Abstract
Glypicans are a family of cell-surface heparan sulfate proteoglycans that regulate growth-factor signaling during development and are thought to play a role in the regulation of morphogenesis. Whole-exome sequencing of the Australian family that defined Keipert syndrome (nasodigitoacoustic syndrome) identified a hemizygous truncating variant in the gene encoding glypican 4 (GPC4). This variant, located in the final exon of GPC4, results in premature termination of the protein 51 amino acid residues prior to the stop codon, and in concomitant loss of functionally important N-linked glycosylation (Asn514) and glycosylphosphatidylinositol (GPI) anchor (Ser529) sites. We subsequently identified seven affected males from five additional kindreds with novel and predicted pathogenic variants in GPC4. Segregation analysis and X-inactivation studies in carrier females provided supportive evidence that the GPC4 variants caused the condition. Furthermore, functional studies of recombinant protein suggested that the truncated proteins p.Gln506∗ and p.Glu496∗ were less stable than the wild type. Clinical features of Keipert syndrome included a prominent forehead, a flat midface, hypertelorism, a broad nose, downturned corners of mouth, and digital abnormalities, whereas cognitive impairment and deafness were variable features. Studies of Gpc4 knockout mice showed evidence of the two primary features of Keipert syndrome: craniofacial abnormalities and digital abnormalities. Phylogenetic analysis demonstrated that GPC4 is most closely related to GPC6, which is associated with a bone dysplasia that has a phenotypic overlap with Keipert syndrome. Overall, we have shown that pathogenic variants in GPC4 cause a loss of function that results in Keipert syndrome, making GPC4 the third human glypican to be linked to a genetic syndrome.
Keywords: Keipert syndrome, glypicans, GPC4, Nasodigitoacoustic syndrome
Main Text
Keipert syndrome, also known as nasodigitoacoustic syndrome (MIM: 255980), is a rare, X-linked disorder characterized by craniofacial and digital abnormalities and variable learning difficulties and sensorineural deafness.1 Keipert et al.2 first described the syndrome in two brothers, and it has subsequently been reported in one other male sibling pair,3 five isolated male individuals,4, 5, 6, 7 and a girl and her less severely affected father.8 The facial appearance is distinctive and comprises a broad forehead, hypertelorism, a prominent nose, a wide mouth, and a prominent upper lip with a cupid’s bow configuration. Changes of the digits are also distinctive: there is widening of all distal phalanges, particularly those of the thumbs and great toes.
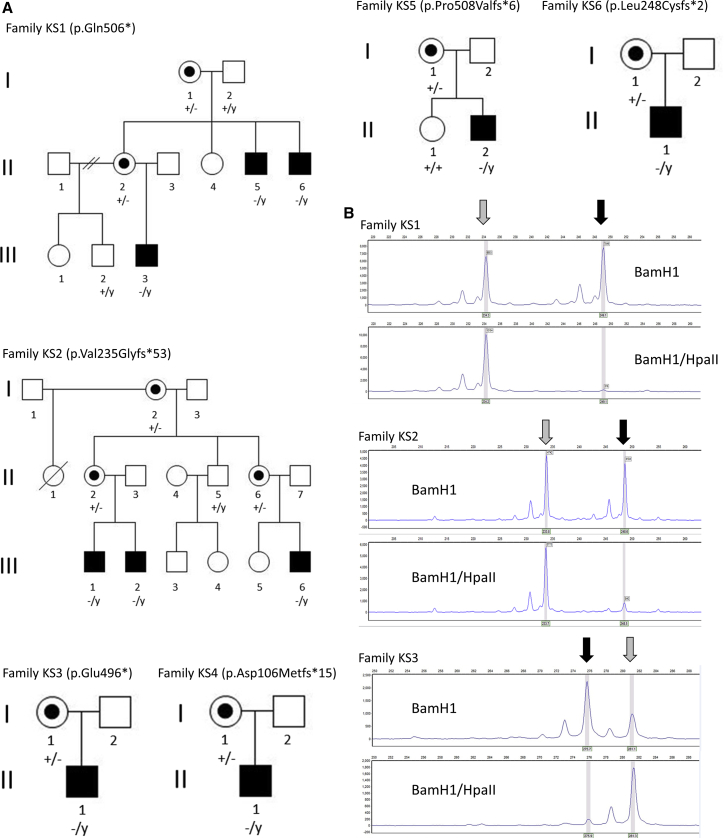
Previously, we described the diagnosis of Keipert syndrome in the maternal nephew of the brothers originally reported by Keipert et al.2 and mapped Keipert syndrome to Xq22.2-Xq28 in this family (KS1).1 To identify the underlying genetic cause of Keipert syndrome, we performed whole-exome capture and massively parallel sequencing of gDNA isolated from the maternal nephew (Figure 1A, KS1 II:5). Institutional ethics approval was provided by the Royal Children’s Hospital (Melbourne), and written informed consent was received from all participants prior to study. Three variants were identified within the linkage region after exclusion of synonymous variants and filtering against population databases (Table S1). The nonsynonymous variant in the gene encoding the arginine vasopressin receptor-2 (AVPR2, c.743G>A [p. Arg248His], GenBank: NM_000054.5) was excluded because it was classified as a variant of uncertain significance (VUS) according to ACMG guidelines, and pathogenic variants in this gene cause diabetes insipidus (MIM: 300538). A truncating variant in the gene encoding melanoma-associated antigen (mutated) 1-like 1 was identified (MUM1L1, c.1404G>A [p.Trp468∗], GenBank: NM_001171020.2). This variant was classified as a VUS and was not considered to be a strong candidate for Keipert syndrome. Moreover, analysis of the Genotype-Tissue Expression (GTEx) database demonstrated negligible expression of MUM1L1 in all tissues except ovary, and dysregulation is associated with ovarian cancers.9 Analysis of the gene in gnomAD revealed moderate evidence of gene intolerance to loss of function10 (pLI = 0.49; observed/expected = 0.2, CI 0.09–0.53). The third variant, which was identified as most likely to be disease causing, was a hemizygous C-to-T transition variant in the ninth and final exon of glypican 4; this variant resulted in premature protein termination (GPC4, c.1516C>T [p.Gln506∗], GenBank: NM_001448.2). GPC4 is classified as extremely intolerant of loss-of-function variants (pLI = 0.94; observed/expected = 0.11, CI 0.04–0.36) and is expressed during development in a variety of tissue types relevant to Keipert syndrome. Notably, GPC4 is a member of the glypican protein family, whose members are cell-surface heparan sulfate proteoglycans that regulate growth-factor signaling during development.11, 12 Sanger sequencing confirmed the variant and segregation within the KS1 pedigree (Figure 1A).
Figure 1.
Pedigree Structure and Segregation of Pathogenic Variants in GPC4
(A) Pedigrees showing the clinical phenotypes and segregation of the WT (+) and mutant (−) GPC4 allele. Affected individuals are shaded, Y indicates the Y chromosome, and carrier females are indicated by a dot.
(B) Representative images of X-inactivation analysis in one carrier female from families KS1 (I:1), KS2 (I:2), and KS3 (I:1) demonstrate extreme skewing of X inactivation, as evidenced by the inability of the methylation-sensitive restriction endonuclease HpaII to digest one allele of the androgen receptor (gray arrow). The second allele, with minimal methylation (black arrow), is almost fully digested by HpaII.
Further interrogation of the gnomAD database identified, from within the ∼182,000 sequenced alleles, two hemizygous alleles predicted to be loss of function with high-confidence (LoF allele frequency = 1.1 × 10−5; c.283C>T, [p.Gln95∗], and c.1150C>T, [p.Arg384∗]), even though the database is considered to be relatively free of sequences from individuals with pediatric disorders. We did not believe this would exclude GPC4 as the gene underlying the disorder in family KS1 because the phenotype within the family is quite variable. For example, KS1 II-5 has subtle phenotypic features and minimal impairment from his condition; he has a job and has fathered a child. We consider it likely that the phenotype of Keipert syndrome in some cases might escape clinical recognition and therefore be included in gnomAD. This situation has been observed for other craniofacial syndromes with phenotypic heterogeneity; for example, gnomAD contains two individuals who have the pathogenic p.Pro250Arg FGFR3 variant that causes Muenke syndrome but presents with variable phenotypes.13
We next searched for additional individuals with pathogenic variants in GPC4 to provide genetic validation of family KS1. Five affected males from three additional kindreds with novel variants in GPC4 were recruited to this study through GeneMatcher (Figure 1A).14 In family KS2, a frameshift variant was identified in exon 3 in two brothers and a male cousin (c.701dup [p.Val235Glyfs∗53]). In family KS3, a truncating variant was identified in exon 9 in one affected male (c.1486G>T [p.Glu496∗]), and in family KS4, a frameshift variant was identified in exon 2 (c.316delG [p.Asp106Metfs∗15]) in one affected male. Notably, no potentially pathogenic variants were identified in either AVPR2 or MUM1L1 in these cases. In addition, two additional families were identified during the course of the study. In family KS5, a frameshift variant (c.1518_1521dupGTGC [p.Pro508Valfs∗6]) was identified in exon 9 in one affected male, and in family KS6, a frameshift variant (c.742delC [p.Leu248Cysfs∗2]) was identified in exon 4 in one affected male. Genotype analysis in all available samples from the six families confirmed segregation of the variants with disease and demonstrated inheritance from a carrier mother (Figure 1A). In the two multigenerational families (KS1 and KS2), this analysis also confirmed that the variant was present in a carrier grandmother. Transmission of the variant was observed through nine segregating meioses in total, providing additional genetic support for the linkage and gene-identification findings. Carrier females were clinically unaffected, although the mother of KS4 I:1 was noted to have possible subtle dysmorphic features such as hypertelorism and a flat nasal bridge. To assess the X chromosome inactivation profile of carrier females, we performed methylation-specific analysis of the (CAG)n repeat of the androgen receptor gene, essentially as described previously.15 In this assay, differential methylation of the X chromosomes is quantitated by methylation-sensitive digestion of genomic DNA. Allelic ratios from 50%–79% were considered to reflect a normal X-inactivation pattern, from 80%–90% were considered to reflect moderate skewing, and >90% were considered to reflect strong skewing. In total, we analyzed six individuals from four families (KS1 I:1 and II:2; KS2 I:2 and II:6; KS3 I:1; and KS4 I:1). Although analysis of the (CAG)n repeat was not informative in family KS4, X-inactivation studies in all the carrier females from KS1, KS2, and KS3 showed extreme skewing of >90% methylation of one allele (representative results are shown in Figure 1B). Collectively, these observations provide significant support for X linkage in the families and strongly suggest that variants in GPC4 underlie Keipert syndrome.
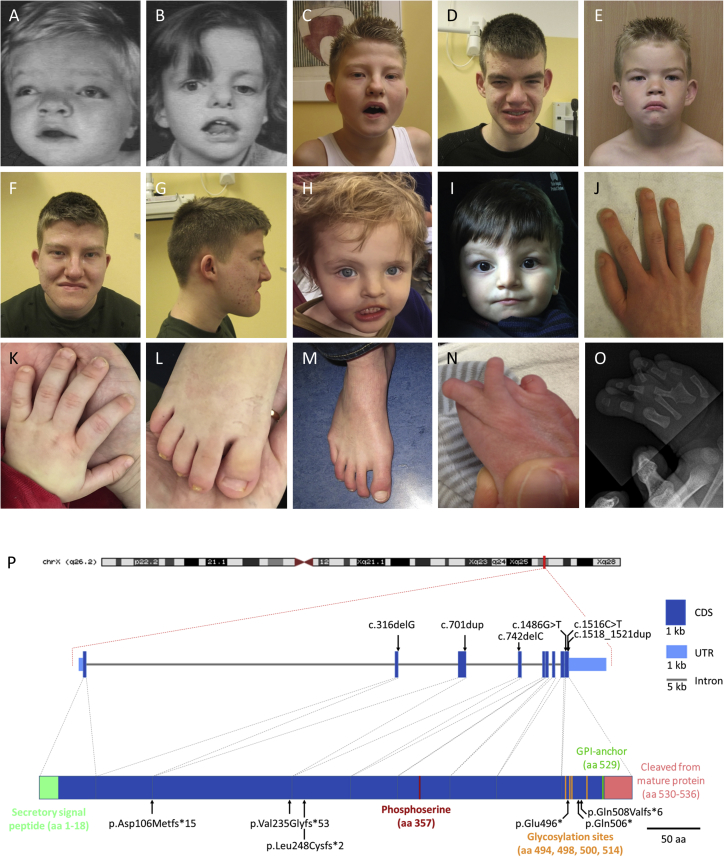
Interestingly, with the exception of the first family (KS1) described with Keipert syndrome, all GPC4 variants were detected via a “genotype-first” approach, yet these individuals share very similar phenotypes with the original kindred (Figures 2A–2O and Table 1). Clinical features present in seven or more of the ten affected individuals were macrocephaly, a prominent forehead, a flat midface, hypertelorism, a broad nose, downturned corners of mouth, and digital abnormalities. The most prominent peripheral skeletal features were brachydactyly, clinodactyly and/or camptodactyly, broad terminal phalanges, and broad thumbs and great toes. Cognitive impairment was variable; intellectual ability ranged from average to moderate intellectual disability, and autism or autistic features were observed in three affected individuals. Identification of these newly reported cases enabled us to extend and refine the phenotype of Keipert syndrome. In particular, the spectrum of digital abnormalities was broadened: absent toenails were observed in individuals KS2 III:2 and KS3 II:1. The latter individual also had synostosis between metatarsals III and IV in the right foot and a missing metatarsal III with hypoplastic phalanx in the left foot. Although sensorineural deafness was described initially as a cardinal feature of Keipert syndrome,1, 2 we did not observe deafness in any of the other families with confirmed GPC4 pathogenic variants, suggesting that deafness might be relatively infrequent in Keipert syndrome. To further test this, we obtained genomic DNA from one female and three male individuals previously described as having a clinical diagnosis of Keipert syndrome5, 7, 8 Three of the four affected individuals were reported to have moderate sensorineural hearing loss. However, GPC4 variants were not detected in any, suggesting that these individuals have an overlapping but distinct disorder.
Figure 2.
Clinical Presentation of Subjects with Keipert Syndrome and Mutations in GPC4
(A–I) Facial features of individuals, demonstrating hypertelorism, a broad forehead, a broad nose, a flat midface, prominent lips, and downturned corners of mouth. Illustrated are (A) KS1 II:5 at age 15 months, (B) KS1 II:6 at age 5 years, (C and D) KS2 III:1 at age 12 years and 19 years, (E, F, and G) KS2 III:2 at ages 7 years and 17 years, (H) KS4 II:1 at age 3 years, and (I) KS5 II:2 at age 2 years.
(J) Right hand of KS2 III:1, showing brachydactyly and broad terminal phalanges.
(K–L) Left hand and right foot of KS5 II:2, showing brachydactyly and a broad great toe.
(M) Right foot of KS2 III:2, showing clinodactyly and broad terminal phalanges.
(N–O) Right foot of KS3 II:1, showing synostosis between metatarsals III and IV.
(P) Schematic representation of the location of pathogenic variants identified in GPC4 and the exon and protein domain structure. The signal peptide sequence (green), phosphoserine (red), glycosylation (orange), lipidation (dark blue), and cleaved GPI anchor (pink) are indicated. The gene structure was derived from the UCSC Genome Bioinformatics database, and protein coordinates were obtained from Uniprot.
Informed consent for publication of photos was obtained from all individuals or their parents.
Table 1.
Clinical Features and GPC4 Variants of Affected Individuals
| Family | KS1 | KS1 | KS1 | KS2 | KS2 | KS2 | KS3 | KS4 | KS5 | KS6 | Summary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pedigree ID | II:5 | II:6 | III:3 | III:1 | III:2 | III:6 | II:1 | II:1 | II:2 | II:1 | |
| Gene variant | c.1516C>T (p.Gln506∗) | c.1516C>T (p.Gln506∗) | c.1516C>T (p.Gln506∗) | c.701dup (p.Val235Glyfs∗53) | c.701dup (p.Val235Glyfs∗53) | c.701dup (p.Val235Glyfs∗53) | c.1486G>T (p.Glu496∗) | c.316delG, p.(Asp106Metfs∗15) | c.1518_1521dupGTGC (p.Pro508Valfs∗6) | c.742delC (p.Leu248Cysfs∗2) | |
| Age at last examination | 40 years | 38 years | 7 months | 19 years | 12 years | 6 years | 3 years | 6 years | 2 years | 3 years | |
| Gender | male | male | male | male | male | male | male | male | male | male | |
| Facial Features | |||||||||||
| Macrocephaly | no | yes | yes | no | no | yes | borderline | yes | yes | yes | 7/10 |
| Prominent forehead | yes | yes | yes | no | yes | unknown | yes | yes | no | yes | 7/9 |
| Flat midface | yes | yes | yes | yes | yes | unknown | yes | yes | no | no | 7/9 |
| Hypertelorism | yes | yes | yes | yes | yes | unknown | yes | yes | yes | yes | 9/9 |
| Broad nose | yes | yes | yes | yes | yes | unknown | yes | yes | yes | yes | 9/9 |
| Downturned corners of mouth | yes | yes | yes | yes | yes | unknown | yes | no | yes | no | 7/9 |
| Prominent lip | yes | yes | yes | yes | yes | unknown | yes | no | no | no | 6/9 |
| Ears simple or low set | yes | no | no | yes | no | unknown | no | yes | yes | yes | 5/9 |
| Skeletal | |||||||||||
| Brachydactyly | yes | yes | no | yes | no | unknown | yes | no | yes | no | 5/9 |
| Clinodactyly | yes | yes | yes | no | yes | unknown | yes | no | no | no | 5/9 |
| Camptodactyly | yes | yes | yes | no | no | unknown | no | no | no | no | 3/9 |
| Broad thumb | yes | yes | yes | no | no | unknown | no | no | yes | yes | 5/9 |
| Broad first toe | yes | yes | yes | yes | no | unknown | yes | no | yes | yes | 7/9 |
| Broad terminal phalanges | yes | yes | yes | yes | yes | yes | yes | no | no | no | 7/10 |
| Other Features | |||||||||||
| Sensorineural hearing loss | moderate unilateral | severe bilateral | moderate bilateral | no | no | unknown | no | no | no | no | 3/9 |
| Cognitive impairment | no | mild intellectual disability | learning difficulties | moderate intellectual disability (IQ 52) | borderline intellectual disability (IQ 76) | no (IQ 92) | borderline intellectual disability (IQ 70) | intellectual disability | probable | yes | 8/10 |
| Additional features | – | unilateral ptosis | unilateral absent kidney | hyperlaxity of joints | hyperlaxity of joints | autism | synostosis of metatarsal bones with hypoplastic phalanx | behavioral difficulties and autistic traits | neonatal cholestasis (self-resolving) | autistic features | – |
| – | strabismus | – | amblyopia | delayed eruption of permanent dentition | – | – | – | neonatal hypotonia | stereotypic movements | – | |
| – | double alveolar margin | – | patent ductus arteriosus | strabismus | – | – | – | speech delay | – | – | |
| – | – | – | hypodontia | absent fifth toenails | – | – | – | gastresophageal reflux | – | – | |
| – | – | – | cryptorchidism (bilateral) | finger-like thumbs | – | – | – | – | – | – | |
| – | – | – | finger-like thumbs | – | – | – | – | – | – | – | |
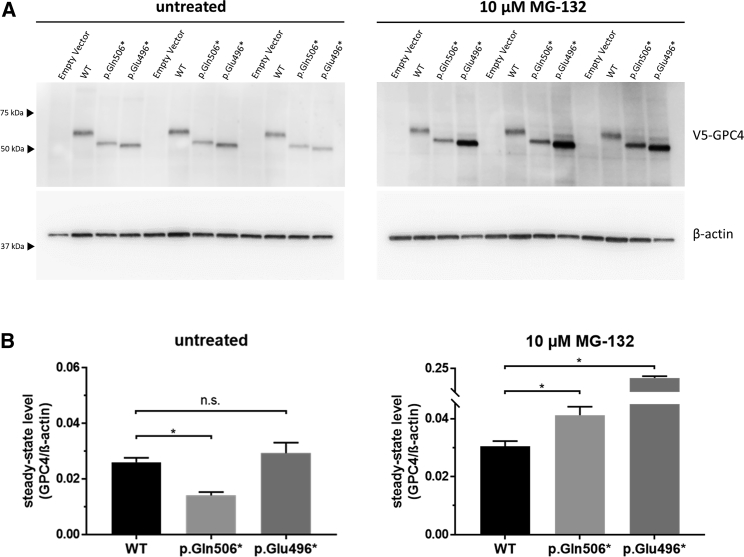
All identified variants were truncating or resulted in a frameshift, suggesting loss of function as the likely disease mechanism. The variants in exon 2 (c.316delG [p.Asp106Metfs∗15]), exon 3 (c.701dup [p.Val235Glyfs∗53]), and exon 4 (c.742delC [p.Leu248Cysfs∗2]) result in loss of >50% of the 556 amino acid protein and were classified as pathogenic. However, the truncating variants in families KS1, KS3, and KS6 (c.1516C>T [p.Gln506∗], c.1486G>T [p.Glu496∗]), and c.1518_1521dupGTGC [p.Pro508Valfs∗6]) are within the last exon and therefore might not be subject to nonsense-mediated decay. Although the resultant proteins lack glycosylation sites and the glycosylphosphatidylinositol (GPI) anchor (Ser529) that are critical for sorting and localization of GPC proteins (Figure 2P), it is possible that the variants do not result in a loss of function. To further test the pathogenic mechanism underlying p.Glu496∗ and p.Gln506∗, we analyzed recombinant N-terminal V5-tagged GPC4 in cultured cells. We have previously utilized similar methodology to investigate the function of glypicans in the formation of active synapses.16 Expression constructs encoding V5-tagged wild-type (WT) or truncated (p.Glu496∗ and p.Gln506∗) recombinant human GPC4 were transfected into HEK293 cells, and stably transfected populations were isolated after 3 weeks of culturing in media supplemented with 400 μg/mL geneticin (G418 sulfate, ThermoFisher Scientific). Qualitative visualization of the cell populations by immunocytochemical analysis with an antibody directed against V5 suggested reduced amounts of the mutant GPC4 proteins compared to WT (data not shown). Therefore, we performed immunoblot analysis to determine steady-state amounts of GPC4 under basal conditions and after inhibition of the ubiquitin proteasome system (UPS), the major intracellular protein degradation pathway for short-lived or damaged proteins. Cells were cultured overnight in basal media or media supplemented with 10 μM MG-132, which we have previously shown is sufficient to inhibit the UPS without causing significant cellular toxicity in HEK293 cells.17 Protein extracts were prepared, and immunoblot analysis, performed essentially as previously described,18 identified an apparent increase in the steady-state amounts of mutant but not WT recombinant V5 GPC4 after proteasome inhibition (Figure 3A). Quantitation and comparison of the recombinant GPC4/β-actin ratio in the untreated cells suggested the steady-state amount of p.Gln506∗ was reduced in comparison to that of the WT (WT 0.026 ± 0.002 vs p.Gln506∗ 0.014 ± 0.002, mean ± SEM, p = 0.042), whereas there was no significant difference in the amounts of p.Glu496∗ compared to WT (WT 0.026 ± 0.002 vs p.Glu496∗ 0.029 ± 0.004, mean ± SEM, p = 0.565). In contrast, after exposure to 10 μM MG-132 for 14 h, both p.Gln506∗ and p.Glu496∗ steady-state amounts were significantly elevated in comparison to steady-state amounts in the treated WT cells (WT 0.030 ± 0.002 vs p.Gln506∗ 0.041 ± 0.003, mean ± SEM, p = 0.024; and WT 0.030 ± 0.002 vs p.Glu496∗ 0.199 ± 0.009, mean ± SEM, p = 0.002) (Figure 3B). Therefore, having an active UPS resulted in a relative decrease in steady-state protein amounts of ∼2.5-fold for p.Gln506∗ and ∼6-fold for p.Glu496∗ compared to those in UPS-inhibited cells. Collectively, the reduced stability and removal of critical protein domains suggest a loss of GPC4 function as the pathogenic mechanism underlying disease in all six families described in this study.
Figure 3.
Truncated GPC4 Proteins Are Unstable and Degraded by the UPS
(A) HEK293 cells transfected with constructs encoding N-terminally tagged WT and truncated GPC4 were grown in the absence or presence of 10 μM MG-132 for 14 hours. Total protein was isolated and analyzed by SDS-PAGE and immunoblotting with antibodies directed against V5 (46-0705, ThermoFisher Scientific, 1:5000) and β-actin (A5441, Sigma; 1:20,000). A representative image is shown, and approximate sizes in kDa are indicated (MultiMark Standard, Invitrogen).
(B) Quantification of three independent experiments performed in triplicate. The immunoreactive signals were quantified with a LAS4000 imager. Steady-state protein amounts are expressed as the ratio of GPC4/β-actin, which is the loading control for normalization.
An asterisk denotes p ≤ 0.05; statistical comparisons were made with a two-tailed t test; error bars represent mean ± SEM.
To date, there has been very limited information about GPC4-associated phenotypes in humans. A hemizygous missense variant (c.1235G>A [p.Arg412Lys]) was reported as likely pathogenic in an individual with a Robinow-syndrome-like phenotype that included some findings seen in Keipert syndrome; such findings included macrocephaly; hypertelorism; broad thumbs and great toes; and camptodactyly. However, this individual had more prominent skeletal features, including mesomelia and cranial sclerosis,19 which were not seen in the Keipert-syndrome-affected individuals with truncating GPC4 mutations in our study. Duplication of GPC4 has also been implicated in one family that is affected by the overgrowth syndrome Simpson-Golabi-Behmel syndrome (SGBS, MIM: 312870), an X-linked disorder caused by pathogenic variation in the gene encoding glypican 3 (GPC3). SGBS is characterized by pre- and postnatal overgrowth, skeletal abnormalities, variable intellectual disability, and congenital anomalies such as congenital heart defects and diaphragmatic hernia.20, 21, 22 This SGBS-affected family, initially reported by Golabi and Rosen,21 was followed up on and was reported in 2010 as not having a GPC3 mutation but as having a duplication of exons 1–9 of GPC4.23 This duplication segregated with a phenotype that included macrosomia, coarse facies, a short nose, a broad nasal bridge, macroglossia, and accessory nipples. The duplication mapped close to the 3′ end of GPC3, and the authors speculated that the duplication might result in decreased expression of GPC3, or alternatively that GPC4 might be a second gene for SGBS.23 However, since that publication, sequencing and deletion analysis of GPC4 in other individuals with SGBS and other overgrowth disorders has not detected any pathogenic variants.20, 24
There have been two reports of SGBS associated with deletions that encompass both GPC3 and GPC4. In a family that included some members with a clinical diagnosis of SGBS and that was originally described by Pilia et al.,22 a deletion that removed the entire GPC4 gene and the last two exons of GPC3 was detected.25 The SGBS phenotype in males in this family was noted to include renal-tract abnormalities and hydrocephalus. The second report was of a 1 Mb deletion encompassing three genes, GPC4, GPC3, and CCDC160, in a male fetus that was terminated at 24 weeks; his phenotype comprised macrosomia, visceromegaly, macroglossia, polyhydramnios, and mild ventriculomegaly.26 In both families, there was insufficient clinical information provided to allow determination of whether an overlapping phenotype, comprising features of both SGBS and Keipert syndrome, might have been present.
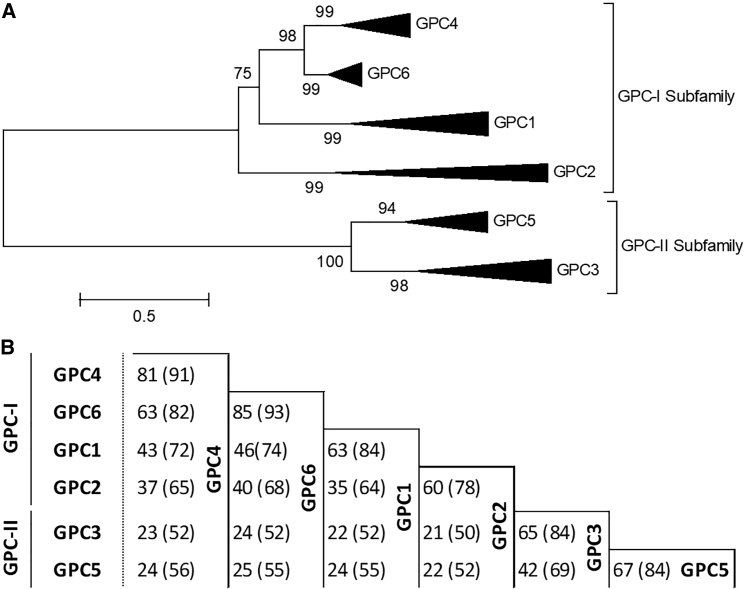
Glypicans are a family of cell-surface heparan sulfate proteoglycans characterized by a GPI anchor that localizes them to the cell surface, where they regulate growth-factor signaling during development and disease.27 They are expressed predominantly during development and are thought to play a role in the regulation of morphogenesis. In mammals, there are six glypicans, GPC1 to GPC6. The genes encoding GPC3 and GPC4 and those encoding GPC5 and GPC6 are clustered on chromosome Xq26 and chromosome 13q32, respectively, suggesting that the glypican family arose as a result of gene duplication. Filmus et al.28 reported evidence of homologs of glypican throughout Eumetazoa but were unable to identify glypican homologs outside the Metazoa linage. To analyze the evolution of the glypican family, we used the maximum likelihood method with Molecular Evolutionary Genetics Analysis (MEGA) software version 6,29 and in doing this, we independently replicated the Eumetazoa result. In addition, evidence of glypicans was identified in the genomes of both the Placozoa Trichoplax adhaerens and the sponge Amphimedon queenslandica. These genomes represent the basal group of multicellular organisms (Figure S1). To determine the orthology of the glypican family, we performed phylogenetic analysis by using the maximum likelihood method. There are two major glypican subfamilies, the GPC-I subfamily, which comprises GPC1, GPC2, GPC4, and GPC6, and the GPC-II subfamily, which comprises GPC3 and GPC5. Robust branch partitioning suggested the likelihood that a single gene ancestor of both subfamilies originated prior to the divergence of the Eumetazoan genome (Figure S2). Notably, in the chordate genome of the lancelet (Branchiostoma floridae, Bflo) the single GPC-I ortholog and the single GPC-II ortholog were identified side-by-side on a single scaffold (BRAFLscaffold_196; GenBank: NW_003101409, data not shown) in an arrangement reflecting both the GPC3-GPC4 and the GPC5-GPC6 genomic clustering. GPC3 and GPC5 are inferred to be descended from a single chordate GPC-I ortholog and GPC4 and GPC6 from a single GPC-II ortholog. Therefore, both loci probably arose from a single locus. To refine the relationship among vertebrate glypicans, we performed phylogenetic analysis by using only glypican proteins from vertebrate genomes that had undergone two rounds of whole-genome duplication. Although glypicans do not appear to encode well-characterized functional protein domains30 and although the amino acid homology of mammalian proteins is as low as 25%, the three-dimensional structure is conserved across the family.11 Among all glypicans, the greatest similarity at the protein level is found between GPC3 and GPC5 and between GPC4 and GPC6 (Figure 4). In addition, this study has demonstrated that GPC4 and GPC6 are the most closely related of all glypicans.
Figure 4.
GPC4 and GPC6 Are the Most Closely Related of All Glypican Proteins
(A) Phylogenetic analysis of vertebrate (2R) glypicans was inferred via the maximum-likelihood method. GPC1, GPC2, GPC4, and GPC6 share a common ancestor; GPC3 and GPC5 share a common ancestor. GPC4 and GPC6 have undergone considerably less diversification than have other glypican proteins. The tree is drawn to scale, and branch lengths are in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
(B) The average amino acid identity and similarity (brackets) of vertebrate (2R) glypican proteins were determined by global pairwise alignment. Sequence accession numbers are indicated in Figure S1.
Apart from GPC4 (this report, Keipert syndrome) and GPC3 (SGBS), only GPC6 has a clear association with Mendelian disorders. Pathogenic variants in GPC6 cause autosomal-recessive omodysplasia (MIM: 258315), a skeletal dysplasia characterized by short limbs and craniofacial abnormalities (frontal bossing, a depressed nasal bridge with a short nose, and a long and prominent philtrum).31 It is notable that there is phenotypic overlap between the craniofacial abnormalities in Keipert syndrome and those in omodysplasia; however, in omodysplasia, limb abnormalities are far more severe than in Keipert syndrome. Pathogenic variants in GPC1, GPC2, and GPC5 have not yet been shown to cause any Mendelian disorder, although common variants in GPC5 have been implicated in acquired nephrotic syndrome32 and in the risk of lung cancer in never-smokers.33 Amplification and overexpression of GPC5 has been observed in rhabdomyosarcoma.34
We have previously identified GPC4 and GPC6 as astrocyte-secreted proteins that induce synapse formation in rodent neurons.16 GPC4 and GPC6 have redundant functions in vitro, but Gpc4 knockout (KO) mice have defective synapse formation in the developing hippocampus, a part of the brain that is enriched for Gpc4.16 In day 7–10.5 embryos, Gpc4 mRNA localizes to a range of sites that include the anterior forebrain neuroepithelium, branchial arches, optic and otic vesicles, limb buds, and somites.12 We performed additional studies in Gpc4-deficient mice to look for the two primary features of Keipert syndrome: craniofacial abnormalities and digital abnormalities. All bone measurement experiments were approved by the Salk Institute Institutional Animal Care and Use Committee (IACUC). In addition, we tested the mice for sensorineural deafness, which was observed in the originally described Keipert-syndrome-affected family but might not be a defining feature of the syndrome. Hearing experiments were approved by Stanford University IACUC.
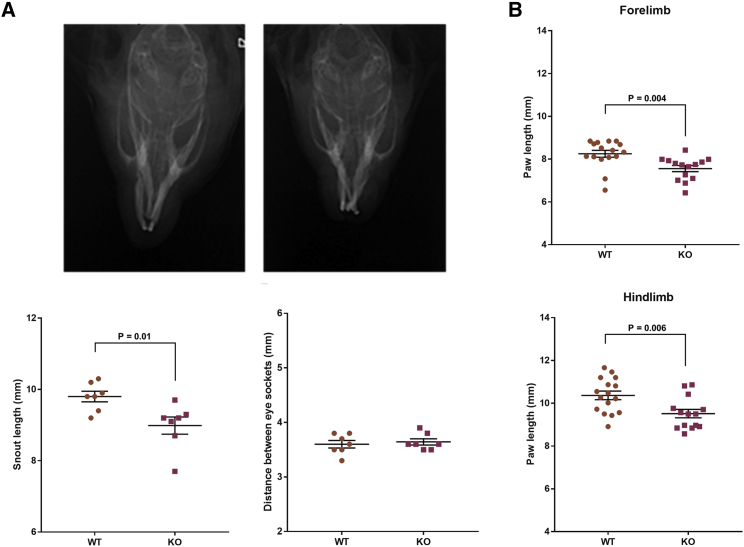
Glypican-4-null (male) mice with targeted KO of exon 3 (strain MMRRC_032331-UCD) were maintained as we previously described,16, 35 and 16- to 20-week-old littermate male Gpc4 KO and WT mice were used for analysis. To determine the effect of Gpc4 loss of function on skull geometry, we took X-rays of the skull by using a CR 7 Digital Dental X-Ray imager and performed two sets of measurements: (1) from the cranial aspect to the tip of the snout and (2) for intra-orbital distance, between the medial-edge of each orbit. There was a significant decrease in the length of the snout in Gpc4 KO mice compared to WT (WT—9.8 ± 0.1 mm, n = 7 versus KO—9.0 ± 0.2 mm, n = 7, mean ± SEM, p = 0.014), but there was no difference in intra-orbital separation (WT—3.6 ± 0.1 mm, n = 7 versus KO—3.6 ± 0.1 mm, n = 7, mean ± SEM, p = 0.64) (Figure 5A). Similar results were obtained when measurements for snout length were taken with an instant-readout precision digital caliper (Figure S3), suggesting that loss of GPC4 causes craniofacial abnormalities in the mouse.
Figure 5.
Gpc4 KO Mice Exhibit Nasodigital Deficits
(A) Example radiographs showing typical skull morphology of adult WT (left) and Gpc4 KO (right) mice. Note the shortened snout in the KO mouse. Quantification of radiograph data demonstrated that Gpc4 KO mice have significantly shorter snouts; snouts were measured from the tip of the snout to the cranial aspect. However, there is no difference in the intra-orbital distance in Gpc4 KO mice compared to WT.
(B) Gpc4 KO mice have significantly shorter front and rear paws.
n = 7 animals per group; data combine right and left paw to give 14 measurements; statistical comparisons were made with a two-tailed t test; error bars represent mean ± SEM.
To determine the effect of Gpc4 loss of function on the digits, mice were placed on a treadmill, and videos of their paws were captured as they walked. We used GaitScan software to analyze both fore and hind paws and analyzed a minimum of five images per paw per mouse. We marked the position of the base of the paw and the tip of each phalanx. The distance between the tip of the longest phalanx (#3 of 5) and base was measured for hind paws. Because the fore paws have only four phalanges, the #3 marker was positioned equidistant between phalange 2 and 4. There was a significant decrease in the length of both the fore and hind paws in Gpc4 KO mice compared to WT (fore paws—WT 8.3 ± 0.16 mm, n = 7 versus KO 7.6 ± 0.14 mm, n = 7, mean ± SEM, p = 0.004; hind paws—WT 10.4 ± 0.2 mm, n = 7 versus KO 9.5 ± 0.2 mm, n = 7, mean ± SEM, p = 0.006) (Figure 5B). We also measured the distance between the phalanges, but there was no difference in the total spread of digits for the forelimb and hindlimbs, as measured from digit 1 to digit 5 (Figure S4). Thus, Gpc4 KO mice recapitulate the skeletal features seen in Keipert syndrome, albeit more subtly.
To test hearing function in the Gpc4 KO mice, we performed auditory brainstem response (ABR) recordings. Mice were anesthetized, and acoustic stimuli were delivered to the ear canals at 8, 16, and 32 kHz. Sound levels were incremented in five dB steps from 10–20 dB below threshold to 80 dB (for 8 and 16 kHz) or 100 dB (for 32 kHz). The threshold for ABR was defined as the lowest stimulus level at which repetitive waves I and V could be identified in the response waveform. The ABR thresholds of P18 Gpc4 KO mice were compared to those of littermate WT control mice. There was no difference in hearing thresholds at the 8, 16, and 32 kHz frequencies measured between the two genotypes (Figure S5). We analyzed the ABR data at 16 kHz, within the most sensitive frequency range for mice for peak 1 amplitudes and latencies. However, we did not observe a shift in first peak amplitude or latency in Gpc4 KO mice compared to controls. In addition, to test the function of outer hair cells, we measured distortion-product otoacoustic emissions (DPOAEs) as previously described.36 No difference in the function of the cochlear amplifier was revealed by DPOAE measurements between the KO and control mice.
In conclusion, we have demonstrated that pathogenic variants in GPC4 underlie Keipert syndrome. The primary and invariant features of the disorder include craniofacial and digital abnormalities. However, despite being described in KS1, the prototypical family, sensorineural deafness does not appear to be consistently associated with loss of GPC4 function. We report that Gpc4 KO mice display morphological abnormalities reminiscent of Keipert syndrome: they have significantly shorter paw and snout lengths than WT mice. In contrast to the relatively subtle physical phenotype in Gpc4-deficient mice, the zebrafish GPC4 mutant knypek does not survive beyond 5–7 days post-fertilization; this is due to severely reduced convergence and extension movements.37 However, when these gastrulation defects are suppressed by gpc4 mRNA injection, knypek embryos display more subtle abnormalities of the craniofacial cartilages; these abnormalities include shortening of the skull and jaw.38 These features are consistent with those observed in individuals with Keipert syndrome and in the Gpc4 KO mice. GPC4 is the third human glypican to be linked to a genetic syndrome, and our data strengthen the evidence linking the family of glypican genes to disorders of cartilage and bone morphogenesis, information that may be relevant to phenotypes associated with dysregulation of glypicans 1, 2, and 5.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the Australian Government National Health and Medical Research Council (program grant 1054618 and fellowship 1002098). P.J.L. was supported by the National Health and Medical Research Council (NHMRC) Career Development Fellowship GNT1032364. M.B. was supported by an NHMRC senior research fellowship (1102971) and an NHMRC program grant (1054618). Additional funding was provided by the Independent Research Institute Infrastructure Support Scheme and the Victorian State Government Operational Infrastructure Program. M.A.M. is a participant in the BIH Charité Junior Clinician Scientist Program funded by the Charité—Universitätsmediz in Berlin and the Berlin Institute of Health. V.R.S. receives support from the National Human Genome Research Institute (NHGRI) UM1 HG006542-07. C.D. and N.J.A. were supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke (NIH-NINDS) R01 NS089791. M.M. was supported by the National Institute on Deafness and other Communicative Disorders (NIDCD) R01 DC09590.
Published: April 11, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.02.026.
Accession Numbers
The ClinVar accession numbers for the GPC4 variants reported in this paper are ClinVar: RCV000659264.1, RCV000659265.1, RCV000659266.1, and RCV000659267.1.
Web Resources
GeneMatcher, https://www.genematcher.org/
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/
Genotype-Tissue Expression (GTEx) project, https://gtexportal.org/home/
Online Mendelian Inheritance in Man, http://www.omim.org/
UCSC Genome Bioinformatics database, https://genome.ucsc.edu/
Uniprot, https://www.uniprot.org/
Varsome, https://varsome.com
Supplemental Data
References
- 1.Amor D.J., Dahl H.H., Bahlo M., Bankier A. Keipert syndrome (nasodigitoacoustic syndrome) is X-linked and maps to Xq22.2-Xq28. Am. J. Med. Genet. A. 2007;143A:2236–2241. doi: 10.1002/ajmg.a.31917. [DOI] [PubMed] [Google Scholar]
- 2.Keipert J.A., Fitzgerald M.G., Danks D.M. A new syndrome of broad terminal phalanges and facial abnormalities. Aust. Paediatr. J. 1973;9:10–13. doi: 10.1111/j.1440-1754.1973.tb02215.x. [DOI] [PubMed] [Google Scholar]
- 3.Balci S., Dagli S. Keipert syndrome in two brothers from Turkey. Clin. Genet. 1996;50:223–228. doi: 10.1111/j.1399-0004.1996.tb02631.x. [DOI] [PubMed] [Google Scholar]
- 4.Cappon S.M., Khalifa M.M. Additional case of Keipert syndrome and review of the literature. Med. Sci. Monit. 2000;6:776–778. [PubMed] [Google Scholar]
- 5.Reardon W., Hall C.M. Broad thumbs and halluces with deafness: a patient with Keipert syndrome. Am. J. Med. Genet. A. 2003;118A:86–89. doi: 10.1002/ajmg.a.10063. [DOI] [PubMed] [Google Scholar]
- 6.Derbent M., Bikmaz Y.E., Agildere M. A patient with Keipert syndrome and isolated fibrous dysplasia of the sphenoid sinus. Am. J. Med. Genet. A. 2011;155A:1496–1499. doi: 10.1002/ajmg.a.34006. [DOI] [PubMed] [Google Scholar]
- 7.Nik-Zainal S., Holder S.E., Cruwys M., Hall C.M., Shaw-Smith C. Keipert syndrome: two further cases and review of the literature. Clin. Dysmorphol. 2008;17:169–175. doi: 10.1097/MCD.0b013e3282f4afc3. [DOI] [PubMed] [Google Scholar]
- 8.Dumic M., Kokic D.D., Matic T., Potocki K. Daughter and her mildly affected father with Keipert syndrome. Am. J. Med. Genet. A. 2006;140:2488–2492. doi: 10.1002/ajmg.a.31489. [DOI] [PubMed] [Google Scholar]
- 9.Emmanuel C., Gava N., Kennedy C., Balleine R.L., Sharma R., Wain G., Brand A., Hogg R., Etemadmoghadam D., George J., Australian Ovarian Cancer Study Group Comparison of expression profiles in ovarian epithelium in vivo and ovarian cancer identifies novel candidate genes involved in disease pathogenesis. PLoS ONE. 2011;6:e17617. doi: 10.1371/journal.pone.0017617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filmus J., Capurro M. The role of glypicans in hedgehog signaling. Matrix Biol. 2014;35:248–252. doi: 10.1016/j.matbio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Ybot-Gonzalez P., Copp A.J., Greene N.D. Expression pattern of glypican-4 suggests multiple roles during mouse development. Dev. Dyn. 2005;233:1013–1017. doi: 10.1002/dvdy.20383. [DOI] [PubMed] [Google Scholar]
- 13.González-Del Angel A., Estandía-Ortega B., Alcántara-Ortigoza M.A., Martínez-Cruz V., Gutiérrez-Tinajero D.J., Rasmussen A., Gómez-González C.S. Expansion of the variable expression of Muenke syndrome: hydrocephalus without craniosynostosis. Am. J. Med. Genet. A. 2016;170:3189–3196. doi: 10.1002/ajmg.a.37951. [DOI] [PubMed] [Google Scholar]
- 14.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spath M.A., Nillesen W.N., Smits A.P., Feuth T.B., Braat D.D., van Kessel A.G., Yntema H.G. X chromosome inactivation does not define the development of premature ovarian failure in fragile X premutation carriers. Am. J. Med. Genet. A. 2010;152A:387–393. doi: 10.1002/ajmg.a.33243. [DOI] [PubMed] [Google Scholar]
- 16.Allen N.J., Bennett M.L., Foo L.C., Wang G.X., Chakraborty C., Smith S.J., Barres B.A. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockhart P.J., Lincoln S., Hulihan M., Kachergus J., Wilkes K., Bisceglio G., Mash D.C., Farrer M.J. DJ-1 mutations are a rare cause of recessively inherited early onset parkinsonism mediated by loss of protein function. J. Med. Genet. 2004;41:e22. doi: 10.1136/jmg.2003.011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephenson S.E.M., Aumann T.D., Taylor J.M., Riseley J.R., Li R., Mann J.R., Tomas D., Lockhart P.J. Generation and characterisation of a parkin-Pacrg knockout mouse line and a Pacrg knockout mouse line. Sci. Rep. 2018;8:7528. doi: 10.1038/s41598-018-25766-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White J.J., Mazzeu J.F., Coban-Akdemir Z., Bayram Y., Bahrambeigi V., Hoischen A., van Bon B.W.M., Gezdirici A., Gulec E.Y., Ramond F., Baylor-Hopkins Center for Mendelian Genomics WNT signaling perturbations underlie the genetic heterogeneity of Robinow syndrome. Am. J. Hum. Genet. 2018;102:27–43. doi: 10.1016/j.ajhg.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cottereau E., Mortemousque I., Moizard M.P., Bürglen L., Lacombe D., Gilbert-Dussardier B., Sigaudy S., Boute O., David A., Faivre L. Phenotypic spectrum of Simpson-Golabi-Behmel syndrome in a series of 42 cases with a mutation in GPC3 and review of the literature. Am. J. Med. Genet. C. Semin. Med. Genet. 2013;163C:92–105. doi: 10.1002/ajmg.c.31360. [DOI] [PubMed] [Google Scholar]
- 21.Golabi M., Rosen L. A new X-linked mental retardation-overgrowth syndrome. Am. J. Med. Genet. 1984;17:345–358. doi: 10.1002/ajmg.1320170128. [DOI] [PubMed] [Google Scholar]
- 22.Pilia G., Hughes-Benzie R.M., MacKenzie A., Baybayan P., Chen E.Y., Huber R., Neri G., Cao A., Forabosco A., Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 23.Waterson J., Stockley T.L., Segal S., Golabi M. Novel duplication in glypican-4 as an apparent cause of Simpson-Golabi-Behmel syndrome. Am. J. Med. Genet. A. 2010;152A:3179–3181. doi: 10.1002/ajmg.a.33450. [DOI] [PubMed] [Google Scholar]
- 24.Veugelers M., Cat B.D., Muyldermans S.Y., Reekmans G., Delande N., Frints S., Legius E., Fryns J.P., Schrander-Stumpel C., Weidle B. Mutational analysis of the GPC3/GPC4 glypican gene cluster on Xq26 in patients with Simpson-Golabi-Behmel syndrome: identification of loss-of-function mutations in the GPC3 gene. Hum. Mol. Genet. 2000;9:1321–1328. doi: 10.1093/hmg/9.9.1321. [DOI] [PubMed] [Google Scholar]
- 25.Veugelers M., Vermeesch J., Watanabe K., Yamaguchi Y., Marynen P., David G. GPC4, the gene for human K-glypican, flanks GPC3 on xq26: deletion of the GPC3-GPC4 gene cluster in one family with Simpson-Golabi-Behmel syndrome. Genomics. 1998;53:1–11. doi: 10.1006/geno.1998.5465. [DOI] [PubMed] [Google Scholar]
- 26.Weichert J., Schröer A., Amari F., Siebert R., Caliebe A., Nagel I., Gillessen-Kaesbach G., Mohrmann I., Hellenbroich Y. A 1 Mb-sized microdeletion Xq26.2 encompassing the GPC3 gene in a fetus with Simpson-Golabi-Behmel syndrome report, antenatal findings and review. Eur. J. Med. Genet. 2011;54:343–347. doi: 10.1016/j.ejmg.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Gupta M., Brand M. Identification and expression analysis of zebrafish glypicans during embryonic development. PLoS ONE. 2013;8:e80824. doi: 10.1371/journal.pone.0080824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filmus J., Capurro M. The role of glypican-3 in the regulation of body size and cancer. Cell Cycle. 2008;7:2787–2790. doi: 10.4161/cc.7.18.6672. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filmus J., Capurro M., Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos-Xavier A.B., Martinet D., Bateman J., Belluoccio D., Rowley L., Tan T.Y., Baxová A., Gustavson K.H., Borochowitz Z.U., Innes A.M. Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am. J. Hum. Genet. 2009;84:760–770. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto K., Tokunaga K., Doi K., Fujita T., Suzuki H., Katoh T., Watanabe T., Nishida N., Mabuchi A., Takahashi A. Common variation in GPC5 is associated with acquired nephrotic syndrome. Nat. Genet. 2011;43:459–463. doi: 10.1038/ng.792. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Sheu C.C., Ye Y., de Andrade M., Wang L., Chang S.C., Aubry M.C., Aakre J.A., Allen M.S., Chen F. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson D., Selfe J., Gordon T., Lu Y.J., Pritchard-Jones K., Murai K., Jones P., Workman P., Shipley J. Role for amplification and expression of glypican-5 in rhabdomyosarcoma. Cancer Res. 2007;67:57–65. doi: 10.1158/0008-5472.CAN-06-1650. [DOI] [PubMed] [Google Scholar]
- 35.Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F., Grant D., Solloway M., Parker L., Ye W. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 36.Xia A., Visosky A.M., Cho J.H., Tsai M.J., Pereira F.A., Oghalai J.S. Altered traveling wave propagation and reduced endocochlear potential associated with cochlear dysplasia in the BETA2/NeuroD1 null mouse. J. Assoc. Res. Otolaryngol. 2007;8:447–463. doi: 10.1007/s10162-007-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topczewski J., Sepich D.S., Myers D.C., Walker C., Amores A., Lele Z., Hammerschmidt M., Postlethwait J., Solnica-Krezel L. The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell. 2001;1:251–264. doi: 10.1016/s1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- 38.LeClair E.E., Mui S.R., Huang A., Topczewska J.M., Topczewski J. Craniofacial skeletal defects of adult zebrafish glypican 4 (knypek) mutants. Dev. Dyn. 2009;238:2550–2563. doi: 10.1002/dvdy.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.