Abstract
We identified individuals with variations in ACTL6B, a component of the chromatin remodeling machinery including the BAF complex. Ten individuals harbored bi-allelic mutations and presented with global developmental delay, epileptic encephalopathy, and spasticity, and ten individuals with de novo heterozygous mutations displayed intellectual disability, ambulation deficits, severe language impairment, hypotonia, Rett-like stereotypies, and minor facial dysmorphisms (wide mouth, diastema, bulbous nose). Nine of these ten unrelated individuals had the identical de novo c.1027G>A (p.Gly343Arg) mutation. Human-derived neurons were generated that recaptured ACTL6B expression patterns in development from progenitor cell to post-mitotic neuron, validating the use of this model. Engineered knock-out of ACTL6B in wild-type human neurons resulted in profound deficits in dendrite development, a result recapitulated in two individuals with different bi-allelic mutations, and reversed on clonal genetic repair or exogenous expression of ACTL6B. Whole-transcriptome analyses and whole-genomic profiling of the BAF complex in wild-type and bi-allelic mutant ACTL6B neural progenitor cells and neurons revealed increased genomic binding of the BAF complex in ACTL6B mutants, with corresponding transcriptional changes in several genes including TPPP and FSCN1, suggesting that altered regulation of some cytoskeletal genes contribute to altered dendrite development. Assessment of bi-alleic and heterozygous ACTL6B mutations on an ACTL6B knock-out human background demonstrated that bi-allelic mutations mimic engineered deletion deficits while heterozygous mutations do not, suggesting that the former are loss of function and the latter are gain of function. These results reveal a role for ACTL6B in neurodevelopment and implicate another component of chromatin remodeling machinery in brain disease.
Keywords: intellectual disability, seizure, ACTL6B, stem cells, genetic engineering, neurodevelopment
Introduction
ACTL6B (MIM: 612458) encodes an actin-related protein (ARP), which are a class of proteins that resemble actin and have roles in chromatin remodeling and histone acetylation.1 Though ACTL6B, known as BAF53B, may interact with multiple complexes in a particular spatiotemporal order, most investigations have focused on its role in the BAF (BRG1/BRM-Associated Factor) or SWI/SNF complex,2 which serves as an important regulator of gene expression by remodeling nucleosomes in an ATP-dependent fashion.3, 4, 5 In order to regulate different sets of genes during development, BAF subunits can be exchanged with homologous alternatives.3 One such switch in BAF subunit composition occurs in developing neural cells as they exit the cell cycle. During this time, the neural progenitor specific BAF (npBAF) complex transitions to the neural-specific BAF (nBAF) complex through the exchange of several subunits, including BAF53A for its paralog BAF53B.6 This is partly achieved through increased expression of miR-9∗ and miR-124 in post mitotic neurons, which repress the expression of the gene that encodes BAF53A, ACTL6A (MIM: 604958).7 nBAF complexes can bind the transactivator CREST and be recruited to genes crucial for dendritogenesis through interactions mediated by BAF53B.8 As a result, loss of BAF53B protein levels during neuronal development results in impaired dendritic outgrowth. An Actl6b knock-out (KO) mouse has previously been generated and found to have deficits in dendritic spine and synapse function, leading to impaired long-term memory and poor survival.9
While different genes that contribute to the BAF complex have been found to be associated with human disease (e.g., Nicolaides-Baraitser syndrome [MIM: 601358] and SMARCA2 [MIM: 600014]; Coffin-Siris syndrome [MIM: 135900] and ARID1B [MIM: 614556]),10, 11 ACTL6B has not been conclusively reported to have a deleterious role in human neurological diseases. In this study, we identified individuals with neurodevelopmental disorders with either inherited recessive mutations or dominantly acting de novo mutations in ACTL6B and sought to understand how mutations in ACTL6B might affect the development of human neurons.
Material and Methods
Description of Studied Individuals
Individuals had whole-exome sequencing as part of local neurodevelopmental studies on developmental delay and intellectual disability, autism, or epilepsy (R1, R2a/b, R3a/b, R4, R5, R7, R9, R10, D2, D3, D7, D8). Informed consent for participating in the genetic studies was obtained on protocols approved by institutional review boards of local hospitals. Individuals D1 and D4 were enrolled in the DDD study and provided informed consent for this study. Other individuals had exome sequencing at GeneDx as part of clinical care (individuals R6, R8a/b, D5, D6, D9), and after ACTL6B was identified as a candidate gene, provided informed consent for the sharing of photographs or samples as applicable.
Experimental Procedures for Sequencing
DNA was extracted from peripheral blood from affected individuals and parents using standard protocols. For individuals who had whole-genome sequencing (R1, R2a/b, R10), the DNA libraries were prepared by using the Illumina TruSeq DNA PCR-Free kits using the manufacturer’s protocol. For individuals who had whole-exome sequencing, the exome libraries were prepared using Agilent SureSelect kits (R3ab, R4, R6, R8ab, R9, D1, D2, D4-D9), Roche-NimbleGen EZ exome kits (R5, D3), and Illumina Nextera kits (R7). More details included in Tables 1 and 2. All libraries were then sequenced on Illumina HiSeq systems.
Table 1.
Pathogenic Variants and Key Clinical Information of Individuals with Bi-allelic Mutations in ACTL6B
| Individual | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8a | R8b | R9 | R10 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inheritance | recessive, homozygous | recessive, compound heterozygous; similarly affected brother passed away at 5 y | recessive, homozygous; similarly affected brother passed away at 4 y | recessive, compound heterozygous | recessive, compound heterozygous | recessive, compound heterozygous | recessive, homozygous | recessive, homozygous | recessive, homozygous; sister of R8a | recessive, compound heterozygous | recessive, homozygous | |||||
| Coding change (NM_016188.4) | c.441_443delCTT | c.695delC | c.1275C>A | c.1279del | c.389G>A | c.556C>T | c.852C>G | c.740G>A | c.1231C>T | c.669+1G>A | c.289C>T | c.1045G>A | c.1045G>A | c.724C>T | c.617T>C | c.1279del |
| Protein change (NP_057272.1) | p.Phe147del | p.Pro232Glnfs∗24 | p.Cys425∗ | p.∗427Aspext∗33 | p.Arg130Gln | p.Gln186∗ | p.Tyr284∗ | p.Trp247∗ | p.Gln411∗ | splicing | p.Arg97∗ | p.Gly349Ser | p.Gly349Ser | p.Gln242∗ | p.Leu206Pro | p.∗427Aspext∗33 |
| gnomAD MAF | 0.00001444, no homozygotes | absent | absent | absent | 0.000008132, no homozygotes | absent | absent | absent | absent | absent | 0.000004064, no homozygotes | 0.00001219, no homozygotes | 0.00001219, no homozygotes | absent | absent | absent |
| Age at assessment | 3 y (F) | 5 y (M) (passed away at age 5) | 11 m (M) (passed away age 5) | 8 y (F) | 5 m (F) | 12 m (M) (passed away age 2) | 4 y (F) | 6 y (F) | 5 y (F) | 14 m (F) | 4.5 y (F) | |||||
| Head circum. (cm) | 43 (−3.5 SD) | NA | 44 (3rd) | 50.3 (10th %ile) | 38.4 (−3.0 SD) | 42 (−2.4 SD) | normal | 41.5 (−3.8 SD) at 18 m | 39 (7th %ile) at 4 m | 43 (−2.5 SD) | 47 (−2.8 SD) | |||||
| ID, DD | +, severe | + | + | +, severe | +, severe | +, severe | +, severe | +, severe | +, severe | +, severe | + | |||||
| Speech | − | − | NA | − | NA | − | NA | − | − | − | − | |||||
| Ambulation | − | − | NA | − | NA | − | NA | − | − | − | +, with support | |||||
| Axial hypotonia | + | + | + | + | + | + | + | + | + | + | + | |||||
| Limb spasticity | + | + | + | + | + | + | + | + | + | + | + | |||||
| Feeding difficulties | + | + | + | − | + | + | + | + | + | + | − | |||||
| Epilepsy | + | + | + | + | + | + | + | + | + | + | + | |||||
| Seizures (age at beginning) | 3 m | 3 y | NA | NA | 2 m | neonatal (25 days) | infancy | infancy | infancy | antenatal | 9 m, infantile spasms | |||||
| Seizure types | myoclonias 2–6 per day | complex partial | NA | NA | tonic and myoclonic | focal onset epilepsy, progressed to infantile spasms | NA | NA | NA | myoclonic and tonic seizures | tonic and myoclonic | |||||
| EEG anomalies | multifocal epileptic activity | NA | multifocal epileptic activity | NA | multifocal epileptic activity | multifocal interictal epileptiform spike discharges, lack of posterior dominant rhythm | multifocal EEG abnormalities | generalized slowing of background rhythms | generalized slowing of background rhythms | multifocal epileptic activity, esp. left hemisphere | multifocal epileptic activity, esp. left hemisphere | |||||
| MRI | prominent subarachnoid spaces and small corpus callosum | normal | mild T2 hyperintensity in frontal periventricular white matter | mild T2 hyperintensity in frontal periventricular white matter | 2 m: symmetric signal changes in the brainstem and in the dorsal medulla oblongata, possibly also around the dentate nucleus | 3 weeks: asymmetric ventricles, cortical dysplasia right parietal lobe; 9 m: cerebral atrophy, hypoplasia of corpus callosum | NA | 5 m: significantly decreased white matter throughout, extremely thin corpus callosum. Normal MR spectroscopy | 10 m: periventricular leukomalacia with white matter volume loss, overall brain volume loss, delayed myelination and thinning of corpus callosum. Normal MR spectroscopy | thin corpus callosum; high signal intensity doral globus pallidus/putamen; some asymmetry gyral patern | 3.5 y: cerebral and cerebellar atrophy, thin corpus callosum | |||||
Table 2.
Pathogenic Variants and Key Clinical Information of Individuals with De Novo Mutations in ACTL6B
| Individual | D1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 | D9 | D10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Coding change (NM_016188.4) | c.1027G>A | c.1027G>A | c.1027G>A | c.1027G>A | c.1027G>A | c.1027G>A | c.1027G>A | c.1027G>A | c.230A>G | c.1027G>C |
| Protein change (NP_057272.1) | p.Gly343Arg | p.Gly343Arg | p.Gly343Arg | p.Gly343Arg | p.Gly343Arg | p.Gly343Arg | p.Gly343Arg | p.Gly343Arg | p.Asp77Gly | p.Gly343Arg |
| gnomAD MAF | absent | absent | absent | absent | absent | absent | absent | absent | absent | absent |
| Age at assessment | 5 y 6 m (M) | 29 y (F) | 6 y 6 m (M) | 5 y 9 m (F) | 4 y 6 m (F) | 3 y (F) | 21 y (F) | 2 y 6 m (F) | 8 y (F) | 12 y (F) |
| Head circum. (cm) | 49 (−2.1 SD) | 53 (11th %ile) | 51 at age 5 year (50th %ile) | 48.6 (−2.2 SD) | 48 (2nd %ile) | 48.0 (20th %ile) | 52.2 (−2.0 SD) | 45.5 (−0.1 SD) | 50th–75th %ile | 52 (−1, −2 SD) |
| Degree of ID/DD | severe | severe | severe | severe | severe | severe | severe | severe | severe | severe |
| Speech | − | − | − | 10–20 words, receptive skills better | − | −; gestures | − | − | −; gestures; receptive skills better | one word |
| Ambulation | − | + | limited | delayed; wide-based gait | − | − | − | − | NA | delayed; wide-based gait |
| Hypotonia | NA | + | + | NA | + | + | NA | - | NA | + |
| Autism spectrum disorder | NA | + | unknown | + | NA | NA | NA | − | + | − |
| Features of ASD | NA | NA | + | NA | stereotypies | NA | handwringing | − | NA | stereotypies |
| Seizure disorder | − | − | − | − | − | − | Infantile spasms and GTCS | − | NA | − |
| MRI | NA | NA | NA | NA | NA | thinning of the corpus callosum | Generalized atrophy at 2y | mild periventricular gliosis | NA | Nl |
| Wide or prominent forehead | + | + | + | + | − | − | + | + | + | − |
| Hypertelorism | + | + | + | − | − | + | − | + | + | − |
| Wide mouth | + | + | + | − | − | − | + | + | NA | + |
| Short phalanges or nails | NA | + | − | + | − | − | + | − | + | − |
Analysis of Sequencing Data
Sequences were aligned using BWA, GATK, Novoalign, Isaac, or LifeScope software. The variants were called using GATK, SAMtools, Annovar, CarpeNovo, Isaac, LifeScope, and in-house pipelines. More details can be found in Tables S1 and S2. After identification of candidate variants in ACTL6B, their segregation was confirmed by Sanger sequencing using standard protocols.
Fibroblast Reprogramming to Induced Pluripotent Stem Cells (iPSCs)
Fibroblasts were obtained from biopsies or from the Coriell Institute (Table S3) and cultured in DMEM (Invitrogen) supplemented with 10% bovine serum albumin (Invitrogen). Fibroblasts were reprogrammed using episomal reprogramming vectors containing Oct4, Sox2, Myc3/4, Klf4, ShRNA P53 (ALSTEM), and a puromycin resistance gene using a Neon Transfection System (Invitrogen). Following transfection, cells were plated on tissue culture plates coated with Matrigel (Corning) in TesR-E7 media (Stem Cell Technologies) supplemented with 2 μg/mL puromycin (Sigma). After 48 h of puromycin selection, fresh TesR-E7 media was exchanged, until distinct and robust iPSC colonies formed, at which point mTESR1 media (Stem Cell Technologies) was used to maintain and proliferate the colonies. Quality control experiments for iPSCs include mycoplasma testing, short tandem repeat profiling to ensure no sample mix-ups, assessment of endogenous pluripotency factor, immunocytochemistry for pluripotency markers, and molecular karyotyping.
Molecular Karyotyping
To ensure no chromosomal abnormalities occurred as a result of iPSC induction or gene editing, DNA from all generated iPSC lines was sent to Prince of Wales Hospital (Shatin, Hong Kong) for sequencing on an Ion Torrent Hi-Q Sequencer (Thermofisher). Samples were sequenced with an average of 4 million 150 bp reads per sample, for an average coverage of 0.0014×. Analysis was preformed using CNV-Seq.12 Positive controls included cells of origin and cells from families with first-degree relationships where we could detect Mendelian inheritance of CNVs > 1 Mb.
Differentiation of iPSCs to Forebrain Neural Progenitor Cells (NPCs)
iPSCs were differentiated to forebrain NPCs according to our previously described methods.13, 14 iPSC colonies were dissociated and resuspended in DMEM/F12 media supplemented with N2 (Invitrogen) B27 (Invitrogen) and BSA (1 mg/mL), Y27632 (10 μM; AdooQ Bioscience), SB431542 (10 mM; Selleckchem), and Noggin (200 ng/mL; GenScript) onto non-adherent plates to form organoids. After 1 week of maintenance as organoids, cells were dissociated and plated on Matrigel-coated plates in DMEM/F12 supplemented with B27, bFGF (20 ng/mL), EGF (20 ng/mL), and laminin (1 μg/mL) for a further 7 days of differentiation, with media exchanged every 3 days. Cells were assessed for NPC morphology and stained for markers of forebrain NPCs (PAX6, SOX2, TUJ1) and OCT4.
Differentiation of NPCs to Post-mitotic Neurons
For short-term (5 days) differentiation, NPCs were plated in DMEM/F12 media supplemented solely with B27. If longer-term (>5 days) differentiation was required, NPCs were plated in BrainPhys SM1 (Stem Cell Technologies) and N2-A supplemented media (Stem Cell Technologies). 50% of this media was exchanged every 3 days. Previous work has shown that neurons generated using this methodology express both GABAergic (∼30%) and glutamatergic (60%) markers14 and are negative for midbrain markers, such as tyrosine hydroxylase. Approximately 5%–10% of cells stain for GFAP, an astrocyte marker.
Whole-Cell Recordings
For whole-cell patch-clamp recordings, individual coverslips containing differentiated hIPSC-derived neurons were transferred into a heated recording chamber and continuously perfused (1 mL/min) with BrainPhys Neuronal Medium (Catalog # 05791; StemCell Technologies) bubbled with a mixture of CO2 (5%) and O2 (95%) and maintained at 25°C. Whole-cell patch clamp recordings were obtained using borosilicate pipettes (3–6 MΩ), filled with intracellular solution that contained the following (in mM): 5 HEPES, 2 KCl, 136 potassium gluconate, 5 EGTA, 5 Mg-ATP, 8 creatine phosphate, and 0.35 GTP. The pH was adjusted to 7.27 with KOH, and the osmolarity adjusted with distilled water or concentrated potassium gluconate if needed to between 295 and 298 mOsm with an osmometer (3320; Advanced Instruments). After a recording was completed, the nominal membrane potential in voltage- and current-clamp recordings was corrected for the calculated 10 mV liquid junction potential. All potential values reported reflect this correction. Once whole-cell recording had been established, neurons were routinely held in voltage clamp at −70 mV except when examining changes in the resting membrane potential and rheobase, which was performed in current clamp. Cells were studied only if they exhibited a stable holding current and access resistance for at least 10 min before experimental manipulations. Data were acquired using a Digidata 1550A/ Multiclamp 700B (Axon Instruments) and Clampex 10.5 (Molecular Devices). Currents were filtered at 2 kHz and digitized at 20 kHz.
CRISPR/Cas9 Gene Editing
A double nickase CRISPR/Cas9 gene editing system with a Paprika RFP (pRFP) reporter and gRNA targeting the first exon of the ACTL6B was used for KO experiments. For ACTL6Bext33 repair experiments, a wild-type CRISPR/CAS9-pRFP gene editing system was used to target the mutation in the stop codon of exon 14 of ACTL6B. 1 μg of construct was added per transfection reaction, and transfection was carried out simultaneously with iPSC induction to ensure clonality, as previously described.14 After transfection, cells were selected for puromycin resistance and RFP visualization as described8 allowing for cell expansion from a single edited fibroblast. Potentially edited colonies were expanded and stored as cell lines after which DNA was extracted and Sanger sequenced at Genome Quebec.
RNA Sequencing
RNA samples with RIN values > 9.0 were submitted to Genome Quebec for RNA sequencing. Eight libraries were run per lane of an Illumina HiSeqV4 2500 flow cell (125 bp paired-end reads), which achieved an average of ∼40 million reads per library. For bioinformatic processing, we used FASTX-Toolkit, TopHat15 Bowtie2,16 and Cufflinks217 with default parameters to preprocess, align, and assemble reads into transcripts, estimate abundance, and test differential expression. More detailed methods can be found in Chen et al.18
Western Blot
Cells were lysed with RIPA buffer (Sigma) supplemented with SIGMAFAST Protease Inhibitor Tablets (Millipore-Sigma). Protein concentrations were determined using a Pierce BCA Protein Assay Kit (ThermoFisher). Approximately 15 μg of protein was loaded per well in Mini-PROTEAN TGX Stain-Free Precast Gels (Biorad). Gels were run at 150V for approximately 75 min, and then transferred to a nitrocellulose membrane using a Trans-Blot Turbo Transfer System (Biorad). Membranes were blocked in 4% non-fat milk dissolved in TBS-T buffer (tris-buffered saline-tritonX; Sigma-Aldrich) for 20 min and then incubated with primary antibodies overnight at 4°C with shaking. Blots were washed three times in TBS-T for 5 min and then incubated with appropriate mouse or rabbit secondary antibodies for 1 hour at room temperature. Blots were washed a further three times in TBST for 5 min, then imaged using a ChemiDoc XRS+ System (Biorad). Blots were imaged and analyzed using ImageLab (Biorad) software, and statistical analysis was preformed using Student’s t tests when two samples conditions were present and a one-way ANOVA when more than two sample conditions were present. Blots were normalized to β-actin. Further details on the antibodies used for WB can be found in Table S5.
Quantitative PCR
Reverse transcriptions were done on the total RNA fraction in order to obtain cDNA in 40 μL volume containing 1 μg of total RNA, 0.5 μg random primers, 0.5 mM dNTPs, 0.01 M DTT, and 400 U M-MLV RT (Invitrogen). The reactions were performed in a total volume of 20 μL on a 384-well plate either using an Applied Biosystems 7900 HT (Applied Biosystems) or a QuantStudio 6 (Thermofisher) PCR Machines. For each well, PCR mix included 10 μL of Power SybrGreen PCR Mastermix (Life Technologies), 1 μL of primers/probe mix, 2 μL of cDNA, H20 up to 20 μL. Serial dilutions of a mix of cDNA ranging between 0.003052 ng and 50 ng were used to generate a calibration curve for an absolute quantification. Protein levels were given as a ratio between the relative quantities of the gene of interest and the endogenous control. GAPDH was used as internal control for normalization. The normalized expression levels were then compared between cell lines using either a Student’s t test or an ANOVA with post hoc t test. Further details on the primers used for qPCR can be found in Table S6.
Immunofluorescence
Adherent cells were washed with PBS, then fixed with 3% paraformaldehyde (Sigma-Aldrich) on slides for 15 min. Samples were permeabilized with 0.5% TX-100 (Sigma-Aldrich) in 0.5% PBS-BSA for 15 min, and then blocked in 0.5% PBS-BSA for an additional 15 min. Primary antibodies were added in appropriate dilutions (Table S5) in 0.5% PBS-BSA and added to samples for 30 min. Samples were washed in 0.5% PBS-BSA containing an appropriate dilution of secondary antibody (Table S5) was added to the samples and incubated for 30 min in the dark. Samples were washed with 0.5% PBS-BSA. Samples were then visualized on an Apotome Florescent microscope (Zeiss). Neurolucida Tracing Software (MBF Bioscience) was used to measure nuclei surface area, soma surface area, and projection length. Images were processed and scale bars added in ImageJ.
Chromatin Immunoprecipitation (ChIP) Sequencing
Samples were prepared for ChIP-Seq and ChIP-qPCR using a Magna ChIP-Seq kit (Millipore-Sigma). Cells were cross-linked at day 0 and day 5 of differentiation by immersion in 37% formaldehyde. Glycine was added at a final concentration of 125 μM to the samples to inactivate cross-linking. DNA was sheared using a S220 Sonicator (Covaris) and precleared using a protein A or G agarose beads. All samples were then probed using both a mouse monoclonal (Santa Cruz, sc-17796) and rabbit polyclonal (Santa Cruz, sc-10768) antibody directed against BRG1 overnight at 4°C. An IGG control was ran for both rabbit polyclonal and mouse monoclonal antibodies using a pooled sample composed of equal parts of all samples used for ChIP. A 0.2 M glycine solution (pH 2.6) was used for elution of cross-linked proteins and DNA from the beads. DNA was purified using Agencourt AMPure XP Beads (Beckman Coulter). Libraries were constructed using an NGS Library Preparation Kit (Millipore-Sigma) and sent to Genome Quebec, where they were sequenced using an Illumina HiSeqV4 2500 flow cell (125 bp paired-end reads) with between 11 and 12 samples sequenced per lane.
ChIP-Seq Analysis
Quality Trimming and Pre-processing
Sequencing adaptors were clipped using Trim Galore. Quality trimming was done with same tool. A phred score cut-off value of 20 was used. Reads shorter than 20 bp were filtered out. Reads were aligned to the Human Reference Genome (hg19) using BWA software v.0.7.10. Resulting bam files were filtered for minimal mapping quality (MAPQ ≥ 20) and all alignments with samflag 4 (read unmapped) were excluded using SAMtools (v.0.1.19). Duplicates reads were removed using the MarkDuplicates module of Picard (v.1.141) with the option REMOVE_DUPLICATES = true.
Peak Calling
The identification of ChIP-seq enriched regions (peaks) was performed using MACS2 (v.2.1.1); (macs2 callpeak --format BAM --broad --nomodel -q 0.05 --broad-cutoff 0.1 --extsize 500). Differential binding-sites analysis were done using the DiffBind Bioconductor R package (v.2.6.6). Diffbind calls some of the DESeq2 (v.1.18.1) functions to perform the contrast analysis between pairwise p.∗427Aspext∗33 or control D5 versus D0 groups (dba.analyze (method = DBA_DESEQ2)). For each comparison, DiffBind generated a set of consensus peaks with the requirement that peaks must be in at least two of the samples (minOverlap = 2). Standardized differential analysis was then performed using the following default settings for dba.analyze: method = DBA_DESEQ2, FDR ≤ 0.05, bSubControl parameter set to TRUE, bFullLibrarySize set to TRUE. Thus, raw number of reads in the control sample was subtracted and the library size was computed for each sample and used for the normalization. sizeFactors is called with the number of reads in the BAM files for each ChIP sample, divided by the minimum of these. The final normalized counts returned are the raw reads (adjusted for control reads) divided by the normalization factors (result of calling sizeFactors()). Significantly different peaks were then annotated with HOMER (v.4.7) using RefSeq annotations (distal_distance = −10000, distance5d_lower = −10000, distance5d_upper = −100000, gene_desert_size = 100000, proximal_distance = −2000).
Results
Identification of French-Canadian Families with Homozygous Mutations in ACTL6B
By exome sequencing of families with neurodevelopmental disorders in Quebec, we identified a family with two children with a homozygous mutation in ACTL6B that eliminates the stop codon (c.1279del, GenBank: NM_016188.4) and extends the reading frame by an additional 33 amino acids (p.∗427Aspext∗33; GenBank: NP_057272.1; individual R3; Figure 1, Tables 1 and S1).
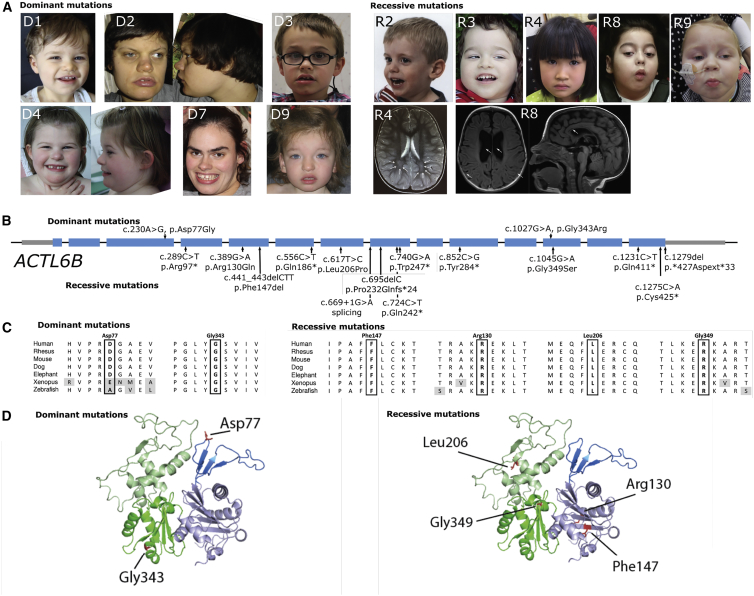
Figure 1.
Location of Mutations in ACTL6B Found in Individuals with Potential Recessive or Dominant Disease-Causing Mutations
(A) Photos of individuals with ACTL6B mutations. Note broad mouth of individuals D1, D2, D3, and D7, diastema in D1, D3, D7, bulbous tip of the nose in all D individuals, and hypertelorism with telecanthi in individual D8. Lower right: MRI images of individuals with recessive ACTL6B mutations. For individual R4, note white matter T2 hyperintensity (arrows). For individual R8, note enlarged lateral ventricles and asymmetric gyral pattern (left, arrows). On the right, note thin corpus callosum (arrows).
(B) Linear graph of mutations in ACTL6B (introns not drawn to scale).
(C) Conservation of the residues affected by amino acid substitutions.
(D) 3D model generated with SWISSMODEL based on S. cerevisiae Arp4 (yeast homolog of ACTL6B), visualized with Swiss-PdbViewer showing that recessive mutations are not focused in one region. Note, however, that the dominant mutations seem to lie at the periphery of the protein and thus they might affect protein-protein interactions.
Sequencing both affected children, their unaffected older brother, and both parents from family R3 revealed that c.1279del was the only mutation identified in the family that was protein altering, followed an autosomal-recessive inheritance model, and was absent from all genomic databases. Both parents and the unaffected brother were carriers, and all are healthy. The phenotype of the disorder is severe: both brothers died early in life (4 and 6 years) of aspiration asphyxiation, were non-verbal, non-ambulatory, and required 24-h care for all needs. Parents reported incessant crying (10+ h per day), seizures beginning at 3 months, and sleep difficulties (Table 1). An MRI of the brain was provided and had no indications from the reviewing radiologist. Careful tracing of the lineage of family R3 using Catholic Church records7 revealed a common ancestor, which we determined to be the most likely origin of the mutation in family R3 (Figure S1A and Supplemental Material and Methods). We genotyped five other members from family R3 across four generations and could identify appropriate inheritance of the mutation from the predicted founder (Figure S1B). The R3 parents of the proband can be traced to a brother and sister going back six generations in the mother and five generations in the father, an event unknown to the R3 parents prior to birth of the probands (Figure S1B).
Another French-Canadian family (defined as both great-grandparents being born in the province of Quebec, Canada) with child R10 (Table 1) also had this same mutation with almost identical phenotype, suggesting that this mutation is not private to the R3 family but rather may be a specific but rare variant in the French-Canadian population. We assume that families R3 and R10 are distantly related but could not identify the branch point at which the pedigrees may overlap.
Bi-allelic Mutations in ACTL6B Cause a Severe Neurodevelopmental Disorder
We were able to identify eight additional families with a similar phenotype (Tables 1 and S1) with bi-allelic mutations. A majority of these identified mutations resulted in premature termination codons and were located in highly conserved sequences (Figures 1B and 1C). We considered it very likely that most of these mutation sites resulted in nonsense-mediated decay (NMD) of the transcript, as they occur well in advance of the penultimate exon,19 and strongly suggests that the disease is due to loss of function of ACTL6B. However, some mutations including the c.1279del mutation were located in the final exon of ACTL6B (Figure 1B) and were therefore not predicted to lead to NMD.19 Heterozygous stop mutations are present in healthy parents, suggesting a recessive disorder. Selected case vignettes can be found in the Supplemental Note.
De Novo Missense Mutations at Specific Loci in ATCL6B Cause a Different, Severe Neurodevelopmental Disorder
Over the course of identifying subjects with mutations in ACTL6B, we found ten individuals with heterozygous de novo missense mutations in ACTL6B with hypotonia, intellectual disability, developmental delay, autism, and Rett-like stereotypies such as handwringing (Figure 1, Tables 2 and S2). Detailed case vignettes of some subjects are presented in the Supplemental Note. This was surprising, given that we observed heterozygous stop/frameshift mutations in healthy individuals arguing for a recessive inheritance model. Nine of ten of these individuals possess the same well-conserved c.1027G>A mutation (p.Gly343Arg) (Figure 1C and Table 2). p.Gly343Arg (GenBank: NM_016188.4; hg19: chr7: 100244258; exon 12) is not seen in the 60,706 ExAC subjects. The same holds true for the other observed variant, p.Asp77Gly (GenBank: NM_016188.4; hg19: chr7: 100253082; exon 3), both of which are likely gain-of-function mutations since heterozygous stop mutation carriers have no disease. 3D modeling of the de novo dominant and the recessive bi-allelic mutations (Figure 1D) shows no spatial clustering of mutation sites. BAF53B has an actin-related domain, which is subdivided into four subdomains.20 Subdomains I and III are structural and also contain residues that interact with ATP.21 Subdomain II is the smallest domain and enables the protein to have polar and non-polar orientations. Previous work has shown that mutations in this subdomain impair dendritic outgrowth.4 Subdomain IV interacts with subdomain 120 and is necessary for the interaction of the protein with actin.21 We mapped these subdomains onto a model of BAF53B derived from S. cervisiae; ARP4 structure and found that the variants occurred in all subdomains. Specifically, p.Phe147del, p.Cys425, p.Arg130Gln, and p.Gln411∗ variants occur in subdomain I, the p.Asp77Gly variant occurred in subdomain II, p.Gly343Arg, p.Gly349Ser, and p.Arg130Gln occur in subdomain III, and p.Gly425∗ occurs in subdomain IV (Tables 1 and 2). We did not find any concentration of mutations in a particular domain.
Two other BAFopathies, Nicolaides-Baraitser syndrome (MIM: 600014) and Coffin-Siris syndrome (MIM: 614556), so called because they affect genes that code for proteins that can be incorporated into the BAF complex, have sparse scalp hair and coarse facial features, though this is a wide spectrum in affected individuals. We obtained images of several probands in this study and did not observe coarse features in the majority of subjects (Figure 1A). However, in individuals with the dominant mutations, we did find common features such as a wide mouth, diastema, and bulbous tip of the nose. In the case of MRI brain structure, this was grossly normal, with subtle but not specific features (common across many MRI scans of children with neurodevelopmental diseases) detected for some individuals (Tables S1 and S2).
Modeling the p.∗427Aspext∗33 Variant in Human Neurons
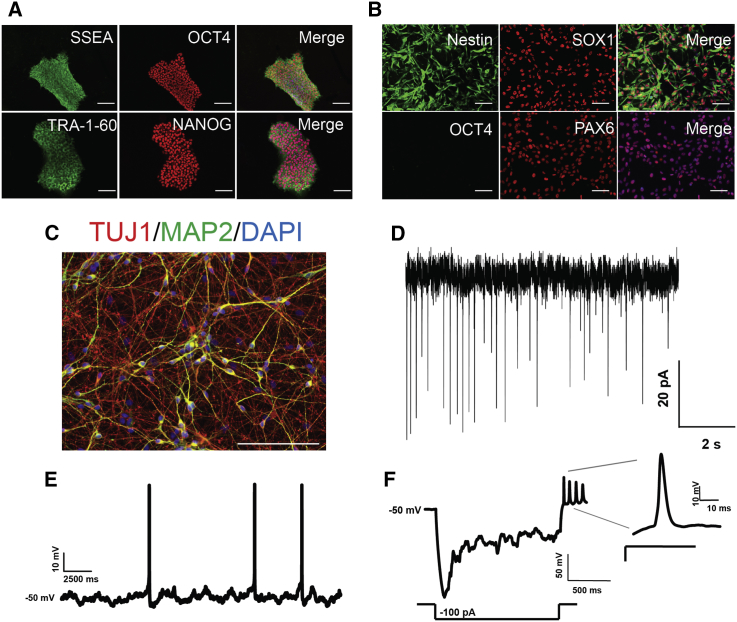
Human stem cells are a powerful model for functional genetic studies as mutations can be assayed on a relevant genetic background and are amenable to genetic engineering. All iPSC lines generated in this study had normal chromosomal integrity, presented typical hallmarks of pluripotency (Figure 2A), including expression of endogenous pluripotency genes (Figures S2A and S2B), and had the capacity to differentiate into all three germ layers (Figure S2C). iPSCs were utilized to generate forebrain neural progenitor cells (NPCs), which expressed neural- and forebrain-specific markers (Figures 2B and S3). Mature neurons generated from wild-type cells expressed markers of cortical neurons and displayed electrophysiological characteristics typical of high-quality iPSC-derived neurons, including spontaneous action potentials and excitatory post-synaptic currents (Figures 2D–2F).
Figure 2.
Generation of iPSC-Derived Neurons for BAF53 Studies
(A) Representative images of quality control staining done on iPSCs.
(B) Representative quality control staining on NPC cultures.
(C) Representative staining of control cells for TUJ1 and MAP2 at D15 of differentiation.
(D) Representative trace of miniature EPSCs from D25 neurons held at −40 mv.
(E) Representative recordings showing spontaneous activity of D25 neurons in current-clamp mode.
(F) Trace of a hyperpolarizing pulse showing a depolarizing sag followed by multiple rebound action potentials. The first action potential is shown at a higher temporal resolution.
All scale bars represent 40 μm.
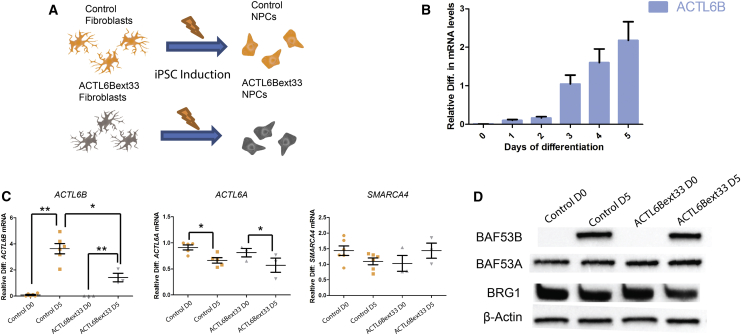
To assess the validity of iPSC-derived neurons to model ACTL6B mutation syndrome, we sought to recapitulate the developmental expression increase2 in ACTL6B in wild-type neurons, where ACTL6B expression is absent from dividing cells but is present in post-mitotic cells.2 We found that ACTL6B increased in expression from day 1 to day 5. To minimize time in culture (which can increase experimental variation), we selected 5 days differentiation as our time point for post-mitotic transcriptomic analysis, where we could be confident ACTL6B would be well expressed (Figures 3A and 3B). To characterize the basic expression pattern of key genes involved in the BAF complex in both p.∗427Aspext∗33 and control cells, we assessed the expression of ACTL6B, ACTL6A, and SMARCA4 (MIM: 603254), a core DNA binding component of the BAF complex. Genes were assessed at day 0 (D0) and day 5 (D5) of differentiation. ACTL6B expression increased significantly in both p.∗427Aspext∗33 and control cells as cells differentiated. We also detected a significant decrease of ACTL6B in p.∗427Aspext∗33 compared to control cells at D5 (Figure 3C). ACTL6A had high expression in proliferating cells with a significant decrease after 5 days, yet was still clearly expressed at day 5 in both control and p.∗427Aspext∗33 cells (Figure 3C). We detected no significant difference in the expression level of SMARCA4 between any cell line or time point (Figure 3C). To confirm and validate these mRNA-based data, we performed western blot on protein extracted from p.∗427Aspext∗33 and control cells at proliferative and post-mitotic time points (Figure 3D). These data suggest that there is no difference in protein level of any of BAF53A, BAF53B, and BRG1 between control and p.∗427Aspext∗33 cells. By developmental period (proliferating and post-mitotic), we observe consistent protein levels of BAF53A and BRG1 and absent BAF53B in proliferating cells.
Figure 3.
Comparison of Control and ACTL6Bext∗33 before and after Expression of ACTL6B
(A) Diagram illustrating the production of control and ACTL6Bext∗33 iPSC-derived NPCs from fibroblasts.
(B) ACTL6B expression normalized to GAPDH expression plotted against number of days of differentiation of NPCs. n = 4, error bars represent standard error around the mean.
(C) Expression of key genes in the SWI/SWF complex in 706 ACTL6Bext∗33 and control NPCs in proliferating and post-mitotic states. Genes are normalized to GAPDH expression. n ≥ 3; Student’s t test, ∗p < 0.05, ∗∗p < 0.01.
(D) Western blots assessing the level of proteins encoded by the genes displayed in (C).
Engineered Homozygous Deletion of ACTL6B in Human Neurons Causes Severe Loss of Dendrites
What is the function of ACTL6B in developing human cells and what is its role in human disease? Our previous experiments suggest that BAF53B is specific to post-mitotic cells, as reported in rodents,22 so we opted to inactivate ACTL6B to determine cellular phenotypes resulting from complete gene loss.
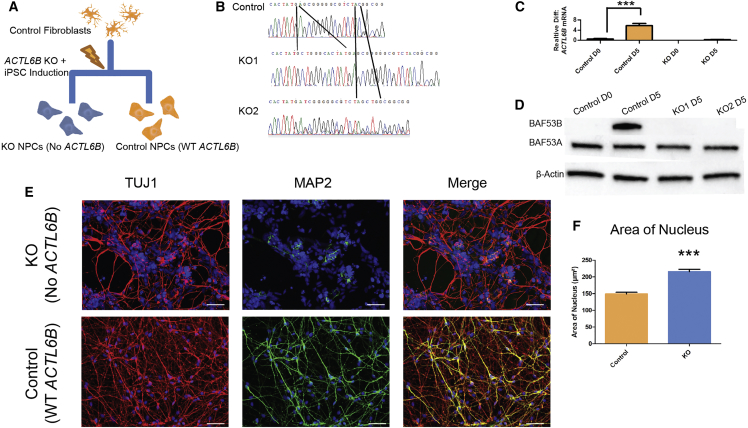
We knocked out ACTL6B from control human cells using a clonal genetic engineering technique23 (Supplemental Material and Methods). We generated two independent ACTL6B knockout cell lines that had different homozygous frameshift mutations in exon 1 (KO1 and KO2; referred to collectively as KO) and compared them to the isogenic cell line that had undergone no editing event (control) (Figures 4A and 4B). The use of two independently edited cell lines with the same outcome (homozygous loss of ACTL6B) is one way to protect againstcell line artifacts, where we do not expect the same artifact to be present in both cell lines. To further ensure this, we Sanger sequenced the five genomic regions most likely to be edited by the gRNAs used, all of which were unedited, suggesting no off-target effects, as has been reported and systematically assessed previously.24 We also performed long-range (1.6 kb) sequencing to ensure that these mutations were in fact homozygous and not due to a large deletion in one allele, in addition to DNA-based qPCR to confirm equal gene dosage between edited and unedited lines (Supplemental Material and Methods). After clonal gene editing and careful genomic integrity assessment, we planned to investigate dendritic length anomalies in a more mature neuronal state, since mouse KO Actl6b neurons show deficits in dendrite development.4 To do this, we differentiated human ACTL6B KO cell lines and their matched isogenic controls for 15 days (D15, a time point where we routinely see extensive neuronal arborization in culture23) and confirmed the loss of ACTL6B expression at the mRNA and protein level (Figures 4C and 4D). We used MAP2 and TUJ1 as dendritic and neuronal markers, respectively, since these are routinely used in neuroscience research for this purpose.25, 26 We observed virtually no MAP2 staining in ACTL6B KO cells, while MAP2 was clearly present in most cells in the isogenic controls (Figure 4E). We also observed a larger nuclear size in the deleted cells, as assayed by DAPI, an effect that is obvious on cell examination (Figure 4F).
Figure 4.
Generation and Characterization of ACTL6B KO Neurons Reveals a Loss of Dendrites
(A) Diagram of the experimental approach taken to generate ACTL6B KO NPCs.
(B) Sanger sequencing traces of two ACTL6B KO lines.
(C) ACTL6B expression in control and ACTL6B KO NPCs at a D0 and D5 time point (n ≥ 3).
(D) Western blots assessing the protein levels of BAF53A/B in ACTL6B KO lines.
(E) Representative TUJ1 and MAP2 staining of control and ACTL6B KO D15 immature forebrain neurons.
(F) Quantification of the surface area of the nucleus in the cell lines shown in (E) (n > 50).
Student’s t test, ∗p < 0.05, ∗∗p < 0.01.
The p.∗427Aspext∗33 Alteration Phenocopies ACTL6B KO Dendritic Deficits and Is Reversible upon Bi-allelic Genetic Repair
From our genotype-phenotype data from affected individuals and their first-degree relatives, we reasoned that recessive mutations cause a loss of function of ACTL6B and thus may mimic the cellular phenotype of the engineered ACTL6B KO cells. To demonstrate this, we reasoned that using these cell lines and comparing them to a clonally repaired version should provide interpretable data.
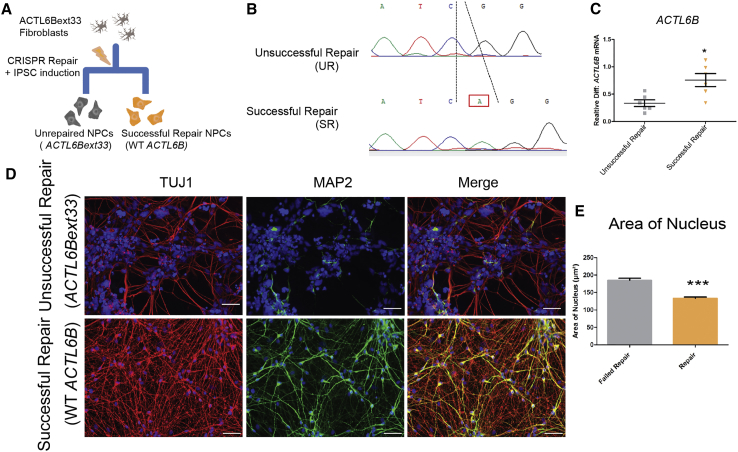
We bi-allelically repaired the ACTL6Bext33 line to a wild-type genotype and simultaneously reprogrammed these edited cell lines, where we had several unsuccessful repairs that could be used as isogenic controls. Homology-directed repair was performed using a wild-type template in p.∗427Aspext∗33 fibroblasts plated at low density and iPSCs colonies derived from a single fibroblast were isolated, ensuring clonality and purity of repair.23 After iPSC expansion of many colonies, we extracted DNA and Sanger-sequenced around the mutation site (Supplemental Material and Methods, sections S2–S4). A colony with c.1279del mutation repaired to a wild-type genotype was identified and labeled as successful repair (SR) and differentiated to NPCs in tandem with an unsuccessful repair (UR) line, which was derived from a colony that received the CRISPR complex and repair template but where no editing event occurred (Figures 5A and 5B). We differentiated these cells from NPCs to D15 neurons and then stained for MAP2 and TUJ1, identical in design to the KO study. As shown in Figures 5D and 5E, affected individual cells recapitulate the loss of MAP2 and increased nuclei size observed in the KO, a result that is reversed on repair of the homozygous base change in ACTL6B. The similar cellular phenotype between affected individual and engineered KO neurons suggest that the ACTL6Bext33 recessive mutation causes similar deficits to the complete KO and thus could be interpreted as causing a loss of function.
Figure 5.
Repair of the ACTL6B c.1279del Mutation Restores Morphological and Dendritic Deficits
(A) Schematic detailing ACTL6B CRISPR repair.
(B) Sanger sequencing traces of a successful repair (SR) and unrepaired (UR) cell line generated from ACTL6Bext∗33 cell line.
(C) ACTL6B expression in SR and UR NPCs at a D5 time point (n = 6).
(D) Representative TUJ1 and MAP2 staining taken from SR and UR forebrain immature neurons at D15. Scale bars represent 40 μm.
(E) Quantification of the surface area of the nucleus and soma and the length of projections in the cell lines shown in (D) (n > 50).
Student’s t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Loss of Dendrites due to Loss of Function of ACTL6B Is Likely due to Delayed Maturation of Young Neurons
Is the observed decrease in MAP2 staining due to immature differentiation, differential differentiation, or a specific deficit in dendrite development? To try to address these questions, we first asked whether the cell types in each condition were equivalent. To investigate this question, we stained D15 cultures from repaired and KO cells and their control with an astrocyte (GFAP) and a neuronal marker (TUJ1), with the hypothesis that perhaps deficits in ACTL6B bias NPCs toward becoming astrocytes. Figure S4A shows the results of this experiment; we could not detect different numbers of cells that stained for GFAP. We include in this experiment a positive control where we add 0.1% BSA which can glialize cell cultures. To support the idea that deficits of ACTL6B do not lead cell cultures to become more glial and to provide more specificity than just GFAP, we assessed the transcriptomic data of p.∗427Aspext∗33 and control cells we had generated in an RNA-seq experiment. We found no consistent pattern in mRNA expression levels of glial markers ALDH1L1, GFAP, and GJA1 and neuronal markers RBFOX3 and TUBB3 to suggest an increase in expression of glial-related genes in ACTL6Bext33 cells compared to control cells (Figure S4B). These data indicate that deficits in ACTL6B do not lead NPCs to become more astrocytic. We therefore ruled out the loss of MAP2 staining being due to cells being pushed toward an astrocytic fate.
As a simple measurement of differentiation, we opted to photograph control, ACTL6Bext.33, ACTL6B KO, and repair cells across several developmental time points. Figure S5 shows that in contrast to repaired and control cell lines, unrepaired and KO ACTL6B cells are not branched prior to day 20, whereas at day 25 through day 50, all lines show branching.
These data suggest that deficits in ACTL6B lead to a delay in differentiation in early post-mitotic states. This delay in differentiation, if true in vivo, may lead to cell connectivity deficits.
The p.∗427Aspext∗33 Variation Alters BRG1 Genomic Binding and Affects Gene Expression
This project began with the index case subject R3 (p.∗427Aspext∗33), who had fibroblasts collected prior to mutation identification, so our study is heavily biased toward this case. To this end, we opted to continue experiments with these cells, with the idea that we might recruit cells from other subjects or design exogenous templates for validation studies.
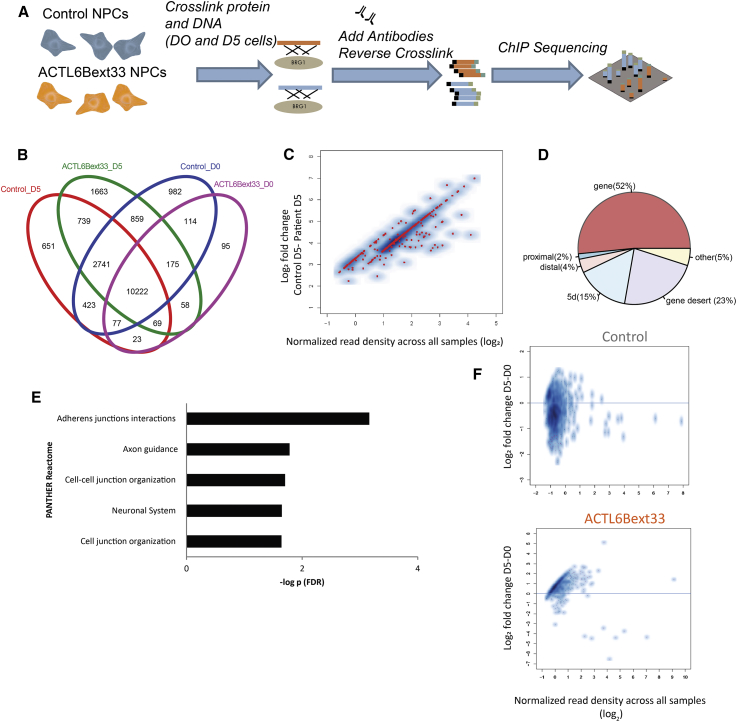
We wanted to understand the molecular consequences of the p.∗427Aspext∗33 variant and how this might lead to neurodevelopmental deficits. Due to a lack of ChIP grade antibodies directed specifically to BAF53B, we chose to perform a ChIP-seq experiment targeting BRG1, a key subunit of the BAF complex with ATPase activity that is found in both BAF53B- and BAF53A-containing BAF complexes.11 We chose the D0 and D5 time points for proliferating and post-mitotic cells, respectively, and performed ChIP in control and ACTL6Bext33 cells using eight replicates per subject per time period (32 ChIP experiments). We performed several QC experiments with different anti-BRG1 antibodies prior to sequencing to ensure appropriate parameters (not shown) and chose two antibodies to provide overlapping datasets of Brg1-containing BAF complex binding (Figure 6A). After sequencing and QC, we analyzed differential binding in D5 cells to understand how the genomic targeting of Brg1-containing BAF complexes may be altered by a mutant BAF53B subunit. ChIP peaks were called in at least 2/8 lines, and differential analysis used reads from all replicates within the peak. 10,222 peaks were common across all data points (Figure 6B), suggesting that BRG1 remains mostly at the same location in the genome, even in mutant ACTL6B cells and irrespective of developmental state.
Figure 6.
ACTL6Bext∗33 Variant Leads to Increased Binding of BRG1-BAF Complex to the Genome
(A) Diagram illustrating the ChIP-seq experiment.
(B) Venn diagram showing overlap of genes that the BRG1 complex is bound to.
(C) Decreased binding at all 382 FDR significant sites in control cells compared to ACTL6Bext∗33 cells (pink dots are significant, while blue dots are not).
(D) Proportion of BRG1 binding sites found in relation to their proximity to a gene.
(E) Gene ontology analysis of differentially bound regions.
(F) Within-subjects differential binding across developmental stages (D0 and D5) showing decreased binding in D0 compared to D5 in ACTL6Bext∗33 cells (pink dots are significant, while blue dots are not). Genes showing a significant difference (FDR-adjusted p values [Benjamini-Hochberg] ≤ 0.05) in D5 relative to D0 using a GLM as implemented in DESeq2.
We focused our primary analysis on D5, since this is when ACTL6B is expressed, and tried to determine whether there was differential binding of BRG1 at peaks called in both p.∗427Aspext∗33 and control cells. Using FDR < 0.05, we found no significant differences; however, using an uncorrected p value of 0.05 revealed 382 common genomic regions that were significantly differentially bound and every one of these showed increased binding in affected individual cells (Figure 6C). Loss of function of BAF53B may lead to increased affinity or stabilization of the BAF complex to its genomic targets, possibly through retention of BAF53A. More than half of the 382 sites that BRG1-containing BAF complexes bound to were associated with genes (Figure 6C). PANTHER GO terms associated with the differentially bound regions were related to cell adhesion and neurodevelopment (Figure 6D). This list included autism-associated genes AUTS2, PTEN, FOXP2, and SMARCA2.
To further assess whether mutant ACTL6B leads to increased binding of BAF to genomic regions, we performed a within-subjects analysis in proliferating (D0) and differentiating (D5) cells, looking for peaks present at both developmental stages using FDR < 0.05 for peak calling. While we did not find the same peaks that were called between case subjects and control subjects (suggesting the experiment was underpowered since we used eight replicates in each block), we found evidence for a general decrease in BRG1 binding in differentiated cells compared to proliferating cells in those genomic regions present at both D0 and D5 in control conditions, in contrast to the p.∗427Aspext∗33 cells where there was a consistent increase in BAF binding at D5 compared to D0 (Figure 6E) at all sites. These data support the notion that a recessive ACTL6B mutation leads to increased association of BRG1 to certain areas of the genome.
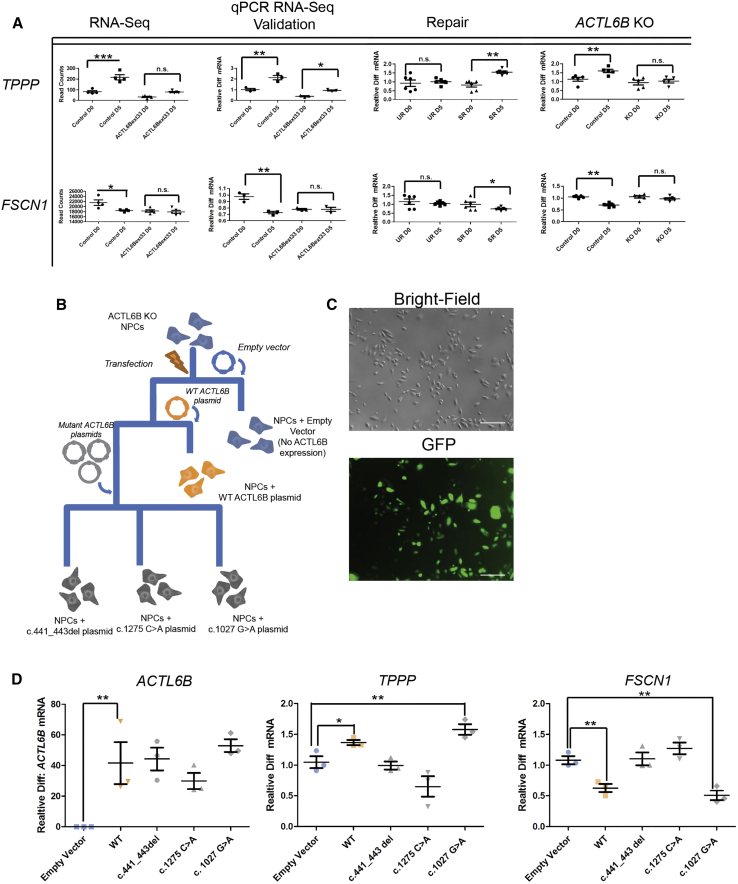
How does genomic BAF binding affect gene expression and how might this differ when ACTL6B is mutated? We performed RNA-seq in affected individual and control cells (n = 4 independent replicates per subject) at D0 and D5 and looked only at those genes that were detected in the ChIP-seq analysis. We were interested in those genes that showed significant changes between D0 and D5 in the ChIP-seq data and which also showed significant change in the RNA-seq data between D0 and D5 (within-subjects); also, we selected those genes that showed RNA-seq differences between mutant cells and control cells at D5 (Table S2). We highlight TPPP (Table S4), a microtubule-binding protein involved in cell process extensions,27, 28 and FSCN1, which has been shown to be involved in neurite shape and trajectory in prior studies in mice.29 Due to their biological function and significance levels in our experiments, we chose to use TPPP and FCSN1 expression levels to assess the external validity of our findings. We note the prevalence (Table S4) of genes that might be implicated in sphingolipid biology or myelin processing (SOX8,30 CERK,31 and A4GALT32), consistent with Wu et al.4 who observed a severe myelination defect in Actl6b KO mice.
We used TPPP and FSCN1 expression as output markers to assess direct versus correlational effects of mutant ACTL6B. We posed two initial questions to test direct versus correlational effects. First, does the ACTL6B KO show a similar pattern of expression to ACTL6Bext33 compared to its isogenic control? Second, do we see the opposite effect in the UR cells compared to the SR cells? We began by validating the RNA-seq data, using the same RNA that was used to make RNA-seq libraries (Figure 7A). Next, we examined the expression of these genes between D0 and D5 time points in ACTL6B KO cells and their isogenic controls, as well as in UR and SR cells. SR cells, when compared to UR cells, showed a significant increase in TPPP and decreased expression of FSCN1 as NPCs mature from D0 to D5. In ACTL6B knock-out cells compared to their isogenic controls, we observed significant and opposite effects to that observed with repaired cells: FSCN1 expression did not decrease, while TPPP expression did not increase as the cells differentiated (Figure 7A). These data provide isogenic evidence that complete loss of ACTL6B and a repair of p.∗427Aspext∗33 recapitulate and reverse, respectively, expression alterations in TPPP and FSCN1 and suggest that expression changes in these genes are directly caused by disruption of ACTL6B.
Figure 7.
External Validity in Multiple ACTL6B Mutant Models in Human Neurons
(A) TPPP and FSCN1 expression in initial RNA-seq (n ≥ 4) and qPCR (n ≥ 3) data (ACTL6Bext∗33 versus control); unrepaired (UR) ACTL6Bext∗33 versus ACTL6Bext∗33 successful repair (SR) (n = 6); and ACTL6B KO versus isogenic control cells (n = 5). Results are represented as mean ± SEM. Student’s t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Experimental plan for generation of multiple human neuronal cell lines expressing various mutant ACTL6B constructs.
(C) Brightfield and GFP images demonstrating high transfection of ACTL6B constructs.
(D) mRNA expression in transfected ACTL6B KO NPCs at D5 time points of ACLT6B, TPPP, and FSCN1 (n = 3). Results are represented as mean ± SEM. Student’s t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
External Validity of TPPP and FSCN1 Expression Levels as Markers of an ACTL6B Recessive, Loss-of-Function Disease in Human Neurons using Constructs Derived from Different ACTL6B Variants
External validity can be provided by KO rescue and by recapitulating expression effects using different mutations in ACTL6B identified in our cohorts (Figures 7B–7D). If TPPP and FSCN1 expression levels are markers of loss of function of ACTL6B, the exogenous re-introduction of ACTL6B on a KO background should help restore their expression toward levels observed in lines with wild-type ACTL6B. Further, expressing mutant ACTL6B to match other variants found in the recessive cohort should re-establish expression changes on an ACTL6B KO background. We therefore made ACTL6B constructs of two recessive mutations, c.441_443del (p.Phe147del) and c.1275C>A (p.Cys425∗), and the most prevalent dominant mutation, c.1027G>A, as well as the WT construct itself. Expressing these variants from trasnsiently delivered vector on an ACTL6B KO background may give us an indication whether the dominant and recessive variants mediate their effects through the same molecular pathways and cause similar effects on the expression of FSCN1 and TPPP. At a D5 time point, cells transfected with WT ACTL6B showed decreased expression of FSCN1 and increased expression of TPPP, consistent with what we observed in the initial KO experiment (Figure 7D), meaning that the WT construct can rescue the expression changes observed in ACTL6B KO cells. We observed that the two recessive variants (c.441_443del and c.1275C>A) mimicked the effects observed in the recessive p.∗427Aspext∗33 variant, while the dominant mutation mimicked wild-type cells (Figure 7D).
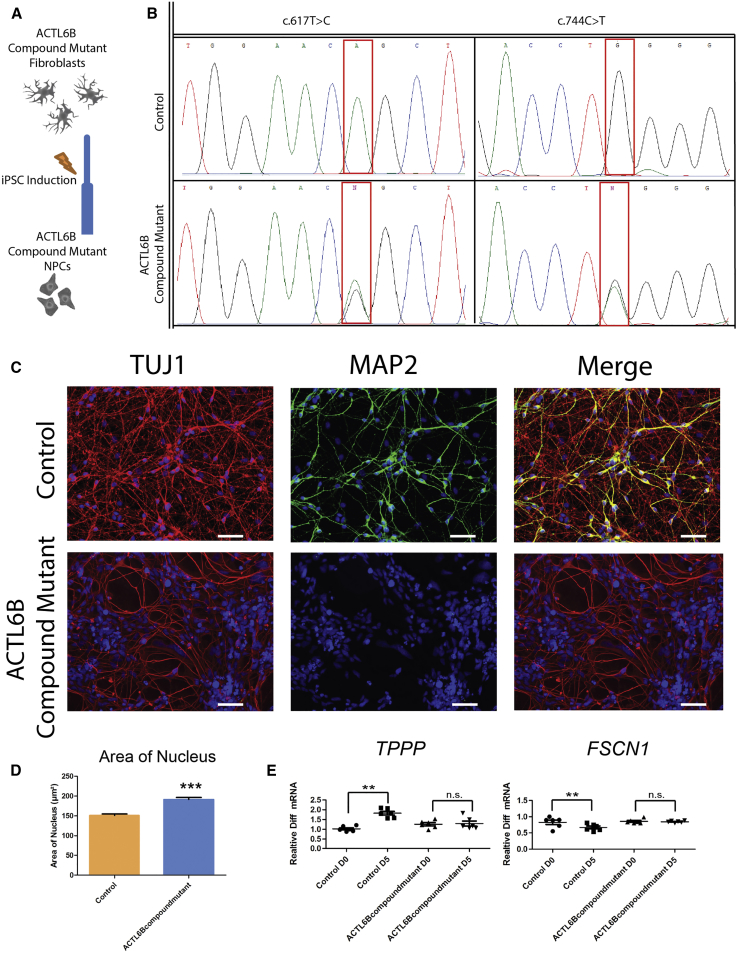
Confirmation of Dendritic Deficits and Gene Expression Markers using Neurons Derived from ACTL6B Mutant c.617T>C/c.724C>T
We obtained fibroblasts from individual R9 with compound heterozygous mutations in ACTL6B (c.617T>C [p.Leu206Pro] and c.724C>T [p.Gln242∗]) (Table 1). We induced the fibroblasts to become iPSCs, differentiated the iPSCs to D15 neurons, and confirmed the mutant genotype of this line (Figures 8A and 8B). We compared this “ACTL6B compound heterozygous mutant” line to healthy control cells differentiated to a day 15 time point and found a similar decrease of MAP2 staining and increased nuclei size as compared to the ACTL6B KO and ACTL6Bext33 lines (Figures 8C and 8D). Assessing the expression of TPPP and FSCN1 at D5 and D0 time points in the ACTL6B compound mutant and control lines also produced results similar to those seen with the ACTL6Bext33 line, with the ACTL6B compound mutant showing a lack of increased expression of TPPP as well as a lack of decreased expression of FSCN1 during differentiation compared to control cells (Figure 8E).
Figure 8.
Neurons Derived from an Individual with a Compound Mutation in ACTL6B Show a Similar Phenotype to ACTL6Bext33 and ACTL6B KO Neurons
(A) Schematic showing generation of ACTL6B compound mutant NPCs.
(B) Sanger sequencing traces of ACTL6B compound mutant and control cell line at both identified point mutations in ACTL6B.
(C) Representative TUJ1 and MAP2 staining of control and ACTL6B compound mutation immature forebrain neurons.
(D) Quantification of the surface area of the nucleus in the cell lines shown in (E).
(E) TPPP and FSCN1 expression in ACTL6B compound mutant versus control cells at mitotic (D0) and post-mitotic (D5) time points (n > 50). Student’s t test, ∗p < 0.05, ∗∗p < 0.01.
Discussion
These data describe two distinct neurodevelopmental diseases caused by dominant or recessive mutations in ACTL6B. This work positions ACTL6B mutations as causing both a recessive neurological disease characterized by severe epileptic encephalopathy and a dominant intellectual disability syndrome with severe speech and ambulation deficits.
Previous studies have identified mutations in genes that code for other subunits of the nBAF and npBAF complexes that are capable of causing disease through dysregulated BAF function, collectively called “Bafopathies.”33 The two foremost diseases among the Bafopathies, Coffin-Siris (CSS) (MIM: 135900) and Nicolaides-Baraitser (NCBRS) (MIM: 601358) syndrome, show interesting parallels and differences to the diseases described here.34, 35 NCBRS is a monogenic disease, caused exclusively by mutations in SMARCA2 (MIM: 600014) that are autosomal dominant, and CSS is a genetically diverse disease and can be caused by mutations in ARID1B (MIM: 614556) and a variety of other genes that play a role in the BAF complex that vary in their inheritance pattern. Nevertheless, common symptoms appear to exist in both these conditions and the diseases described here. Common symptoms reported across conditions include intellectual disability, developmental delay, hypotonia, and some form of dysmorphic facial features.34 Like individuals with recessive mutations in ACTL6B, individuals with NCBRS also show early-onset seizures,36 and seizures are also reported in individuals with CSS, although they are not necessarily early onset.34, 37, 38 Individuals with NCBRS also show short phalanges,36 as observed in some individuals with dominant mutations in ACTL6B. However, some of the specific developmental symptoms observed in these diseases, such as sparse scalp hair39 or an absent fifth digit,36 do not appear in individuals with either recessive or dominant mutations in ACTL6B. This could suggest that while a general disruption of the BAF complex in a variety of protein subunits will inevitably lead to intellectual disability and developmental delay, the specific protein subunit that is affected will determine the presence and nature of dysmorphisms and epilepsy.
To understand why mutations in ACTL6B cause disease, we modeled the disease in human NPCs and neurons. We first confirmed that ACTL6B expression was induced upon neural cells becoming post-mitotic. We then went on to make several different cell models in the hope of reducing variation across experimental variables. We made forebrain progenitor cells from a healthy individual with experimentally induced knock-out of ACTL6B to understand the effects of complete loss of the gene, in addition to cells derived from individuals R9 and R3 with an isogenic engineered repaired cell line.
While we cannot precisely determine the mechanism of the disease that appears to be caused by bi-allelic mutations in ACTL6B, our results do illuminate several key features of the etiology of the disease. First, the presence of BAF53B in the ACTL6Bext33 cell line at D5 eliminates the possibility that the symptoms are caused by an absence of BAF53B, as is likely the case in other variants of BAF53B where NMD is predicted to occur.19 Instead, it seems plausible that the symptoms are the result of a loss of function of BAF53B stemming from changes in the structure of the protein. This hypothesis is supported by our observations in the ACTL6B KO model, which shows similar deficits in both MAP2 staining and the expression of key genes identified in the ACTL6Bext33 that are regulated by BAF. ACTL6B KO cells expressing the recessive mutations in ACTL6B observed in our recessive cohort fail to rescue aberrant expression of genes, whereas reintroduction of wild-type ACTL6B does, strongly suggesting that the phenotype is the result of a loss of function.
However, how the recessive mutations render BAF53B non-functional could be due to one of several possibilities. Perhaps the most intuitive answer is that mutations disrupt the ability of BAF53B to bind to the BAF complex. If this fails to occur, this might allow BAF53A to remain bound in the complex. Given that there are a great many BAF complexes that dynamically exchange parts to affect cell differentiation at any one time, a high proportion of BAF53A in BAF might cause increased affinity of BAF to the genome in a differentiating cell state compared to both a proliferating cell state when BAF53B is absent or a differentiated cell state where BAF53B is present but not functional and/or when the interaction with the complex is impaired. The increased presence of BAF53A in the BAF complex, associated with a more proliferative neuronal cell state, might also explain the delayed differentiation we observed in disease cells. Another explanation could be that recessive mutations do not prevent BAF53B from binding to the BAF complex, but instead prevent the various components of the complex from interacting properly and thus prevent the BAF complex from interacting with the genome and other proteins in yet unknown ways. Finally, there is the possibility that recessive mutations do not disrupt BAF complex function significantly at all, but instead prevent BAF53B from properly interacting with the other complexes such as SRCAP,40 TIP60/NuA4,19 and INO80.41 Future work will need to look at how mutant BAF interacts with different proteins.
With respect to the dominant mutation identified in eight unrelated individuals, we failed to observe the transcription effects we found in the KO, ACTL6B compound heterozygous mutant, or the ACTL6Bext33 cells, suggesting that dominant and recessive mutations in ACTL6B cause disease through distinct molecular pathways. This is also consistent with the observation of different symptoms in individuals with recessive or dominant mutations in ACTL6B. Based on the limited molecular information we have for dominant mutations, there exist many potential explanations for how a point mutation in ACTL6B might cause the observed symptoms. Given that these subjects all have a functioning copy of BAF53B that presumably incorporates normally into BAF or other complexes, it is reasonable to suggest that the ACTL6B dominant mutations identified here may be gain of function. One of the few clues that we have at this stage of investigation is that the highly specific nature of the p.Gly343Arg variant suggests that a very precise interaction is being disrupted or created. Previous work in mice9 has shown that deletion of the hydrophobic domain of BAF53b results in a dominant-negative form of the protein, causing deficits in memory, LTP, and gene expression. These deficits were likely caused by altering the ability BAF53B to interact with other proteins.9 It is possible that the dominant mutations observed in this study cause disease by altering the hydrophobic domain of BAF53B through a similar mechanism.
Several genes of note may be important targets of the BAF complex at different developmental stages. FSCN1 is one of these and is strongly associated with the formation of actin, particularly in early neurodevelopment.29, 43 The lack of increase of FSCN1 observed in our models of ACTL6B dysfunction as cells differentiate may therefore be the result of neuronal cells remaining in a more proliferative or immature state due to an impairment in the ability of ACTL6B to transition the BAF complex from promoting genes associated with proliferation to those associated with neuronal outgrowth.44
The stable expression levels of TPPP observed in models of ACTL6B dysfunction as NPCs differentiate may reflect the cytoskeletal changes observed both within our own models and in the deficits in dendritic spine and synapse function observed in mouse models of Actl6b KO. It should also be noted that our ChIP-seq data that initially highlighted these genes as being dysregulated are based upon only BRG1-containing BAF complexes. As BRG1 and BRM are mutually exclusive components of the BAF complex that are both found in dividing and post-mitotic neural cells,11 it is possible that the dataset we generated is only a partial picture of the regions of the genome where BAF may bind in these cells.
This work brings together extensive clinical samples and human stem cell modeling to demonstrate that mutations in ACTL6B in human cause severe neurological disorders. Substantially more work will need to be done to understand the precise mechanisms of how recessive or dominant mutations in ACTL6B affect incorporation into the BAF complex and how this incorporation can alter differential genomic binding and gene expression patterns.
Declaration of Interests
A.G.B., I.M.W., and A.C. are employees of GeneDx. C.E. is president of ManuStem.com and has commercial interests with Stem Cell Technologies. The other authors declare no conflict of interest.
Acknowledgments
We acknowledge funding by FRQS doctoral (S.B.); Indonesian Endowment Fund for Education PhD award (M.J.); CONACYT (Mexico) and MITACS (K.M.V.); Genome Canada and Génome Québec (J.L.M. and E.R.); Canada Research Chairs program (G.T. and C.E.); AMED, MEXT, JST, MHLW, and Takeda Science Foundation (N. Matsumoto); and CIHR grant (C.E. and P.M.C.). We are grateful to Gerald Crabtree who provided a custom antibody to BAF53B. C.E. is grateful to family R3 who collaborated with his lab from mutation detection to human neuron modeling.
Published: April 25, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.022.
Contributor Information
Carl Ernst, Email: carl.ernst@mcgill.ca.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Web Resources
Bioconductor, http://bioconductor.org/packages/release/bioc/html/DiffBind.html
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
HOMER, http://homer.ucsd.edu/homer/
Imagelab, http://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z
Neurolucida, https://www.mbfbioscience.com/neurolucida
Online Mendelian Inheritance in Man, http://www.omim.org
Trim Galore, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
VarSome, https://varsome.com/
Supplemental Data
References
- 1.Meagher R.B., Kandasamy M.K., Deal R.B., McKinney E.C. Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 2007;17:325–332. doi: 10.1016/j.tcb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Biggar S.R., Crabtree G.R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessard J., Wu J.I., Ranish J.A., Wan M., Winslow M.M., Staahl B.T., Wu H., Aebersold R., Graef I.A., Crabtree G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J.I., Lessard J., Olave I.A., Qiu Z., Ghosh A., Graef I.A., Crabtree G.R. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Peterson C.L. Multiple SWItches to turn on chromatin? Curr. Opin. Genet. Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 6.Sudarsanam P., Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 7.Yoo A.S., Staahl B.T., Chen L., Crabtree G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staahl B.T., Crabtree G.R. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr. Opin. Neurobiol. 2013;23:903–913. doi: 10.1016/j.conb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel-Ciernia A., Matheos D.P., Barrett R.M., Kramár E.A., Azzawi S., Chen Y., Magnan C.N., Zeller M., Sylvain A., Haettig J. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santen G.W.E., Kriek M., van Attikum H. SWI/SNF complex in disorder: SWItching from malignancies to intellectual disability. Epigenetics. 2012;7:1219–1224. doi: 10.4161/epi.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokpor G., Xie Y., Rosenbusch J., Tuoc T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00243. 243–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakshi R., Hassan M.Q., Pratap J., Lian J.B., Montecino M.A., van Wijnen A.J., Stein J.L., Imbalzano A.N., Stein G.S. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J. Cell. Physiol. 2010;225:569–576. doi: 10.1002/jcp.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell S., Hettige N., Silveira H., Peng H., Wu H., Jefri M., Antonyan L., Zhang Y., Zhang X., Ernst C. Differentiation of human induced pluripotent stem cells (iPSCs) into an effective model of forebrain neural progenitor cells and mature neurons. Bio-Protoc. 2019;9 doi: 10.21769/BioProtoc.3188. Published online March 5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell S., Peng H., Crapper L., Kolobova I., Maussion G., Vasuta C., Yerko V., Wong T.P., Ernst C. A rapid pipeline to model rare neurodevelopmental disorders with simultaneous CRISPR/Cas9 gene editing. Stem Cells Transl. Med. 2017;6:886–896. doi: 10.1002/sctm.16-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds B.A., Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen E.S., Gigek C.O., Rosenfeld J.A., Diallo A.B., Maussion G., Chen G.G., Vaillancourt K., Lopez J.P., Crapper L., Poujol R. Molecular convergence of neurodevelopmental disorders. Am. J. Hum. Genet. 2014;95:490–508. doi: 10.1016/j.ajhg.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khajavi M., Inoue K., Lupski J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez R., Holmes K.C. Actin structure and function. Annu. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes K.C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 22.Yoo M., Choi K.Y., Kim J., Kim M., Shim J., Choi J.H., Cho H.Y., Oh J.P., Kim H.S., Kaang B.K., Han J.H. BAF53b, a neuron-specific nucleosome remodeling factor, is induced after learning and facilitates long-term memory consolidation. J. Neurosci. 2017;37:3686–3697. doi: 10.1523/JNEUROSCI.3220-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell S., Maussion G., Jefri M., Peng H., Theroux J.F., Silveira H., Soubannier V., Wu H., Hu P., Galat E. Disruption of GRIN2B impairs differentiation in human neurons. Stem Cell Reports. 2018;11:183–196. doi: 10.1016/j.stemcr.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada A., Teng J., Takei Y., Oguchi K., Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J. Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goedert M., Crowther R.A., Garner C.C. Molecular characterization of microtubule-associated proteins tau and MAP2. Trends Neurosci. 1991;14:193–199. doi: 10.1016/0166-2236(91)90105-4. [DOI] [PubMed] [Google Scholar]
- 27.Skjoerringe T., Lundvig D.M., Jensen P.H., Moos T. P25alpha/Tubulin polymerization promoting protein expression by myelinating oligodendrocytes of the developing rat brain. J. Neurochem. 2006;99:333–342. doi: 10.1111/j.1471-4159.2006.04073.x. [DOI] [PubMed] [Google Scholar]
- 28.Mino R.E., Rogers S.L., Risinger A.L., Rohena C., Banerjee S., Bhat M.A. Drosophila Ringmaker regulates microtubule stabilization and axonal extension during embryonic development. J. Cell Sci. 2016;129:3282–3294. doi: 10.1242/jcs.187294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraft R., Escobar M.M., Narro M.L., Kurtis J.L., Efrat A., Barnard K., Restifo L.L. Phenotypes of Drosophila brain neurons in primary culture reveal a role for fascin in neurite shape and trajectory. J. Neurosci. 2006;26:8734–8747. doi: 10.1523/JNEUROSCI.2106-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolt C.C., Lommes P., Friedrich R.P., Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- 31.Boggs J.M., Gao W., Zhao J., Park H.J., Liu Y., Basu A. Participation of galactosylceramide and sulfatide in glycosynapses between oligodendrocyte or myelin membranes. FEBS Lett. 2010;584:1771–1778. doi: 10.1016/j.febslet.2009.11.074. [DOI] [PubMed] [Google Scholar]
- 32.Kaczmarek R., Mikolajewicz K., Szymczak K., Duk M., Majorczyk E., Krop-Watorek A., Buczkowska A., Czerwinski M. Evaluation of an amino acid residue critical for the specificity and activity of human Gb3/CD77 synthase. Glycoconj. J. 2016;33:963–973. doi: 10.1007/s10719-016-9716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bögershausen N., Wollnik B. Mutational landscapes and phenotypic spectrum of SWI/SNF-related intellectual disability disorders. Front. Mol. Neurosci. 2018;11:252. doi: 10.3389/fnmol.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mari F., Marozza A., Mencarelli M.A., Lo Rizzo C., Fallerini C., Dosa L., Di Marco C., Carignani G., Baldassarri M., Cianci P. Coffin-Siris and Nicolaides-Baraitser syndromes are a common well recognizable cause of intellectual disability. Brain Dev. 2015;37:527–536. doi: 10.1016/j.braindev.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Pretegiani E., Mari F., Renieri A., Penco S., Dotti M.T. Nicolaides-Baraitser syndrome: defining a phenotype. J. Neurol. 2016;263:1659–1660. doi: 10.1007/s00415-016-8194-0. [DOI] [PubMed] [Google Scholar]
- 37.Kosho T., Miyake N., Carey J.C. Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:241–251. doi: 10.1002/ajmg.c.31415. [DOI] [PubMed] [Google Scholar]
- 38.Bender H.A., Zaroff C.M., Karantzoulis S., Nakhutina L., MacAllister W.S., Luciano D. Cognitive and behavioral functioning in Coffin-Siris syndrome and epilepsy: a case presentation. J. Genet. Psychol. 2011;172:56–66. doi: 10.1080/00221325.2010.506604. [DOI] [PubMed] [Google Scholar]
- 39.Santen G.W., Aten E., Sun Y., Almomani R., Gilissen C., Nielsen M., Kant S.G., Snoeck I.N., Peeters E.A., Hilhorst-Hofstee Y. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 40.Vogel-Ciernia A., Wood M.A. Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology. 2014;80:18–27. doi: 10.1016/j.neuropharm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S., Shi Y., Mulligan P., Gay F., Landry J., Liu H., Lu J., Qi H.H., Wang W., Nickoloff J.A. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laeremans A., Van de Plas B., Clerens S., Van den Bergh G., Arckens L., Hu T.-T. Protein expression dynamics during postnatal mouse brain development. J. Exp. Neurosci. 2013;7:61–74. doi: 10.4137/JEN.S12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Son E.Y., Crabtree G.R. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:333–349. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.