (2000) who performed a biochemical fractionation of Xenopus egg extracts in search of protein complexes containing ISWI. After ion-exchange column chromatography, the majority of XISWI was recovered in a fraction called ISWI-C, which contained XACF1 and an unidentified polypeptide whose molecular weight was close to that of XWSTF. It is therefore likely that the ISWI-C fraction is a mixture of the two separate complexes we report here. Full description of the minor fractions containing XISWI remains to be reported.
Are ISWI Complexes Involved in Higher-Order Chromosome Dynamics?
Despite considerable progress in the biochemical analyses of ISWI remodeling complexes in vitro, their physiological role in nucleosome dynamics remains elusive (reviewed by Längst and Becker, 2001). Our immunodepletion experiments show that XISWI is not required for the deposition of core histones onto protein-free DNA or sperm chromatin in Xenopus egg extracts. It is very likely that other factors, such as nucleoplasmin and N1/N2, are primarily responsible for histone deposition in this cell-free system (e.g., Philpott and Leno, 1992). In contrast, as judged by a micrococcal nuclease assay, the major nucleosome spacing activity in the extracts does indeed reside in the XISWI remodeling complexes. This conclusion is further strengthened by our finding that XISWI is among the most abundant polypeptides associated with interphase chromatin and mitotic chromosomes. The “division of labor” between histone deposition and subsequent nucleosome rearrangement is also consistent with a recent reconstitution experiment using purified Drosophila nucleosome assembly protein-1 (NAP-1) and ACF1 (Nakagawa et al., 2001).
Previous studies have shown that the mobilization of nucleosomes mediated by ISWI complexes can activate transcription by allowing a transcription factor to bind to a specific promoter region in vitro (Ito et al., 1997; Mizuguchi et al., 1997). However, Xenopus eggs are capable of undergoing multiple rounds of cell division with virtually no new protein synthesis, until the midblastula transition (Newport and Kirschner, 1982). The abundance of ISWI on embryonic chromosomes therefore argues that it may play a more global role in chromatin dynamics outside the control of gene expression. One attractive possibility is that nucleosomal “fluidity” catalyzed by ISWI may enhance or help proper interactions between chromatin and its cell cycle–specific regulators such as condensin and cohesin. Nevertheless, we have been unable to detect any obvious defects in DNA replication, sister chromatid cohesion, or chromosome condensation in the XISWI-depleted cell-free extracts. It remains possible that the in vitro conditions used in this study, although highly physiological, may not be sensitive enough to detect potential contributions of ISWI to higher-order chromosome dynamics.
A recent genetic study has shown that ISWI function is essential for cell viability in Drosophila (Deuring et al., 2000). Interestingly, the global structure of the X chromosome is compromised in ISWI mutants in a male-specific manner. In Drosophila, the dosage compensation machinery specifically targets the male X chromosome and upregulates transcription of many X-linked genes. This functional change is accompanied by hyperacetylation of histone H4 on the X chromosome, leading to the hypothesis that the hyperacetylated chromosome is more sensitive to a loss of ISWI function. In Saccharomyces cerevisiae, neither of the two ISWI-encoding genes, ISW1 and ISW2, is essential for cell viability under normal growth conditions although their absence affects transcription of a subset of genes (Tsukiyama et al., 1999; Goldmark et al., 2000; Kent et al., 2001). The currently available genetic data point out the diversity and complexity of ISWI functions in vivo, demanding further analysis of this class of remodeling ATPases in different organisms from many different angles.
Histone H3 Phosphorylation and Mitotic Chromosome Dynamics
Murnion et al. (2001) has recently shown that a histone H3 kinase activity is associated with mitotic chromosomes in Xenopus egg extracts. This activity can be eluted from the chromosomes and immunoprecipitated with an antibody specific to aurora B. Our current results complement these observations and further demonstrate that immunodepletion of the INCENP-aurora B complex from Xenopus egg extracts greatly reduces the level of histone H3 phosphorylation on mitotic chromosomes. Thus, aurora B is likely to be the major histone H3 kinase present in the extracts, consistent with genetic studies in yeast, Caenorhabditis elegans and Drosophila (Hsu et al., 2000; Speliotes et al., 2000; Adams et al., 2001; Giet and Glover, 2001). Remarkably, mitotic dissociation of XISWI complexes is partially suppressed in the absence of XINC-XAUB. Because none of XISWI, XACF1, or XWSTF appears to be a direct target of the aurora B kinase, it is likely that phosphorylation of histone H3 itself or of other substrates regulates the cell cycle–dependent association of the XISWI complexes with chromatin. We have also found that depletion of the XINC-XAUB complex has little effect on the targeting of condensin or chromosome condensation in the cell-free extracts. Immunofluorescence staining with a phospho-specific H3 antibody detected a residual level of signals on chromosomes assembled in the XINC-XAUB–depleted extract (our unpublished results). We cannot exclude the possibility therefore that a kinase(s) other than aurora B plays a minor yet significant role in H3 phosphorylation, condensin targeting, and chromosome condensation in Xenopus egg extracts. Candidates for such kinases might include aurora A, another member of the aurora kinase family. A recent study has demonstrated, however, that neither H3 phosphorylation nor chromosome condensation is interfered by depletion of aurora A from Xenopus egg extracts (Scrittori et al., 2001). In addition, uncoupling of H3 phosphorylation and chromosome condensation has been observed in an extract treated with a phosphatase inhibitor, microcystin (Murnion et al., 2001). Finally, our previous reconstitution experiments showed that phosphorylation of histone H3 has little impact on the interaction between purified condensin and nucleosomes in vitro (Kimura and Hirano, 2000). Taken all the results together, we conclude that, at least in Xenopus egg extracts, phosphorylation of histone H3 at serine 10 is unlikely to play a direct role in recruiting condensin to chromosomes in mitosis.
Our results are in apparent contradiction to the generally accepted view that histone H3 phosphorylation is important for chromosome condensation. In particular, Giet and Glover (2001) reported that chromosomal targeting of a condensin subunit is compromised when aurora B is inactivated by RNA interference in Drosophila. The reason for the discrepancy between this work and our current study is unknown. It is important to note, however, that the extent of condensation defects observed in the absence of H3 phosphorylation is variable between different studies or different organisms (Wei et al., 1999; Speliotes et al., 2000; Adams et al., 2001; Giet and Glover, 2001). For example, Adams et al. (2001) found no tight correlation between the level of histone H3 phosphorylation and chromatin compaction in Drosophila. What is then the role of this highly specific phosphorylation event in mitotic chromosome dynamics? We suspect that histone H3 phosphorylation may constitute part of the regulatory mechanism that coordinates the multiple structural changes of chromosomes that occur during prometaphase. These would include unloading of interphase chromatin components (such as the ISWI complexes), partial release of sister chromatid cohesion, and mitotic maturation of kinetochore structures. Linear compaction of chromatin fibers is only one of the many events taking place during this stage and may not be the primary consequence of H3 phosphorylation. This idea would provide a reasonable explanation for the complex phenotypes observed in the absence of aurora B function (Speliotes et al., 2000; Adams et al., 2001; Giet and Glover, 2001). Clearly, future work is required to determine the exact roles of H3 phosphorylation in higher-order chromosome dynamics. It will also be important to identify physiological substrates of aurora B other than histone H3.
Extending the List of Structural Components of Mitotic Chromosomes
The original motivation of the current work was to identify novel proteins essential for mitotic chromosome dynamics. As one such candidate, we found XISWI to be a major component of mitotic chromosomes. It was therefore rather disappointing to find that depletion of the ISWI complexes had little impact on the morphology of chromosomes assembled in Xenopus egg cell-free extracts. However, there are several precedents of abundant chromosomal proteins that have no direct role in chromosome assembly. For example, immunodepletion of the embryonic linker histone B4 results in little defect in chromosome condensation (Ohsumi et al., 1993). XCAP-D/Xklp1, a kinesin-like protein localizing to chromosomal arms, is required for chromosome positioning and bipolar spindle organization (Vernos et al., 1995) but plays no apparent role in the formation of mitotic chromosomes (T. Hirano, unpublished results). After the identification of the two ISWI complexes in this study, only a few polypeptides remain to be characterized on chromosomes assembled in this cell-free system. The current results, taken together with the previous ones, argue that the number of structural components essential for chromosome condensation may be very limited, being much smaller than generally assumed. This idea further emphasizes the central roles of the condensin complex (Hirano and Mitchison, 1994; Hirano et al., 1997) and topoisomerase II (Adachi et al., 1991; Hirano and Mitchison, 1993) in this process that can be reconstituted in Xenopus egg cell-free extracts. We anticipate that continued characterization of these extracts will provide additional insights into our understanding of higher-order chromosome structure and function and eventually help reconstitute a whole chromosome structure from purified components in vitro.
ACKNOWLEDGMENTS
The authors thank M. H. Jones (Chugai Research Institute for Molecular Medicine, Japan) for the BAZ plasmids. We are also grateful to members of the Hirano laboratory for critically reading the manuscript, P. J. Gillespie for instruction in the DNA replication assay, and M. Hirano for instruction in molecular cloning. This work was supported by grants from the National Institutes of Health (to T.H. and R.K.) and the Pew Scholars Program in the Biomedical Sciences (to T.H.). D. MacCallum was the recipient of an Andrew Seligson Memorial Fellowship. A. Losada was supported by the Robertson Research Fund and the Leukemia and Lymphoma Society.
Abbreviations used:
- ACF1
ATP-utilizing chromatin assembly factor 1
- AUB
aurora B
- CHRAC
chromatin accessibility complex
- HSS
high-speed supernatant
- INCENP/INC
inner centromere protein
- ISWI
imitation switch
- LSS
low-speed supernatant
- NURF
nucleosome remodeling factor
- SMC
structural maintenance of chromosomes
- WSTF
Williams syndrome transcription factor
- XCAP
Xenopus chromosome–associated polypeptide
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09-0441. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09-0441.
REFERENCES
- Aalfs JD, Kingston RE. What does ‘chromatin remodeling’ mean? Trends Biochem Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Luke M, Laemmli UK. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophilainner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopuseggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci USA. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Chong JP, Thommes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopusreplication licensing system. Methods Enzymol. 1997;283:549–564. doi: 10.1016/s0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- Corona DF, Eberharter A, Budde A, Deuring R, Ferrari S, Varga-Weisz P, Wilm M, Tamkun J, Becker PB. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC) EMBO J. 2000;19:3049–3059. doi: 10.1093/emboj/19.12.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun JW. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophilaaurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Guschin D, Geiman TM, Kikyo N, Tremethick DJ, Wolffe AP, Wade PA. Multiple ISWI ATPase complexes from Xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J Biol Chem. 2000;275:35248–35255. doi: 10.1074/jbc.M006041200. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Cell cycle control of higher-order chromatin assembly around naked DNA in vitro. J Cell Biol. 1991;115:1479–1489. doi: 10.1083/jcb.115.6.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopusegg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the DrosophilaBarren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, Allis CD. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu J-i, Shimane M. A novel family of bromodomain genes. Genomics. 2000;63:40–45. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci USA. 2000;97:11972–11977. doi: 10.1073/pnas.220326097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Längst G, Becker P. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci. 2001;114:2561–2568. doi: 10.1242/jcs.114.14.2561. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of XenopusSMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopusand human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, Chapman L, Rhodes D, Higgs DR. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromain. Mol Cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- Murnion ME, Adams RR, Callister DM, Allis CD, Earnshaw WC, Swedlow JR. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J Biol Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nakagawa T, Bulger M, Muramatsu M, Ito T. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J Biol Chem. 2001;276:27384–27391. doi: 10.1074/jbc.M101331200. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner MW. A major developmental transition in early Xenopusembryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- Ohsumi K, Katagiri C, Kishimoto T. Chromosome condensation in Xenopusmitotic extracts without histone H1. Science. 1993;262:2033–2035. doi: 10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopusegg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandaltzopoulos R, Becker PB. A solid-phase approach for the analysis of reconstituted chromatin. Methods Mol Biol. 1999;119:195–206. doi: 10.1385/1-59259-681-9:195. [DOI] [PubMed] [Google Scholar]
- Scrittori L, Hans F, Angelov D, Charra M, Prigent C, Dimitrov S. pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopusegg extract. J Biol Chem. 2001;276:30002–30010. doi: 10.1074/jbc.M102701200. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Uren A, Vaux D, Horvitz HR. The survivin-like C. elegansBIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–106. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopuskinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]