Abstract
Purpose
The aim of this study is to investigate which ADAMTS genes play a major role in the development of primary hip osteoarthritis, by comparing the tissue and blood samples in patients with hip osteoarthritis and a control group.
Material and methods
Human articular cartilage was obtained from femoral heads of 15 patients with end stage osteoarthritis undergoing total hip replacement. As the control group, the cartilages was obtained from femoral heads of 15 patients, who did not have osteoarthritis or degenerative changes in hip joint, undergoing hip replacement following the fracture of the femoral neck. After the cartilage samples were taken from the resection materials, the DNA polymorphisms in the patients' cartilage samples were tested by Polymerase Chain Reaction (PCR), the serum levels of aggrecanase genes were analyzed with Enzyme-Linked ImmunoSorbent Assay (ELISA).
Results
The level of ADAMTS5 and ADAMTS9 genes were found significantly lower as a result of ELISA analysis degenerative arthritis group than the control group (p < 0,05). ADAMTS 1, 4, 8, 15 were similar between the two groups in ELISA analysis (p > 0,05). As a result of quantitative real time RT-PCR analysis, the level of ADAMTS8 mRNA increased 3.5 fold in hip degenerative arthritis group when compared with femoral neck fractures group. ADAMTS1, ADAMTS4 and ADAMTS5 expression levels in hip degenerative arthritis group were decreased 2.5, 2 and 2.5 fold, respectively. ADAMTS9, 15 were found to be similar between two groups.
Concluson
As a result of this study on hip osteoarthritis, the ADAMTS8 levels was found to be significantly higher in the end stage of hip osteoarthritis. Unlike similar studies on knee osteoarthritis, ADAMTS1,4,5 levels were found to be lower.
Keywords: ADAMTS genes, Osteoarthritis, Genetic, Hip, PCR, ELISA
Introduction
Osteoarthritis is a degenerative joint disease and characterized by the erosion of the articular cartilage, osteophytes at the edges of the joint, subchondral sclerosis, biochemical and morphological changes in the synovial membrane and joint capsule and its frequency increases with age. The etiology and pathophysiology of osteoarthritis are not fully understood. However, it is known that genetic, metabolic, biochemical and biomechanical factors may cause osteoarthritis via the destruction of cartilage and initiating a chain of destruction.1, 2
ADAMTS enzymes, a Disintegrin and Metalloproteinase with Thrombospondin Motifs, are responsible for cartilage matrix destruction in diseases such as osteoarthritis and rheumatoid arthritis, and they are a family of extracellular protease.3 As terminological, the protease (aggrecanase) enzymes encoded by ADAMTS genes are called ADAMTS enzymes. The ADAMTS family contains 20 members and the studies have put forward that the ADAMTS 1, 4, 5, 8, 9 and 15 genes have aggrecanase activity.4 Nevertheless, which ADAMTS member is responsible for the development of primary osteoarthritis is not clear. It is suggested that these genes have similar aggrecanase activities in the development of osteoarthritis. Aggrecans are made of proteoglycan, the main substance of cartilage consisting of a great number of chondroitin sulphate chains. With the negative electric charge, these chondroitin sulphates cause the cartilage to swell by interacting with water molecules. Therefore, the cartilage tissue gains absorption feature against mechanic stress. Aggrecans have a vital role in shock absorption. The destruction of aggrecans by aggrecanase enzymes may have a critical process seen in degenerative diseases such as osteoarthritis.4, 5 Aggrecanases are the main enzymes responsible for aggrecan destruction.6, 7
The aim of this study is to investigate which ADAMTS genes play a major role in the development of primary hip osteoarthritis, by comparing the tissue and blood samples in patients with hip osteoarthritis and a control group. The data gathered from this study will form as the basis information for the identification of the functions of gene products to be discovered in the future for the treatment of primer osteoarthritis and for the development of antiproteolytic therapies.
Material and methods
The study protocol was approved by the local ethics committees at our institutions (March 2014). 30 patients, mean age was 67 ± 7,8 (range 60–77), were included in the study (15 study and 15 control group) and all of the patients were selected from females. The study group was selected among patients who received total hip prosthesis due to hip osteoarthritis. The control group was selected among the patients who underwent bipolar hemiarthroplasty due to femoral neck fracture. All patients in the study group had end stage osteoarthritis. On the contrary, there is no evidence of osteoarthritis in the hip of the control group of patients. Osteoarthritis was diagnosed by clinical history and physical examination with radiological findings,8 and the diagnosis was confirmed by gross pathologic changes in the femoral head at the time of joint removal. The exclusion criteria of both groups in the study were sequelae of avascular necrosis, developmental hip dysplasia, Perthes disease, posttraumatic lesion, history using steroid and smoking. The patients in both groups had no complaints to suggest osteoarthritis in other joints.
Tissue samples
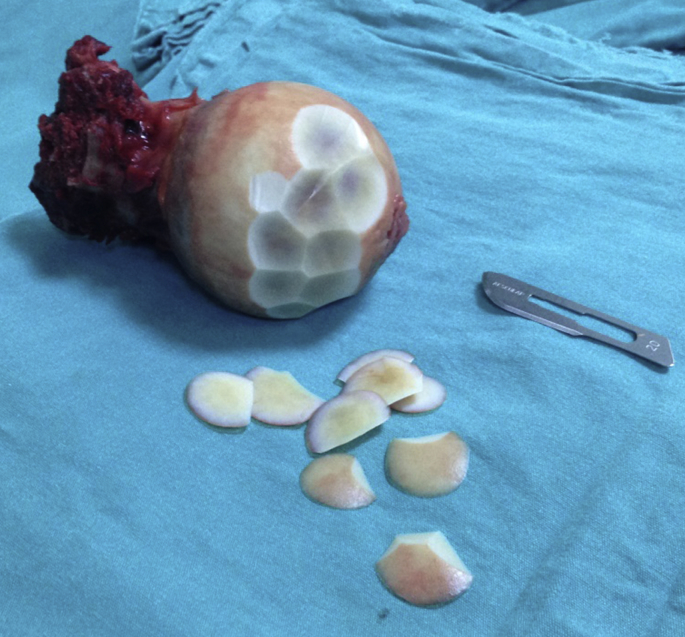
Human articular cartilage was obtained from femoral heads of 15 patients with end stage osteoarthritis undergoing total hip replacement. As the control group, the cartilages were obtained from femoral heads of 15 patients, who did not have osteoarthritis or degenerative changes in hip joint, undergoing hip replacement following the fracture of the femoral neck. After the cartilage samples were taken from the resection materials as shown in Fig. 1, they were transferred according to the rules of the cold chain and were stored at −80 °C. Then, the DNA polymorphisms in the patients' cartilage samples were tested by PCR. Besides, the serum levels of aggrecanase genes were analyzed with ELISA.
Fig. 1.
Obtaining the cartilage tissue from femoral neck fracture.
Detection of ADAMTS by ELISA
Six genes (ADAMTS 1, 4, 5, 8, 9 and 15) were analyzed and ADAMTS were measured in serum using a human ELISA kit (Bioassay Technology Laboratory; Shanghai, China) according to manufacturer's instructions. Plates with 96-slots which were pre-coated with appropriate antibodies were used. Then serum samples were added followed by horseradish peroxidase HRP-conjugated secondary mAb. Tetramethyl benzidine was subsequently added to the reaction, a colored product was formed in proportion to the amount of human ADAMTS presence in the sample or standard. The reaction was terminated by addition of acid and optical density was measured by microtiter plate reader at 450 nm. Serum levels of ADAMTS (catalog number): (ADAMTS1, E1155Hu; ADAMTS4, E1977Hu; ADAMTS8, E3481Hu; ADAMTS9, E3558Hu; ADAMTS15, E3486Hu) were read off from a standard curve according to the manufacturer's instruction. The intra- and inter-assay coefficients of the variation of all kits were <8% and <10%, respectively. Assay range is 1 ng/ml to 300 ng/ml.
Total RNA isolation
Total RNA was extracted with TRIzol (Ambion/RNA by Life Technologies, Carlsbad, CA, USA) according to the previously reported technique.9 RNA (1 microgram) was used for reverse transcription using with Reverse Transcriptase (Thermo Scientific, Waltham, MA, USA) with random hexamers (Thermo Scientific) with random primers according to the manufacturer's instruction (Table 1). The human housekeeping genes, beta actin (β-actin) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were used as a control for the Polymerase Chain Reaction (PCR). To prevent genomic DNA contamination, reverse transcriptase-deficient samples were amplified as controls. Elution of the total RNA from the samples was accomplished by using RNase-free water. To determine RNA concentration and to check the purity of samples, UV spectrophotometry was utilized. For that purpose, the purity was decided from the samples that required 260/280 ratio of 2.0 and 260/230 ratio of 1.7.
Table 1.
The forward and reverse primers used in the real-time polymerase chain reaction analyses for ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS8, ADAMTS9, ADAMTS15, Beta actin and GAPDH. (ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; β-actin, beta actin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase).
| β-actin | Forward | 5′- TTCCTGGGCATGGAGTCCT -3′ |
| Reverse | 5′- AGGAGGAGCAATGATCTTGATC -3′ | |
| GAPDH | Forward | 5′- CCTGCACCACCAACT -3′ |
| Reverse | 5′- TCTTCTGGGTGGCAGTGATG -3′ | |
| ADAMTS1 | Forward | 5′- GGACAGGTGCAAGCTCATCTG -3′ |
| Reverse | 5′- TCTACAACCTTGGGCTGCAAA -3′ | |
| ADAMTS4 | Forward | 5′- GGGATAGTGACCACATTGTT -3′ |
| Reverse | 5′- AGGCACTGGGCTACTACTAT -3′ | |
| ADAMTS5 | Forward | 5′- TATGACAAGTGCGGAGTATG -3′ |
| Reverse | 5′- TTCAGGGCTAAATAGGCAGT -3′ | |
| ADAMTS8 | Forward | 5′- GTTCCCATCGTTCTGCACAC -3′ |
| Reverse | 5′- ACCATGTGGTGGACTCGCCT -3′ | |
| ADAMTS9 | Forward | 5′- TTCCACAGGTCACTGAGCA -3′ |
| Reverse | 5′- GTTTGCAACCCTGCGAGTAT -3′ | |
| ADAMTS15 | Forward | 5′- GGTACTTGCCTTGGCTGTTC -3′ |
| Reverse | 5′- GTGGGGGAGACAATAAGAGC -3′ |
Real-time PCR
Real-time PCR was performed on cDNA samples obtained (Thermo Scientific Revert Aid First Strand cDNA Synthesis Kit) as described in the previous report.9 The PCR mixture consisted of SYBR Green PCR Master Mix, which includes DNA polymerase, SYBR Green I Dye, dNTPs (deoxynucleotide triphosphates), PCR buffer, forward and reverse primers (10 pmol each) and cDNA of samples in a total volume of 20 μl/mL of PCR mixture. The amplified β-Actin and GAPDH were used for normalizing the efficiency of cDNA synthesis and the load of RNA applied. PCR conditions were as follows: initial denaturation at 95 °C for 5 minutes, followed by amplification for 40 cycles, each cycle consisting of denaturation at 95 °C for 10 seconds, annealing at 58 °C for 30 seconds, polymerization at 72 °C for 1 minute, and the last stage, polymerization at 72 °C for 5 minutes. The findings related to ADAMTS1, 4, 5, 8, 9 and 15 were presented as graphics, in which the bars represent means of measurements and error bars represent standard error of means (SEM).
Results
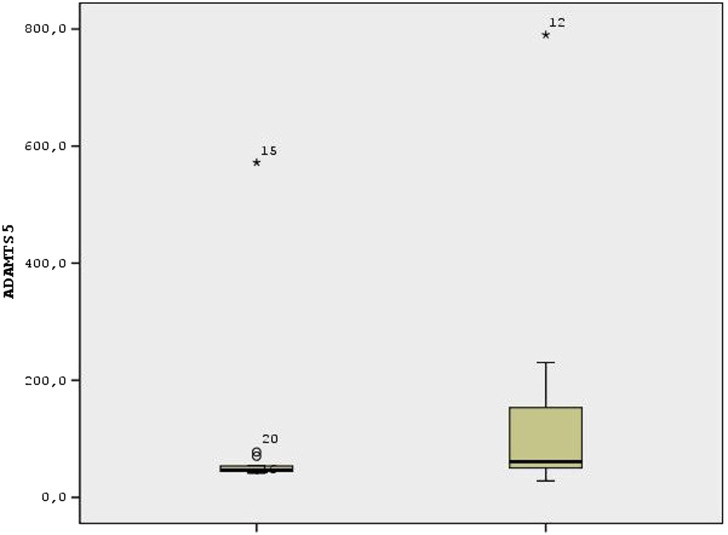
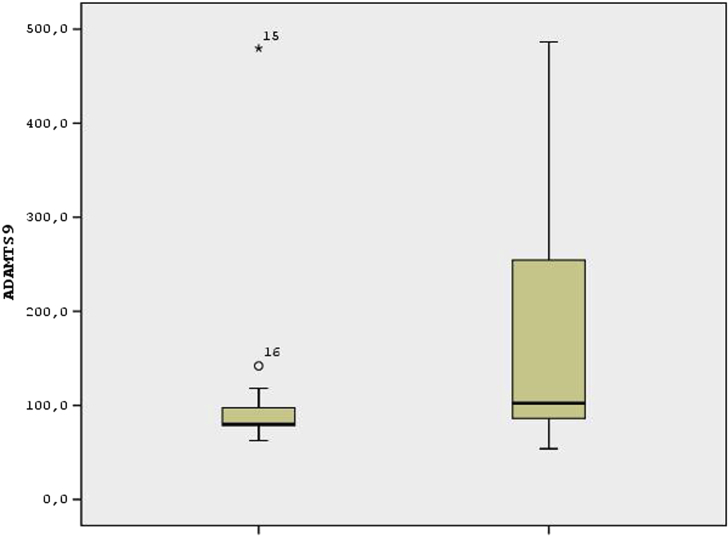
The level of ADAMTS5 and ADAMTS9 genes were found significantly lower as a result of ELISA analysis in the last stage of the hip degenerative arthritis group than the control group (p < 0,05), as shown in Fig. 2, Fig. 3. ADAMTS 1, 4, 8, 15 were similar between the two groups (p > 0,05).
Fig. 2.
According to the ADAMTS5 variable, the data of the experimental group were collected at low values and more homogeneous according to the data of the control group. There are three outliers in the experimental group and one outlier in the control group.
Fig. 3.
According to the ADAMTS9 variable, the data of the experimental group were collected at low values and more homogeneous according to the data of the control group. There are two outliers in the experimental group.
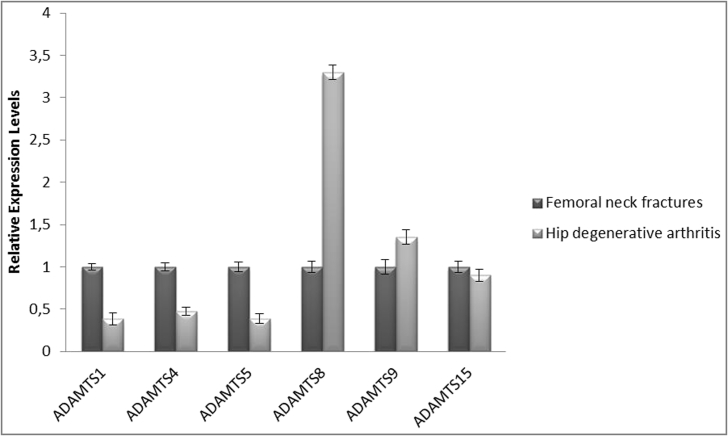
Expression levels for ADAMTS 1, 4, 5, 8, 9 and 15 RNA were analyzed in the cartilage tissue of the subjects and the Results were represented as graphics. The error bars represent the standard deviation of means. As shown in Fig. 4, the ADAMTS1, ADAMTS4 and ADAMTS5 mRNA expression significantly decreased in hip degenerative arthritis group, as a result of quantitative real time RT-PCR analysis (p < 0,05). ADAMTS1, ADAMTS4 and ADAMTS5 expression levels in hip degenerative arthritis group were decreased 2.5, 2 and 2.5 fold when compared with femoral neck fractures group, respectively. The level of ADAMTS8 mRNA increased 3.5 fold in hip degenerative arthritis group when compared with femoral neck fractures group. In addition, there was no significant difference between the femoral neck fractures group and the hip degenerative arthritis group in terms of ADAMTS9 and ADAMTS15 mRNA expression (Fig. 4).
Fig. 4.
Graphical representation of mRNA expression levels in RT-PCR analysis.
Discussion
The etiology of osteoarthritis is still not fully understood. However, the mechanical, biochemical and enzymatic factors are involved in pathogenesis. Determining the roles of these factors in the pathogenesis of osteoarthritis is important for the prevention of the disease and the development of other treatment modalities.10 The result of all these factors is the loss of the balance between the construction-destruction of the cartilage,11 and ADAMTS genes are considered as one of the contributing factors.12 Nevertheless, which ADAMTS member is responsible for the development of primary osteoarthritis is not clear. There are many studies in the literature regarding knee osteoarthritis,11, 13 yet there is only one publication about the relationship between hip osteoarthritis and ADAMTS genes.6 According to the literature, this study is the second one regarding hip osteoarthritis to our knowledge. This research was carried out on ADAMTS 1, 4, 5, 8, 9 and 15 genes which are thought to be the cause of cartilage degradation.13, 14, 15
Malfait et al demonstrated in an animal study conducted in 2002 that the enzymes ADAMTS4 and ADAMTS5 have aggrecanase enzyme activity by showing the mRNA and protein expression levels of these genes in the cartilage tissue and the aggrecan degradation products in the joint fluid.16 Moreover, it has been demonstrated in ex vivo studies done on animal models that a decrease in the expression levels of the ADAMTS-4 and ADAMTS-5 genes protects against proteoglycan degradation and reduces the severity of osteoarthritis in animals. In an animal study conducted by Glasson et al in 2005, the catalytic domain of the ADAMTS5 (aggrecanase-2) of mice was firstly suppressed and knee instability was created surgically. Thus, the severity of cartilage degeneration was decreased significantly, compared to the control group.4
Human studies have been undertaken based on the information obtained from animal studies, particularly on the roles of the ADAMTS4 and ADAMTS5 genes in the development of osteoarthritis. In a human study conducted in 2007, the ADAMTS1, -4 and -5 genes in cartilage samples with and without knee osteoarthritis were analyzed using the real-time PCR and Western Blot methods. It was demonstrated that ADAMTS-4 and ADAMTS-5 play a role in the cartilage degeneration in human osteoarthritis and that ADAMTS1 does not.17 All of these studies show us that both ADAMTS-4 and ADAMTS-5 are a significant mediator of aggrecan loss in cytokine-stimulated normal and osteoarthritic knee cartilage in which the degeneration continues. However, it is not entirely clear at present, which aggrecanase plays the main role in cartilage degeneration.11
There are studies comparing osteoarthritic cartilages obtained from the patients who underwent total knee prosthesis implantation due to knee osteoarthritis and normal cartilages obtained from autopsies using RT-PCR analysis,18, 19 and that ADAMTS5 is the strongest aggrecanase was emphasized in these studies. Naito et al also compared osteoarthritic cartilages which were obtained from the knee joint with normal cartilages which were obtained from the hip joint using RT-PCR analysis and emphasized that the main factor causing osteoarthritic cartilage is ADAMTS4.20 In another study conducted by rheumatologists on the knee joint, 144 patients were divided into three groups as mild-moderate-severe and the ADAMTS-4 and -5 levels and their relationships with degradation products were analyzed in the synovial fluid samples using the ELISA and Western Blot methods. They stated that the level of ADAMTS-4 was found high particularly in the early stage of the disease and that it is more correlated with the degradation products compared to ADAMTS-5.21 The Results of these studies differ from the findings of this research. The reason for these contradictory results may be that the cartilage samples were taken from the knee joint, that they were taken at different stages of osteoarthritis or that exclusion criteria for primary osteoarthritis were not taken into consideration in previous studies.s a result of RT-PCR tests, it was observed that while the mRNA expression level of ADAMTS8 statistically increased compared to the control group, the mRNA expression levels of ADAMTS1, 4 and 5 decreased. There are studies in the literature that support the Results of this research. ADAMTS8 mRNA has been shown to be expressed in osteoarthritic human cartilages.13 In a study conducted by Demircan et al on knee osteoarthritis, patients with early and late stage of osteoarthritis were compared with a group with no osteoarthritis. ADAMTS8 was found to be significantly high in the group with early and late stage of osteoarthritis. In the same study, ADAMTS4 and 5 were found to be higher in the group with early stage osteoarthritis compared to the control group, while no significant difference was found in the late stage.22 In a similar manner to the present study, Kevorkian et al conducted RT-PCR analysis on the cartilage samples that they obtained from the patients who developed degenerative arthritis and the patients with femoral neck fracture.6 They demonstrated that the most significant decrease in the expression levels in osteoarthritic cartilages was in ADAMTS-1 (p < 0.001) and that the expression levels of ADAMTS5 (p < 0.05) and ADAMTS9 (p < 0.01) genes had decreased compared to normal cartilages, which is similar to our study. In addition to Kevorkian et al's study, the reflection of these genes on protein was analyzed with ELISA in this research. As a result of PCR and ELISA, only the finding of ADAMTS5 supported each other. This result is attributed to the fact that ADAMTS mRNA expressions did not reflect on the protein level to because it underwent post-translational modification. Since ELISA results depend on threshold values and ready-to-use kits, PCR results may provide more reliable information. Therefore, PCR analyses may have a superior asset to this study.
Most of the human and animal studies that were based on knee osteoarthritis investigated the relationship of ADAMTS genes and osteoarthritis and emphasized that ADAMTS 4 and 5 in particular play a role in the development of knee osteoarthritis. In this study, the expression level of ADAMTS5 was found to have statistically decreased in the osteoarthritic group, while no difference was found for ADAMTS4 between the groups. In another study on hip osteoarthritis, the mRNA expression levels of ADAMTS1 and 5 were found to have decreased in the hip osteoarthritic group.6 Aggrecanase activities have been stated to show difference at different stages of the disease,21 and aggrecan loss is greater in the early stage of osteoarthritis. It is thought that the reason why the expression levels of the genes are low in this study may be because it was conducted on the final stage of osteoarthritic cartilages.
In addition, studies revealed that there is an aggrecanase activity in the synovium as well, and that there may be a transmission from cartilage tissue to the synovium. It is thought that these transmissions may cause the decrease in the expression levels of ADAMTS4 and 5 genes.21, 23 Nevertheless, ADAMTS8 was found to be as high as significant and it may be more important in the end stage of hip osteoarthritis.
As a conclusion, ADAMTS genes may have an important role in the end stage of hip osteoarthritis. According to this study, ADAMTS8 levels increased and ADAMTS1,4,5 levels decreased in the end stage of hip osteoarthritis. Besides this, ADAMTS9 and ADAMTS15 were found to be similar between two groups. Further studies on early and mid-stage osteoarthritic cartilages will provide more detailed and reliable information about primary hip osteoarthritis.
The data gathered from this study will form as the basis information for the identification of the functions of gene products. It is important for early diagnosis, monitoring prognosis and development of treatment options. In previous similar studies have shown that the ADAMTS 4 and 5 genes were primarily responsible for knee osteoarthritis. On the basis of these studies, therapeutic human researches have been made and ADAMTS-5-selective monoclonal antibody have been developed.24 As a result of this study on hip osteoarthritis, the ADAMTS8 gene was found to be significantly higher. Likewise, the result of this study will be the basis for therapeutic studies on hip osteoarthritis.
Acknowledgments
This study was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) 3001 Program. (Project number: 114Z544).
Footnotes
Peer review under responsibility of Turkish Association of Orthopaedics and Traumatology.
References
- 1.Steinberg J., Zeggini E. Functional genomics in osteoarthritis: past, present, and future. J Orthop Res. 2016;34:1105–1110. doi: 10.1002/jor.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atik O.S., Erdoğan D., Seymen C.M. Is there crosstalk between subc hondral bone, cartilage, and meniscus in the pathogenesis of osteoarthritis? Eklem Hastalık Cerrahisi. 2016;27:62–67. doi: 10.5606/ehc.2016.14. [DOI] [PubMed] [Google Scholar]
- 3.Apte S.S. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284:31493–314977. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glasson S.S., Askew R., Sheppard B. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 5.Stanton H., Rogerson F.M., East C.J. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 6.Rogerson F.M., Chung Y.M., Deutscher M.E. Cytokine-induced increases in ADAMTS-4 mRNA expression do not lead to increased aggrecanase activity in ADAMTS-5-deficient mice. Arthritis Rheum. 2010;62:3365–3373. doi: 10.1002/art.27661. [DOI] [PubMed] [Google Scholar]
- 7.Fosang A.J., Rogerson F.M. Identifying the human aggrecanase. Osteoarthritis Cartilage. 2010;18:1109–1116. doi: 10.1016/j.joca.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Kellgren J.H., Lawrence J.S. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kevorkian L., Young D.A., Darrah C. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50:131–141. doi: 10.1002/art.11433. [DOI] [PubMed] [Google Scholar]
- 10.Atik O.S. The role of metabolomics in osteoarthritis for early diagnosis, monitoring prognosis and treatment. Eklem Hastalık Cerrahisi. 2015;26:1. doi: 10.5606/ehc.2015.01. [DOI] [PubMed] [Google Scholar]
- 11.Verma P., Dalal K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J Cell Biochem. 2011;112:3507–3514. doi: 10.1002/jcb.23298. [DOI] [PubMed] [Google Scholar]
- 12.van den Berg W.B. Osteoarthritis year 2010 in review: pathomechanisms. Osteoarthritis Cartilage. 2011;19:338–341. doi: 10.1016/j.joca.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Collins-Racie L.A. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2014;23:219–230. doi: 10.1016/j.matbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Somerville R.P.T. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct. ADAMTS subfamily related to Caenorhabditis elegans GON-1. JBiol Chem. 2003;278:9503–9513. doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- 15.Rodrıquez-Manzaneque J.C. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem Biophys Res Commun. 2002;293:501–508. doi: 10.1016/S0006-291X(02)00254-1. [DOI] [PubMed] [Google Scholar]
- 16.Malfait A.M., Liu R.Q., Ijiri K. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem. 2002;277:22201–22208. doi: 10.1074/jbc.M200431200. [DOI] [PubMed] [Google Scholar]
- 17.Ilic M.Z., East C.J., Rogerson F.M. Distinguishing aggrecan loss from aggrecan proteolysis in ADAMTS-4 and ADAMTS-5 single and double deficient mice. J Biol Chem. 2007;282:37420–37428. doi: 10.1074/jbc.M703184200. [DOI] [PubMed] [Google Scholar]
- 18.Bau B., Gebhard P.M., Haag J. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 2002;46:2648–2657. doi: 10.1002/art.10531. [DOI] [PubMed] [Google Scholar]
- 19.Song R.H., Tortorella M.D., Malfait A.M. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 2007;56:575–585. doi: 10.1002/art.22334. [DOI] [PubMed] [Google Scholar]
- 20.Naito S., Shiomi T., Okada A. Expression of ADAMTS4 (aggrecanase-1) in human osteoarthritic cartilage. Pathol Int. 2007;57:703–711. doi: 10.1111/j.1440-1827.2007.02167.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang E., Yan X., Zhang M. Aggrecanases in the human synovial fluid at different stages of osteoarthritis. Clin Rheumatol. 2010;32:797–803. doi: 10.1007/s10067-013-2171-0. [DOI] [PubMed] [Google Scholar]
- 22.Demircan K., Gürlü G., Güven K.N. The potential link of COMP-ADAMTS and aggrecanase-ADAMTS genes with osteoarthrit. FASEB J. 2016;30(Supp):645–651. [Google Scholar]
- 23.Vankemmelbeke M.N., Holen I., Wilson A.G. Expression and activity of ADAMTS-5 in synovium. Eur J Biochem. 2001;268:1259–1268. doi: 10.1046/j.1432-1327.2001.01990.x. [DOI] [PubMed] [Google Scholar]
- 24.Larkin J., Lohr T.A., Elefante L. Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification. Osteoarthritis Cartilage. 2015 Aug;23(8):1254–1266. doi: 10.1016/j.joca.2015.02.778. [DOI] [PMC free article] [PubMed] [Google Scholar]