Abstract
Colony stimulating factor 1 receptor (CSF1R) plays key roles in regulating development and function of the monocyte/macrophage lineage, including microglia and osteoclasts. Mono-allelic mutations of CSF1R are known to cause hereditary diffuse leukoencephalopathy with spheroids (HDLS), an adult-onset progressive neurodegenerative disorder. Here, we report seven affected individuals from three unrelated families who had bi-allelic CSF1R mutations. In addition to early-onset HDLS-like neurological disorders, they had brain malformations and skeletal dysplasia compatible to dysosteosclerosis (DOS) or Pyle disease. We identified five CSF1R mutations that were homozygous or compound heterozygous in these affected individuals. Two of them were deep intronic mutations resulting in abnormal inclusion of intron sequences in the mRNA. Compared with Csf1r-null mice, the skeletal and neural phenotypes of the affected individuals appeared milder and variable, suggesting that at least one of the mutations in each affected individual is hypomorphic. Our results characterized a unique human skeletal phenotype caused by CSF1R deficiency and implied that bi-allelic CSF1R mutations cause a spectrum of neurological and skeletal disorders, probably depending on the residual CSF1R function.
Keywords: CSF1R, mutation, skeletal dysplasia, osteosclerosis, dysosteosclerosis, Pyle disease, leukoencephalopathy, HDLS, osteoclast
Main Text
Colony stimulating factor 1 (CSF1) regulates survival, proliferation, differentiation, and phagocytic and chemotactic activity of cells from the monocyte/macrophage lineage, which could be specialized as microglia in brain and as osteoclasts in bone.1 The effects of CSF1 are mediated by its receptor, CSF1R, which triggers multiple signal transduction pathways.2 CSF1R is expressed not only in cells of hematopoietic origin, but also in non-hematopoietic cells including Paneth cells,3 renal proximal tubule epithelial cells,4 oocyte and female reproductive tract,5 as well as decidual cells and trophoblastic cells.6 The pleiotropic effects of CSF1R deficiency have been under study extensively.
In mouse, bi-allelic Csf1r deficiency is reported to cause sclerosing skeletal dysplasia and gross anatomical abnormalities in developing brains, leading to death within 6 weeks.7, 8, 9 In contrast, Csf1r+/− mice gradually develop cognitive and sensorimotor deficits, depression with anxiety-like behavior, and enlarged ventricles from 6 months of age.10 In humans, CSF1R mono-allelic mutations, which are considered to cause haploinsufficiency or dominant-negative effects, lead to hereditary diffuse leukoencephalopathy with spheroids (MIM: 164770), an adult-onset rapidly progressive neurodegenerative disorder characterized by variable clinical manifestations, including behavioral changes, dementia, depression, Parkinsonism, and seizures.11 Recently, Monies et al. reported two deceased babies who possibly had a homozygous CSF1R mutation.12 The parents had a likely pathogenic CSF1R variant, c.1620T>A (p.Tyr540∗), in a heterozygous state, although DNA of the two babies were not available. The affected individuals are reported to have structural anomalies of central nervous system (CNS) and osteopetrosis.
In this study, we identified five different CSF1R mutations in seven affected individuals from three unrelated families with diverse ethnicity. They had bi-allelic mutations and developed a syndrome of recessive inheritance characterized by brain malformation with calcifying leukoencephalopathy and skeletal dysplasia compatible with dysosteosclerosis (DOS [MIM: 224300])13 or Pyle disease (MIM: 265990).14 All the mutations led to functional deficiency of CSF1R. Our findings establish a link between CSF1R signaling deficiency and a unique skeletal and brain phenotype in humans, suggesting that the effect of CSF1R loss in triggering human diseases is dose sensitive and tissue specific.
The study protocol was approved by the ethical committee of RIKEN and participating institutions. Peripheral blood was obtained after the informed consent. All study procedures were performed in accordance with the principles of the Declaration of Helsinki.
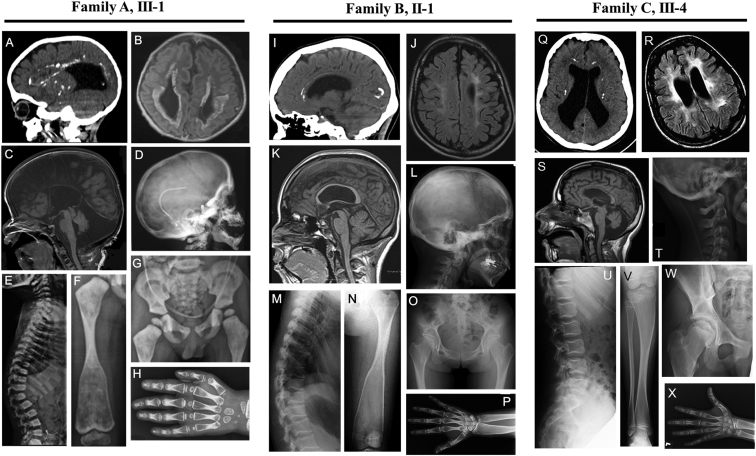
The initial blood samples were collected from a 5-year-old boy (Figure 1A, A-III-1) who was the only child of healthy and non-consanguineous Brazilian parents. The boy was born at 37 5/7 weeks with normal birth weight and length (Tables 1 and S1; Supplemental Note). Brain imaging studies showed a Dandy-Walker malformation, scattered periventricular calcifications, corpus callosum agenesis, and abnormal signal in the periventricular white matter (Figures 2A–2C). Ventriculo-peritoneal shunt was placed at 3 months of age. He was hypotonic and developed focal seizures. He sat without support at 13 months of age and had his first steps at 20 months of age, but since age 2.5 years he regressed motorically. At the last visit at the age of 4.5 years, he was severely dysarthric with partial head control and unable to sit without support. A skeletal survey in the first months of life showed bone abnormalities. He had poor vision with nystagmus secondary to optic nerve atrophy. Physical examination at age 4 years and 4 months disclosed facial dysmorphisms (long eyelashes, bilateral epicanthus, bulbous nose, dysplastic ears, etc.); narrow, bell-shaped thorax with pectus carinatum; joint restrictions at the elbow and ankle joints; and dorsal kyphosis. Complete blood count (CBC) and serum markers for bone metabolism showed normal results (Table 1), except for decreased ALP level. A skeletal survey showed diffuse osteosclerosis of the cranio-facial bones, most prominent in the skull base (Figure 2D). The vertebral bodies and the neural arches were mildly flat with sclerotic margins (Figure 2E). Pelvic bones showed sclerosis (Figure 2G). The tubular bones showed diaphyseal sclerosis and metaphyseal radiolucency with metaphyseal under-modeling (Figures 2F and 2H). The skeletal changes were compatible with that of DOS. The parents and grandparents denied bone fractures and had normal height. The father had mild cortical hyperostosis in the long tubular bones (Figures S1A–S1C and S1E–S1G). Cranial computed tomography (CT) scan of the parents revealed no brain anomaly (Figures S1D and S1H). The 76-year-old paternal grandfather showed short-term memory loss from the age of 70. His cranial CT scan showed one parietal parenchymal calcification (Figure S1I).
Figure 1.
Transmission of the CSF1R Variants in the Three Families with the Syndrome
(A and B) Electropherograms of Sanger sequencing showing that the two variants, c.1441C>T (p.Gln481∗) (V1) and c.395C>T (p.Pro132Leu) (V2), were compound heterozygous for CSF1R in the affected child in family A (III-1). The father (II-1) and the paternal grandfather (I-1) were heterozygotes for c.1441C>T, while the mother (II-2) and the maternal grandfather (I-3) are heterozygotes for c.395C>T.
(C and D) Electropherograms of Sanger sequencing showing that the two variants, c.1879_1881del (p.Lys627del) (V3) and c.1859−119G>A (V4), were compound heterozygous for CSF1R in the affected individual in family B (II-2).
(E and F) Electropherograms of Sanger sequencing showing that the variant, c.1969+115_1969+116del (V5), was homozygous for CSF1R in four affected individuals in family C.
Abbreviation: W, wild-type.
Table 1.
Clinical and Radiographic Findings of the Three Families with Bi-allelic CSF1R Mutations
| Individual | III-1 | II-2 | III-4 | III-6 |
|---|---|---|---|---|
| Family | family A | family B | family C | family C |
| Mutation | ||||
| Allele 1 | c.395C>T (p.Pro132Leu) | c.1859−119G>A (p.Ser620delins40) | c.1969+115_1969+116del (p.Pro658Serfs∗24) | |
| Allele 2 | c.1441C>T (p.Gln481∗) | c.1879_1881del (p.Lys627del) | = Allele 1 | |
| Demographics | ||||
| Sex | male | female | female | male |
| Age at the last visit | 4 years 10 months | 37 years | 23 years | 14 years |
| Ethnic background | Brazilian | Japanese | Chaldean | Chaldean |
| Consanguinity | – | – | + | + |
| Gestational/Perinatal Record | ||||
| Fetal ultrasound | Dandy-Walker complex | normal | N/A | hydrocephalus |
| Gestational age | 37 5/7 weeks | N/A | full term | full term |
| Birth weight (g) | 4,270 (>90th centile) | N/A | N/A | 3,300 |
| Birth length (cm) | 50 (75–90th centile) | N/A | N/A | N/A |
| OFC (cm) | 40 (>98th centile) | N/A | normal | normal |
| Skeletal Radiograph | ||||
| Fracture | – | + | – | – |
| Skull sclerosis | + | + | + | + |
| Optic canal narrowing | + | + | – | – |
| Pelvic bone sclerosis | + | + | – | – |
| Vertebral sclerosis | + | + | – | – |
| Platyspondyly | + | + concaved at posterior thirds | + | + |
| Tubular bones | ||||
| Under-modeling | + | + | + | + |
| Widened metaphysis | + | + | + | + |
| Radiolucent metaphysis | + | + | + | + |
| Constricted diaphysis | + | + | + | + |
| Sclerotic diaphysis | + | + | + | + |
| Brain Imaging | ||||
| Multiple calcifications | + | + | + | + |
| Stepping stone appearance (peri-callosal region) | + | + | − | − |
| Corpus callosum agenesis | + | – | – | – |
| Dilated ventricle | + | + | + | + |
| Hydrocephalus | + congenital | – | – | + congenital |
| Dandy-Walker complex | + | – | large cysterna magna | large cysterna magna |
| Abnormal white matter signal | + | + | + | + |
| Laboratory Examination | ||||
| Complete blood count | normal | normal | normal | normal |
| Serum marker for bone metabolism | ||||
| TRACP-5b | 240 (170–590 mU/dl)a | 262 (120–420 mU/dl)b | N/A | N/A |
| CTX | 282 (155–873 pg/mL)c | N/A | WNL | WNL |
| NTX | N/A | 9.1 (7.5–16.5 nmol BCE/l)d | WNL | WNL |
Abbreviations: N/A, not available; OFC, occipitofrontal circumference; TRACP-5b, tartrate-resistant acid phosphatase-5b; CTX, C-terminal telopeptide; NTX, type I collagen cross-linked N-telopeptides; BCE, bone collagen equivalents; WNL, within normal limits.
Normal range for male.
Normal range for female.
Normal range for male at age 18–30, the index at age less than 18 is not established.
Normal range for premenopausal female.
Figure 2.
Radiological Features of the Affected Individuals with Bi-allelic CSF1R Mutations
(A–H) Individual 1 (family A, III-1).
(A) Sagittal cranial CT at age 2 years displays multiple calcifications mainly in the pericallosal and periventricular areas and enlarged ventricles.
(B and C) Brain MRI at age 1 month shows agenesis of the corpus callosum, cerebellar hypoplasia, communication of the IV ventricle with enlarged posterior fossa.
(D–H) X-rays showing severe osteosclerosis.
(D) Lateral view of the skull showing sclerosis of the cranial base.
(E) The margins of the vertebral bodies and the neural arches of the thoracic spine are sclerotic. The vertebral bodies are mildly flat with anterior pointing of D12-L1.
(F) The right femur shows sclerosis of mid-diaphysis with wide, radiolucent ends (Erlenmeyer flask deformity).
(G) Pelvic bones and proximal femora show marked sclerosis.
(H) Under-modeling, sclerosis of mid-diaphysis, and sub-metaphyseal radiolucency similar to the long tubular bone are seen.
(I–P) Individual 2 (family B, II-2) at age 36 years.
(I) Sagittal CT image displays linear calcifications of corpus callosum and marked dilated ventricle.
(J) MRI-FLAIR weighted image shows confluent hyperintensities in periventricular white matter and malformation of frontal lobe.
(K) MRI-T1 weighted sagittal image shows hypogenesis of cerebral cortex and corpus callosum with enlarged ventricle.
(L–P) X-rays.
(L) Diffuse osteosclerosis in craniofacial bones, particularly in the skull base.
(M) The neural arches of the thoracolumbar spine are mildly sclerotic. The end plates of the vertebral bodies are sclerotic and concave at their posterior thirds.
(N) The femur is under-modeled with radiolucent ends and sclerotic diaphysis.
(O) Pelvic bones and proximal femora show sclerosis, most prominently in the femoral shafts and iliac bodies.
(P) The right hand. Under-modeling, sclerosis of mid-diaphysis, and sub-metaphyseal radiolucency.
(Q–X) Individual 3 (family C, III-4) age 14 years.
(Q) Brain CT. Multiple calcification in the corona radiata and corpus callosum. Enlargement of both lateral ventricles.
(R and S) Brain horizontal and sagittal MRIs. Enlargement of lateral ventricles and cisterna magna, and patchy hyperintensity signal in extensive area of white matter.
(T–X) X-rays showing similar but milder osteosclerosis than in individual 1 and 2.
(T) Skull. Mild sclerosis of the cranial base.
(U) Vertebral bodies are concaved at their posterior thirds with mild sclerosis of the end plates.
(V) Metaphyseal under-modeling and radiolucency in the tibia.
(W) Increased bone density of pelvic bones and proximal femora.
(X) The left hand. Under-modeling, sclerosis of mid-diaphysis, and sub-metaphyseal radiolucency.
To identify the causal mutation, a whole-exome sequencing (WES) was performed for the affected individual-parents trio as described previously.15, 16, 17 The detailed information is available in Supplemental Methods and Tables S2 and S3. Variants were filtered by frequency (<0.005 in population databases, including gnomAD and ExAC). Individual A-III-1 was a compound heterozygote for c.1441C>T (p.Gln481∗) and c.395C>T (p.Pro132Leu) in CSF1R (GenBank: NM_005211) (Figure 1A). c.395C>T was reported once in gnomAD in a heterozygous state and c1441C>T was present in a control from 1,200 elderly, healthy Brazilian individuals in AbraOM. Mutations in the genes related to the known skeletal dysplasia were not identified in the dataset. We confirmed these variants by Sanger sequencing in the affected individual and their transmission in the families (Figures 1A and 1B). Considering that the human phenotypes are similar to the mice with bi-allelic Csf1r mutation,7, 8, 9, 10 we focused our attention on this gene. Notably, osteosclerosis in the affected individual is milder than those of the bi-allelic Csf1r mutant mice, suggesting that the severity of skeletal phenotype may depend on the remaining functional alleles.
Subsequently, individuals were recruited via a private network of skeletal dysplasia experts (J.S. and G.N.) based on their having a very unique skeletal and brain features not reported previously as a definite disease entity. From this recruitment strategy, CSF1R mutations were identified in two additional families from Japan and USA.
Individual B-II-2, a 37-year-old Japanese female, was one of the two daughters of healthy and non-consanguineous parents (Figure 1C). Her birth and development were uneventful. At age 5, she fell down and suffered coccyx fracture. A skeletal survey revealed bone abnormalities and she was diagnosed with osteopetrosis. After she graduated from college, she worked as an office clerk. When she was 28, her visual acuity gradually deteriorated in the left eye. She was found to have increased intra-cranial pressure due to type 1 Chiari malformation and received foramen magnum decompression. At age 31, she started to drag her right foot when walking. At age 33, she started to experience difficulties in vocalization, which made her speak only a few words. Her walking ability gradually worsened until at age 35 when she could walk only for short distances with support. At age 36, she visited our clinic with a chief complaint of walking difficulties. Her height and weight were normal (Tables 1 and S1, Supplemental Note). Neurological examinations disclosed impaired cognitive function. She could walk only one or two steps holding onto others. She was severely hypophonic and had generalized spastic rigidity. Cranial CT scan showed dilated lateral ventricle and periventricular calcifications (Figure 2I). MRI studies disclosed multiple malformation of frontal lobe, hypogenesis of cerebral cortex, and corpus callosum with enlarged ventricle (Figures 2J and 2K). Bone survey disclosed diffuse osteosclerosis in craniofacial bones, particularly of the skull base (Figure 2L). The neural arches of the thoracolumbar spine were sclerotic. The vertebral bodies were concaved at their posterior thirds with endplate sclerosis (Figure 2M). The tubular bones showed metaphyseal under-modeling with metaphyseal radiolucency and diaphyseal sclerosis (Figures 2N and 2P). The iliac bodies and proximal femoral diaphyses were sclerotic (Figure 2O). Laboratory examination including CBC and serum markers for bone metabolism were normal (Table 1). The skeletal and neurologic changes were similar to those of individual A-III-1 and the mice with bi-allelic Csf1r mutation, but osteosclerosis and brain malformation were milder. The proband’s father, aged 70, did not show any abnormalities in neurological examination including brain MRI and bone survey. The proband’s mother, aged 69, showed scoliosis and mild short stature; however, brain MRI and bone survey did not show any obvious abnormalities. The older sister, aged 38, has no bone abnormality. She was diagnosed with multiple sclerosis, but no obvious lesions suggesting HDLS were seen.
WES analysis on the affected individual (B-II-2) revealed c.1859−119G>A and c.1879_1881del (p.Lys627del) in CSF1R. Neither variant was deposited in any available database. We confirmed these variants by Sanger sequencing in the affected individual and their transmission in the families (Figures 1C and 1D). Notably, the affected female had a child, suggesting the CSF1R mutations in the affected individual would not affect the reproductive system, which was previously reported to be associated with CSF1R.5, 6
Individual C-III-4 was a 14-year-old male who had hydrocephalus diagnosed by fetal US. He was from an extended Chaldean family of double first cousins (Figure 1E). He was a full-term baby with a birth weight of 3,300 g (+0.8 SD). He was hypotonic and developed focal seizures. He had developmental delay and intellectual disability (IQ 50) with superimposed progressive deterioration in cognitive function and vocalization. He showed rigido-spasticity with hyperactive tendon reflexes and pathological reflexes. His height was 148 cm (−2.4 SD) and body weight was 50 kg (−1.1 SD). Brain CT and MRI showed dilated ventricles and multiple punctuate calcifications (Figures 2Q–2S). A skeletal survey showed bone sclerosis similar to but milder than those in individual A-III-1 and B-II-2 (Figures 2T–2X), which resembled those of Pyle disease. Iliac crest biopsies showed formation of calcified cartilage islands in areas of woven bone (Figure S4). CBC and serum markers for bone metabolism were within normal limits (Table 1).
Individual C-III-6 was a 23-year-old female, a cousin of individual C-III-4 (Figure 1E). Her birth and development were uneventful. She had a mild limitation in intellectual functioning (IQ 73) and presented with a progressive deterioration in cognitive function. She had a progressive disability in vocalization. Rigido-spasticity and hyperactive tendon reflexes were evident, and pathological reflexes were elicited. Her height and body weight were normal (Table 1). Neuroimaging disclosed periventricular calcifications, dilated ventricle, and signal hyperintensities in the white matter (Figures S2A and S2B). Skeletal imaging showed sclerotic skull, mild platyspondyly, and failure of metaphyseal modeling of tubular bones (Figures S2C–S2F), similar to those of individual C-III-4. CBC and serum markers for bone metabolism were also within normal limits (Table 1).
The Chaldean family had total five affected individuals (four females and one male) (Figure 1E). The course of the disease and clinical characteristics were similar in the five affected individuals with variable clinical severity. The phenotypes represented a rapidly progressive encephalopathy with intellectual decline, pyramidal, extrapyramidal and cerebellar features, ataxia, dysarthria, seizures, and psychiatric symptoms, culminating in spastic quadriplegia, mutism, and persistent vegetative state. The onset of symptomatic neurological and skeletal defects were variable. Of the five, three attended special education and two were delayed in walking and language milestones. The premorbid cognitive and intellectual abilities of two affected females were remarkable in that one completed college and one completed her first year before the manifestations of ataxia and dysarthria. The phenotypes were similar to those in family B and C, but osteoscleorosis was milder.
Whole-genome sequencing (WGS) for the affected individuals (C-III-4 and C-III-6) were performed as previously described.18 CSF1R variant screening on the WGS dataset revealed a homozygous variant c.1969+115_1969+116del, which was not deposited in any available database. We confirmed the variant by Sanger sequencing in the affected individuals and their transmission in the families (Figures 1E and 1F).
In total, the present analysis identified five different CSF1R variants in seven affected individuals from three unrelated families with diverse ethnicity. We further characterized the functional consequence of these variants by a series of experiments. The c.1441C>T variant in family A causes a premature stop codon (p.Gln481∗) in the extracellular domain of CSF1R. Using RNAs extracted from the individuals’ blood, we checked the potential effect of nonsense-mediated mRNA decay (NMD) by RT-PCR. The result indicated that mRNA harboring this mutation was nearly absent (Figure 3A), suggesting the presence of a highly efficient NMD.
Figure 3.
Functional Analysis of CSF1R Mutations
(A) Electropherograms of Sanger sequencing for genomic DNA (gDNA) and complementary DNA (cDNA) fragments from the individuals in family A (I-1, II-2, III-1), who carried the CSF1R nonsense mutation, c.1441C>T (p.Gln481∗). Compared with the gDNA, the red T peak presenting c.1441C>T (arrow) was hardly absent in the cDNA. The primers for PCR and Sanger sequencing for gDNA were the same as those for cDNA.
(B and C) Aberrant splicing caused by c.1859−119G>A.
(B) A schematic diagram showing that the mutation generates a new splicing acceptor site in the deep intron and causes aberrant splicing to result in the adjacent exon extension. Purple box: the wild-type exon 13 (e13) and exon 14 (e14); red box: the extended part of e14.
(C) RT-PCR for the affected individual-derived lymphoblastoid cell line. An extra band (837 bp) was detected in the individual (B-II-2). Panel, a human cDNA from the multiple tissue cDNA panel (Clontech); Ctrl, a cDNA from the lymphoblastoid cell line of an unaffected individual.
(D–F) Aberrant splicing caused by c.1969+115_1969+116del.
(D) ESEfinder suggests that c.1969+115_1969+116del affects a splicing regulatory site. The colored blocks show the binding scores for different splicing factors. The shown genomic regions (x axis) are 154 and 152 nucleotides of intron 14 for the wild-type (top) and mutant (bottom) alleles, respectively. The mutation eliminates several potential binding sites for the splicing factors, including SRSF1 (red) and SRSF5 (green). SRSF, serine/arginine-rich splicing factor.
(E) A schematic diagram showing splicing of a cryptic exon (e14b; red box). The variant activates e14b by utilizing cryptic splice sites. The purple boxes indicate the wild-type exon 14 (e14) and exon 15 (e15).
(F) RT-PCR for the affected individual-derived fibroblasts. Cyclohexamide (CHX; 100 μg/mL) was added to inhibit nonsense mutation induced mRNA decay (NMD). An extra band (468 bp) larger than the expected size band (369 bp) was detected in the affected individuals (C-III-4 and C-III-6) when NMD was inhibited. The 468-bp segment has a 99-bp intronic sequence insertion between e14 and e15. c.1969+115_1969+116del was detected in the 99-bp insertion sequence, which has an in-frame stop codon resulting in a truncated protein p.Pro658Serfs∗24. The truncated protein is actually absent due to NMD. Control 1 and 2, cDNA from two unaffected individuals.
(G) Western blot of lysates from the wild-type (WT) CSF1R or variant CSF1R (p.Pro132Leu, p.Lys627del, p.Ser620delins40) transfected HEK293 cells treated with M-CSF. ACTB was used as a loading control.
(H) Graphic representation of the ratio of phosphorylated JNK to total JNK. Data are expressed as arbitrary units indicating mean ± SEM of three independent western blot experiments. Statistical significance was tested with one-way ANOVA, ∗p < 0.05.
The c.1859−119G>A variant identified in family B is located distant from the intron-exon junction and the branch point. Generation of a cryptic splice acceptor site was predicted by Human Splicing Finder (Figures 3B and S3). RT-PCR using affected individual-derived lymphoblastoid cell line indicated an extra band in the affected individual’s cDNA (Figure 3C). Sanger sequencing proved that the band had a 117-bp insertion into the open reading frame, thus leading to an elongated protein p.Ser620delins40 (Figure 3C). The inserted fragment is an extension of exon 14 to intron 13 and begins just after c.1859−119G>A. Notably, another variant (c.1879_1881del) in the opposite allele of c.1859−119G>A, which causes an in-frame deletion of lysine in the intracellular kinase domain of CSF1R, was absent in the large band presenting the aberrant splicing but was detected in the small band that does not show any aberrant splicing (Figure 3C). Together, we can conclude that the c.1859−119G>A is a splicing mutation generating a novel cryptic splice acceptor site, resulting in a 1-residue deletion and 40-residue insertion in CSF1R. The variant identified in family C, c.1969+115_1969+116del, is located within intron 14, distant from the intron-exon junction and the branch point. ESEfinder suggests that the intronic variant may eliminate several potential binding sites for the splicing factors (Figure 3D). RT-PCR analysis using affected individual-derived fibroblasts indicated that an additional transcript approximately 100 bp larger than the wild-type transcript was detected in the affected individuals (C-III-4 and C-III-6) when NMD was inhibited (Figures 3E and 3F). Sanger sequencing of the larger amplicon revealed insertion of a 99-bp intronic sequence between exons 14 and 15. The cryptic exon contains c.1969+115_1969+116del and an in-frame stop codon resulting in a truncated open reading frame (p.Pro658Serfs∗24) (Figure 3F). In the absence of cycloheximide, the mis-spliced transcript was not present, indicating that the truncated CSF1R would not be translated due to NMD (Figure 3F). The bands corresponding the wild-type mRNA in the affected individuals were significantly weaker than those of the control subjects. Together, we can conclude that c.1969+115_1969+116del is a splicing mutation activating a 99-bp cryptic-exon (Figure 3E) and decreasing the normal splicing simultaneously. The inclusion of the cryptic-exon leads to an in-frame stop codon, inducing NMD.
It has previously been shown that phosphorylation of CSF1R-tyrosine triggered by the ligand binding to the receptor activates multiple kinase pathways including PI3K, ERK1/2, and JNK.19, 20 To evaluate the functional impacts of three CSF1R variants (c.395C>T [p.Pro132Leu], c.1879_1881del [p.Lys627del], c.1859−119G>A [p.Ser620delins40]), we examined the JNK phosphorylation level activated by CSF1 treatment in the cells expressing these mutated proteins. The results indicated that all the variant CSF1Rs had decreased phosphorylation in JNK, compared to the wild-type CSF1R (Figures 3G and 3H). These results demonstrate that all the variants identified in our affected individuals are mutations causing functional deficiency of CSF1R. The milder phenotypes of the affected individuals than those observed in mice with bi-allelic Csf1r mutation suggest that at least one of the mutant alleles in each affected individual would be hypomorphic rather than amorphic.
CSF-1 is the main regulator of mononuclear phagocyte production and function.1 The effects of CSF-1 are mediated by binding to CSF1R, which is expressed in cells including in primitive multipotent hematopoietic cells, mononuclear phagocyte progenitor cells, tissue macrophages, osteoclasts, and microglia.21 CSF1R is activated through its dimerization stabilized by ligand binding and auto-phosphorylation of the tyrosine in intracellular domain, which initiates a series of membrane-proximal kinase cascades.22 Targeted ablation of Csf1r in mice causes severe decrease of macrophage populations, including microglia in brain and osteoclasts in bone;7, 8 however, the precise CSF1R-dependent mechanism in the regulation of osteoclastogenesis and microglial development remains unknown due to the great complexity of CSF1R downstream pathways. Some in vitro studies suggested that the activation of JNK, one of the CSF1R downstream kinases, is critical for macrophage development20 and anti-apoptotic functions of microglial cells.23 In our study, the CSF1R mutations in the affected individuals caused decreased JNK activation, consistent with the reported JNK roles in osteoclastogenesis and microglia development.
The skeletal changes of our affected individuals were variable but corresponded to the DOS-Pyle disease spectrum. DOS is a rare sclerosing bone disease belonging to the group 23 of genetic skeletal disorders (osteopetrosis and related disorders).24 DOS was first defined by Spranger et al. in 1968 to distinguish a syndrome characterized by osteosclerosis and platyspondyly.13 DOS shows sclerotic skull, flat and diffusely dense vertebral bodies, and expanded and radiolucent sub-metaphyseal portions of the long tubular bones with sclerotic diaphysis.25 The skeletal changes of DOS, particularly in the tubular bones and vertebrae, are distinct from osteopetrosis, a main type of sclerosing bone disease. Affected individuals in family A and B presented with these DOS phenotypes (Table 1, Figure 2). DOS is genetically heterogeneous. Mutations in two genes have been reported to cause DOS. One is SLC29A3 (MIM: 612373), where homozygous and compound heterozygous missense mutations were identified in two unrelated individuals.26 The other is TNFRSF11A ([MIM: 603499] alias RANK), where a homozygous splice-site mutation that caused skipping of an exon was found by us in a Turkish individual.16 In this study we found the third gene associated to the DOS phenotype. Notably, DOS caused by the mutations of SLC29A3 and TNFRSF11A do not exhibit the neural degeneration and brain malformations.
In contrast, Pyle disease is an autosomal-recessive condition characterized by striking metaphyseal broadening.14 It belongs to the group 24 of genetic skeletal disorders (other sclerosing bone disorders).24 Although DOS and Pyle disease are classified into different groups, they have considerable phenotypic overlaps. The radiological hallmark of Pyle disease is massive metaphyseal expansion extending into meta-diaphyseal junction of the long tubular bones. Cortices of the expanded meta-diaphyseal segments are very thin, while the mid-diaphyseal cortices are preserved. The vertebral bodies may show mild platyspondyly. There may be very mild sclerosis in the calvarium and skull base. Bone is potentially brittle, but spontaneous fractures are not common. Affected individuals in family C presented with these Pyle disease phenotypes (Table 1, Figures 2T–2X and S2). SFRP4 mutations have been found in Pyle disease27 but not in DOS. Neurological disorders are not reported in the SFRP4-associated Pyle disease. sFRP4 plays a critical role in bone development and remodeling by regulating both osteoblasts and osteoclasts,28 suggesting that the unique phenotype may be associated with both osteoblasts and osteoclasts. The potential link between sFRP4 and CSF1R remains to be established.
Mice deficient for Csf1r recapitulate skeletal and neural involvements found in our affected individuals. Bi-allelic inactivation of Csf1r causes severe sclerosing skeletal dysplasia and gross anatomical abnormalities in developmental brains, leading to early death.7, 8 The skeletal phenotypes of Csf1r−/− mice include increased bone density, flattened and deformed vertebrae, expanded epiphyseal and metaphyseal region with significantly reduced bone mineralization, and poorly formed cortical bone.9 All the skeletal characters are corresponded to the DOS-Pyle disease spectrum, although the severity is different. The affected individuals with bi-allelic CSF1R mutation in our study presented milder skeletal phenotypes than those of Csf1r−/− mice, suggesting at least one partially functioning allele exists in these affected individuals. The brain abnormalities of Csf1r−/− mice are characterized by severe microglial depletion and enlarged ventricles, probably attributing to the early lethality.7 In contrast, Csf1r+/− mice do not show significant phenotypes before adulthood but develop late-onset neural dysfunctions including cognitive and sensorimotor deficits, depression with anxiety-like behavior, and enlarged ventricles. Skeletal abnormalities are not found in Csf1r+/− mice.10 In human, dominant CSF1R mutations cause HDLS, an adult-onset, rapidly progressive neurodegenerative disorder without any skeletal dysplasia. Brain imaging studies disclose a high prevalence of dilated lateral ventricles, white matter lesions, cortical atrophy, thinning of the corpus callosum, and white matter calcifications showing a “stepping stone appearance.”29 The neural phenotypes in the affected individuals in family B and C are highly compatible to HDLS, while the affected individual in family A presents far more severe neural involvements associated with brain malformation. The babies reported by Monies et al., who possibly harbor CSG1R c.1620T>A (p.Tyr540∗), have CNS structural anomalies overlapping with those observed in family A.12 Their skeletal phenotypes are not well described and characterized, but some osteosclerosis is definite. These neonatal case subjects further support our idea that bi-allelic CSF1R mutations produce a syndrome characterized by osteosclerosis with modeling failure and neurological disorder with structural CNS abnormalities.
In this study, we report that CSF1R mutations induce unique skeletal phenotypes in addition to neurologic disorders in humans. Our results suggest that bi-allelic mutations which result in severe decrease of CSF1R dosage produce the skeletal phenotypes, while heterozygous mutations, even those that are suspected to have dominant-negative effects, do not have definite skeletal phenotypes. Thus, severe loss of function (LoF) could be necessary to cause skeletal phenotypes. It is hypothesized that skeletal tissue is more tolerant than brain tissue to CSF1R LoF, although tissue-specific studies of function would be necessary to explore this consideration. In the central nervous system, the amount of microglia and/or its function are likely to be highly regulated. Almost all the dominant CSF1R mutations reported in HDLS are restricted to the intracellular kinase domain.30 These heterozygous mutations were suggested to have a dominant-negative effect via ligand-dependent dimerization and autophosphorylation.31 Unlike previously reported dominant mutations, p.Gln481∗ identified in our study completely abolishes the intracellular part of the receptor that starts at amino acid 537. The RT-PCR analysis for the individual-derived cells demonstrated that the nonsense mutation is subjected to complete NMD (Figure 3A). Thus, p.Gln481∗ cannot exert a dominant-negative effect. Its heterozygotes including the father and the paternal grandfather of family A do not show any severe neural phenotypes corresponding to HDLS. Similarly, another null mutation (p.Tyr540∗) reported by Monies et al. did not cause phenotypes in several heterozygotes.12 These results suggest that the mechanism of action of the CSF1R mutations in HDLS is not haploinsufficiency, but dominant-negative effect causing more than 50% LoF.
Together, our findings provide evidence of a continuous phenotypic spectrum caused by different degrees of CSF1R LoF in humans and imply that the effect of CSF1R inactivation would be sensitive to dosage and be variable among human organs and tissues. Anti-CSF1 or anti-CSF1R drugs are currently in development for treatment of Alzheimer disease, brain injuries, and malignant tumors, in which increased levels of CSF1R or activated CSF1R-dependent inflammation were found.32, 33 Biological insights from this study could give important insights to guide dosage and route of administration of these new drugs. Additionally, hematopoietic stem cell transplantation may be considered as a treatment approach in this condition, since amelioration after the transplantation has been reported in a case subject with HDLS,34 and the phenotype of Csf1r-deficient mouse femurs could be rescued in femoral anlage transplantation experiments.9
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the affected individuals and their family for their help to the study. This study was supported by research grants from Japan Agency for Medical Research and Development (AMED) (contract no. 14525125), the Japan Society for the Promotion of Science (WAKATE B, no. 17K16710), RIKEN-MOST, FAPESP 2015/21783-9/CEPID 2013/08028-1; CNPq 304130/2016-8, and the Australian National Health and Medical Research Council (NHMRC 1068278). The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program.
Published: April 11, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.004.
Contributor Information
Débora Romeo Bertola, Email: debora.bertola@usp.br.
Shiro Ikegawa, Email: sikegawa@ims.u-tokyo.ac.jp.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
AbraOM, http://abraom.ib.usp.br/
ESE finder 3.0, http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home
gnomAD Browser, https://gnomad.broadinstitute.org/
Human Gene Mutation Database (HGMD), https://portal.biobase-international.com/hgmd/pro/start.php
Human Splicing Finder, http://www.umd.be/HSF3/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Stanley E.R., Berg K.L., Einstein D.B., Lee P.S., Pixley F.J., Wang Y., Yeung Y.G. Biology and action of colony--stimulating factor-1. Mol. Reprod. Dev. 1997;46:4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Pixley F.J., Stanley E.R. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Huynh D., Dai X.M., Nandi S., Lightowler S., Trivett M., Chan C.K., Bertoncello I., Ramsay R.G., Stanley E.R. Colony stimulating factor-1 dependence of paneth cell development in the mouse small intestine. Gastroenterology. 2009;137:136–144, 144.e1–144.e3. doi: 10.1053/j.gastro.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menke J., Iwata Y., Rabacal W.A., Basu R., Yeung Y.G., Humphreys B.D., Wada T., Schwarting A., Stanley E.R., Kelley V.R. CSF-1 signals directly to renal tubular epithelial cells to mediate repair in mice. J. Clin. Invest. 2009;119:2330–2342. doi: 10.1172/JCI39087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arceci R.J., Pampfer S., Pollard J.W. Expression of CSF-1/c-fms and SF/c-kit mRNA during preimplantation mouse development. Dev. Biol. 1992;151:1–8. doi: 10.1016/0012-1606(92)90207-w. [DOI] [PubMed] [Google Scholar]
- 6.Arceci R.J., Shanahan F., Stanley E.R., Pollard J.W. Temporal expression and location of colony-stimulating factor 1 (CSF-1) and its receptor in the female reproductive tract are consistent with CSF-1-regulated placental development. Proc. Natl. Acad. Sci. USA. 1989;86:8818–8822. doi: 10.1073/pnas.86.22.8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erblich B., Zhu L., Etgen A.M., Dobrenis K., Pollard J.W. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS ONE. 2011;6:e26317. doi: 10.1371/journal.pone.0026317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai X.M., Ryan G.R., Hapel A.J., Dominguez M.G., Russell R.G., Kapp S., Sylvestre V., Stanley E.R. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 9.Dai X.-M., Zong X.-H., Akhter M.P., Stanley E.R. Osteoclast deficiency results in disorganized matrix, reduced mineralization, and abnormal osteoblast behavior in developing bone. J. Bone Miner. Res. 2004;19:1441–1451. doi: 10.1359/JBMR.040514. [DOI] [PubMed] [Google Scholar]
- 10.Chitu V., Gokhan S., Gulinello M., Branch C.A., Patil M., Basu R., Stoddart C., Mehler M.F., Stanley E.R. Phenotypic characterization of a Csf1r haploinsufficient mouse model of adult-onset leukodystrophy with axonal spheroids and pigmented glia (ALSP) Neurobiol. Dis. 2015;74:219–228. doi: 10.1016/j.nbd.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rademakers R., Baker M., Nicholson A.M., Rutherford N.J., Finch N., Soto-Ortolaza A., Lash J., Wider C., Wojtas A., DeJesus-Hernandez M. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 2011;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monies D., Maddirevula S., Kurdi W., Alanazy M.H., Alkhalidi H., Al-Owain M., Sulaiman R.A., Faqeih E., Goljan E., Ibrahim N. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: implications in variant interpretation. Genet. Med. 2017;19:1144–1150. doi: 10.1038/gim.2017.22. [DOI] [PubMed] [Google Scholar]
- 13.Spranger J., Albrecht C., Rohwedder H.J., Wiedemann H.R. [Dysosteosclerosis--a special form of generalized osteosclerosis] Fortschr. Geb. Rontgenstr. Nuklearmed. 1968;109:504–512. [PubMed] [Google Scholar]
- 14.Edwin P. A case of unusual bone development. JBJS. 1931;13:874–876. [Google Scholar]
- 15.Wang Z., Horemuzova E., Iida A., Guo L., Liu Y., Matsumoto N., Nishimura G., Nordgren A., Miyake N., Tham E. Axial spondylometaphyseal dysplasia is also caused by NEK1 mutations. J. Hum. Genet. 2017;62:503–506. doi: 10.1038/jhg.2016.157. [DOI] [PubMed] [Google Scholar]
- 16.Guo L., Elcioglu N.H., Karalar O.K., Topkar M.O., Wang Z., Sakamoto Y., Matsumoto N., Miyake N., Nishimura G., Ikegawa S. Dysosteosclerosis is also caused by TNFRSF11A mutation. J. Hum. Genet. 2018;63:769–774. doi: 10.1038/s10038-018-0447-6. [DOI] [PubMed] [Google Scholar]
- 17.Guo L., Elcioglu N.H., Mizumoto S., Wang Z., Noyan B., Albayrak H.M., Yamada S., Matsumoto N., Miyake N., Nishimura G., Ikegawa S. Identification of biallelic EXTL3 mutations in a novel type of spondylo-epi-metaphyseal dysplasia. J. Hum. Genet. 2017;62:797–801. doi: 10.1038/jhg.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons C., Dyment D., Bent S.J., Crawford J., D’Hooghe M., Kohlschütter A., Venkateswaran S., Helman G., Poll-The B.T., Makowski C.C., Care4Rare Consortium A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain. 2017;140:3105–3111. doi: 10.1093/brain/awx314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morandi A., Barbetti V., Riverso M., Dello Sbarba P., Rovida E. The colony-stimulating factor-1 (CSF-1) receptor sustains ERK1/2 activation and proliferation in breast cancer cell lines. PLoS ONE. 2011;6:e27450. doi: 10.1371/journal.pone.0027450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himes S.R., Sester D.P., Ravasi T., Cronau S.L., Sasmono T., Hume D.A. The JNK are important for development and survival of macrophages. J. Immunol. 2006;176:2219–2228. doi: 10.4049/jimmunol.176.4.2219. [DOI] [PubMed] [Google Scholar]
- 21.Byrne P.V., Guilbert L.J., Stanley E.R. Distribution of cells bearing receptors for a colony-stimulating factor (CSF-1) in murine tissues. J. Cell Biol. 1981;91:848–853. doi: 10.1083/jcb.91.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung Y.-G., Stanley E.R. Proteomic approaches to the analysis of early events in colony-stimulating factor-1 signal transduction. Mol. Cell. Proteomics. 2003;2:1143–1155. doi: 10.1074/mcp.R300009-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Svensson C., Part K., Künnis-Beres K., Kaldmäe M., Fernaeus S.Z., Land T. Pro-survival effects of JNK and p38 MAPK pathways in LPS-induced activation of BV-2 cells. Biochem. Biophys. Res. Commun. 2011;406:488–492. doi: 10.1016/j.bbrc.2011.02.083. [DOI] [PubMed] [Google Scholar]
- 24.Bonafe L., Cormier-Daire V., Hall C., Lachman R., Mortier G., Mundlos S., Nishimura G., Sangiorgi L., Savarirayan R., Sillence D. Nosology and classification of genetic skeletal disorders: 2015 revision. Am. J. Med. Genet. A. 2015;167A:2869–2892. doi: 10.1002/ajmg.a.37365. [DOI] [PubMed] [Google Scholar]
- 25.Houston C.S., Gerrard J.W., Ives E.J. Dysosteosclerosis. AJR Am. J. Roentgenol. 1978;130:988–991. doi: 10.2214/ajr.130.5.988. [DOI] [PubMed] [Google Scholar]
- 26.Campeau P.M., Lu J.T., Sule G., Jiang M.M., Bae Y., Madan S., Högler W., Shaw N.J., Mumm S., Gibbs R.A. Whole-exome sequencing identifies mutations in the nucleoside transporter gene SLC29A3 in dysosteosclerosis, a form of osteopetrosis. Hum. Mol. Genet. 2012;21:4904–4909. doi: 10.1093/hmg/dds326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiper P.O.S., Saito H., Gori F., Unger S., Hesse E., Yamana K., Kiviranta R., Solban N., Liu J., Brommage R. Cortical-bone fragility--insights from sFRP4 deficiency in Pyle’s disease. N. Engl. J. Med. 2016;374:2553–2562. doi: 10.1056/NEJMoa1509342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haraguchi R., Kitazawa R., Mori K., Tachibana R., Kiyonari H., Imai Y., Abe T., Kitazawa S. sFRP4-dependent Wnt signal modulation is critical for bone remodeling during postnatal development and age-related bone loss. Sci. Rep. 2016;6:25198. doi: 10.1038/srep25198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konno T., Broderick D.F., Mezaki N., Isami A., Kaneda D., Tashiro Y., Tokutake T., Keegan B.M., Woodruff B.K., Miura T. Diagnostic value of brain calcifications in adult-onset leukoencephalopathy with axonal spheroids and pigmented glia. AJNR Am. J. Neuroradiol. 2017;38:77–83. doi: 10.3174/ajnr.A4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson J.L., Suh E., Wood E.M., Lee E.B., Coslett H.B., Raible K., Lee V.M., Trojanowski J.Q., Van Deerlin V.M. Common neuropathological features underlie distinct clinical presentations in three siblings with hereditary diffuse leukoencephalopathy with spheroids caused by CSF1R p.Arg782His. Acta Neuropathol. Commun. 2015;3:42. doi: 10.1186/s40478-015-0219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pridans C., Sauter K.A., Baer K., Kissel H., Hume D.A. CSF1R mutations in hereditary diffuse leukoencephalopathy with spheroids are loss of function. Sci. Rep. 2013;3:3013. doi: 10.1038/srep03013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cannarile M.A., Weisser M., Jacob W., Jegg A.M., Ries C.H., Rüttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer. 2017;5:53. doi: 10.1186/s40425-017-0257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olmos-Alonso A., Schetters S.T., Sri S., Askew K., Mancuso R., Vargas-Caballero M., Holscher C., Perry V.H., Gomez-Nicola D. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain. 2016;139:891–907. doi: 10.1093/brain/awv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichler F.S., Li J., Guo Y., Caruso P.A., Bjonnes A.C., Pan J., Booker J.K., Lane J.M., Tare A., Vlasac I. CSF1R mosaicism in a family with hereditary diffuse leukoencephalopathy with spheroids. Brain. 2016;139:1666–1672. doi: 10.1093/brain/aww066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.