Abstract
Glioblastoma (GBM) is one of the most aggressive and lethal tumor types. Evidence continues to accrue indicating that the complex relationship between GBM and the brain microenvironment contributes to this malignant phenotype. However, the interaction between GBM and neurotransmitters, signaling molecules involved in neuronal communication, remains incompletely understood. Here we examined, using human patient-derived xenograft lines, how the monoamine dopamine influences GBM cells. We demonstrate that GBM cells express dopamine receptor 2 (DRD2), with elevated expression in the glioma-initiating cell (GIC) population. Stimulation of DRD2 caused a neuron-like hyperpolarization exclusively in GICs. In addition, long-term activation of DRD2 heightened the sphere-forming capacity of GBM cells, as well as tumor engraftment efficiency in both male and female mice. Mechanistic investigation revealed that DRD2 signaling activates the hypoxia response and functionally alters metabolism. Finally, we found that GBM cells synthesize and secrete dopamine themselves, suggesting a potential autocrine mechanism. These results identify dopamine signaling as a potential therapeutic target in GBM and further highlight neurotransmitters as a key feature of the pro-tumor microenvironment.
SIGNIFICANCE STATEMENT This work offers critical insight into the role of the neurotransmitter dopamine in the progression of GBM. We show that dopamine induces specific changes in the state of tumor cells, augmenting their growth and shifting them to a more stem-cell like state. Further, our data illustrate that dopamine can alter the metabolic behavior of GBM cells, increasing glycolysis. Finally, this work demonstrates that GBM cells, including tumor samples from patients, can synthesize and secrete dopamine, suggesting an autocrine signaling process underlying these results. These results describe a novel connection between neurotransmitters and brain cancer, further highlighting the critical influence of the brain milieu on GBM.
Keywords: cancer stem cell, cellular plasticity, dopamine, glioblastoma
Introduction
Glioblastoma (GBM) is the most common and aggressive primary malignant brain tumor afflicting adults, with standard of care treatment leading to a median survival of only 15 months (Alifieris and Trafalis, 2015). Recent clinical trials have shown increased survival to ∼21 months (Stupp et al., 2017). A key factor limiting the efficacy of treatment is the remarkable plasticity exhibited by GBM cells, which allows them to effectively adapt to changes in the microenvironment induced by conventional radiation and chemotherapy (Olmez et al., 2015; Safa et al., 2015). Our group and others have shown that cellular plasticity processes enable GBM cells to adopt many phenotypes, including the glioma initiating cell (GIC) state, characterized by a neural stem cell-like expression profile and heightened resistance to therapy (Dahan et al., 2014; Olmez et al., 2015; Safa et al., 2015; Lee et al., 2016). Given the dynamic nature of this process, a key question is what mechanisms regulate and control cellular plasticity. Several factors have been shown to activate GBM plasticity, including hypoxia, acidity, and metabolic changes (Heddleston et al., 2009; Soeda et al., 2009; Hjelmeland et al., 2011; Hardee et al., 2012; Tamura et al., 2013). All of these factors are influenced by the dynamic microenvironmental milieu occupied by GBM cells. Therefore, further analysis of how environmental factors specific to the CNS affect tumors are critical for devising novel therapies for patients with GBM.
The GBM microenvironment is unique for a multitude of reasons, including the range of CNS-specific factors, such as neurotransmitters and neurotrophins, known to influence cell proliferation and differentiation. Given the evidence that plasticity can be induced in GBM tumor cells by a variety of microenvironmental factors, including oxygen availability and acidity (Evans et al., 2004; Soeda et al., 2009; Hjelmeland et al., 2011), it is logical that CNS-specific cues like neurotransmitters may also influence GBM cellular behavior. Indeed, evidence is accumulating that CNS-specific cues contribute to the onset and progression of GBM (Charles et al., 2011). It has been shown, for example, that factors secreted from neurons such as neuroligin-3 directly augment GBM cell growth (Venkatesh et al., 2015, 2017). Further, Villa et al. (2016) have demonstrated that GBM cells use cholesterol secreted by healthy astrocytes to enhance their growth. In addition, glutamate, a neurotransmitter, has been shown to augment GBM growth via effects on epidermal growth factor receptor (EGFR) signaling (Schunemann et al., 2010).
One neurotransmitter family likely to contribute to the pro-tumor brain microenvironment in GBM is the monoamines (dopamine, serotonin, and norepinephrine), which are critical to behavior, emotion, and cognition (Beaulieu and Gainetdinov, 2011; Lammel et al., 2014). A recent review highlighted the potential influence of monoamines on GBM tumors (Caragher et al., 2018). Critically, multiple studies have shown that monoamine signaling influences the behavior and phenotype of healthy neural stem cells (NSCs), which have similar expression profiles and functional attributes to GICs (Galli et al., 2004; Singh et al., 2004). In light of these reports, we set out to investigate how dopamine receptor 2 (DRD2) activation influences GBM signaling and phenotype.
Materials and Methods
Cell culture.
U251 human glioma cell lines were procured from the American Type Culture Collection. These cells were cultured in DMEM (HyClone, ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and 2% penicillin-streptomycin antibiotic mixture (CellGro, Mediatech). Patient-derived xenograft (PDX) glioma specimens (GBM43, GBM12, GBM6, GBM5, and GBM39) were obtained from Dr. C. David James at Northwestern University and maintained according to published protocols (Hodgson et al., 2009). Notably, these cells are known to represent different subtypes of GBM. GBM43 and GBM12 are proneural, GBM6 and GBM39 are classical, and GBM5 is mesenchymal, according to the subtypes established by Verhaak et al. (2010).
Determination of dopamine levels.
Cell culture supernatant was collected, filtered, and flash frozen. The details of the mass spectrometric/ultra performance liquid chromatography (UPLC) procedure are described previously (Huang et al., 2014). The LC system (Waters Acquity UPLC) and a Waters Acquity UPLC HSS T3 1.8 μm column were used for separation. The UPLC system was coupled to a triple-quadrupole mass spectrometer (TSQ Quantum Ultra, ThermoFisher Scientific), using ESI in positive mode. All data were processed by Waters MassLynx 4.0 and ThermoFisher Scientific Xcaliber software. Data were acquired and analyzed using LCquan 2.5 software (ThermoFisher Scientific).
Animals.
Athymic nude mice (ν/nu; Charles River Laboratories) were housed according to all Institutional Animal Care and Use Committee guidelines and in compliance with all applicable federal and state statutes governing the use of animals for biomedical research. Briefly, animals were housed in shoebox cages with no >5 mice per cage in a temperature and humidity-controlled room. Food and water were available ad libitum. A strict 12 h light/dark cycle was maintained.
Microarray.
RNA was extracted from samples using RNeasyI kit according to the manufacturer's instructions (Qiagen). One thousand nanograms of RNA were used for Microarray analysis according to the manufacturer's directions (Illumina). HumanHT12 (48,000 probes, RefSeq plus EST) was used for all microarrays. All microarrays were performed in triplicate.
Glycolytic measurement.
GBM 39, 12, and 5 were adhered at a concentration of 0.5–1.5 × 105 cells per well to XFe96 well culture plates (Agilent) using CellTak tissue adhesive (Corning). We then performed a Mito-Stress test as described in the manufacturer's protocol using a Seahorse XFe96 well extracellular flux analyzer. Briefly, cells were attached and suspended in XF Base medium supplemented with 2 mm l-glutamine (Fisher). After baseline extracellular acidification rate (ECAR) reading, glucose was injected to a final concentration of 10 mm to determine glucose stimulated ECAR rate. Next, oligomycin (1 μm final concentration) was injected to determine maximal glycolytic capacity. Last, a competitive inhibitor of glycolysis (2-deoxyglucose) was injected (50 mm) to validate ECAR as a measure of glycolytic flux. Data were analyzed using Agilent's proprietary Wave software.
ChIP sequencing.
The PDX line GBM 43 was subject to DMSO or temozolomide (TMZ) treatment for 4 d, after which cells were washed, collected, and pelleted. Cells were incubated with anti-H3K27ac antibody, after which crosslinking of protein binding occurred. DNA was sheared and crossing linking was reversed. Resultant DNA was sent to ZYMO Research Facility where it underwent next-generation sequencing according to the manufacturer's established protocol.
ChIPSeq reads underwent FastQC quality analysis after sequencing and no abnormalities were detected. Alignment was done using Bowtie2 software and peak calling was performed using the MACS2 CallPeak function with p value set to 0.05. All ChIPSeq data were visualized using Integrated Genomics Viewer.
Electrophysiology.
Whole-cell current-clamp recordings of GBM cells were obtained under visual guidance with a Zeiss Axioskop FS (Zeiss); fluorescence was used to target GICs. DRD2 agonist was delivered by localized superfusion of targeted cells using a Picospritzer device (Parker Hannifin). Recordings were made with a MultiClamp 700B (Molecular Devices) and Digidata data acquisition devices (Molecular Devices) and digitized at 20,000 samples/s. The baseline membrane potential was corrected for the liquid junction potential calculated using pClamp 10 Software (Molecular Devices). Patch-clamp electrodes were manufactured from filamented borosilicate glass tubes (Clark G150F-4, Warner Instruments). Current-clamp electrodes were filled with an intracellular solution containing the following (in mm): 140 K-gluconate, 1 CaCl2 × 6 H2O, 10 EGTA, 2 MgCl2 × 6 H2O, 4 Na, 2 ATP, and 10 HEPES with a resistance of ∼3 MΩ. Cells were recorded in artificial CSF (ACSF) containing the following (in mm): 118 NaCl, 3 KCl, 1.5 CaCl2, 1 MgCl2 × 6 H2O, 25 NaHCO3, 1 NaH2PO4, and 30 d-glucose, equilibrated with medical grade carbogen (95% O2 plus 5% CO2, pH 7.4). Chemicals for ACSF were obtained from Sigma-Aldrich. Cells were submerged under ACSF, saturated with carbogen in recording chambers (flow rate, 12 ml/min). Experiments were performed at 30 ± 0.7°C using a TC-344B Temperature Regulator with an in-line solution heater (Warner Instruments).
Statistical analysis and study design.
All statistical analyses were performed using the GraphPad Prism Software v8.0. In general, data are presented as mean (with SD) for continuous variables, and number (percentage) for categorical variables. Differences between two groups were assessed using Student's t test or Wilcoxon rank sum test as appropriate. For tumorsphere formation assay a score test for heterogeneity between two groups was used and reported as p value and χ2 for each group. Difference among multiple groups were evaluated using ANOVA with post hoc Tukey's test, or Mann–Whitney U test followed by Bonferroni correction as appropriate. Survival curves were graphed via Kaplan–Meier method, and compared by log-rank test. All tests were two-sided and p < 0.05 was considered statistically significant. Experiments such as Western blots, microarray, fluorescence activated cell sorting analysis (FACS), glycolytic measurements, and microarray were performed in biological triplicate. HPLC was performed as a biological duplicate across separate experimental conditions data were averaged and then student's t tests were performed to determine significance. All in vivo experiments contained at least three animals per group, with males and females equally represented, and each animal was treated as a technical replicate.
Results
Therapeutic stress induces epigenetic modifications and subsequent increased expression of DRD2
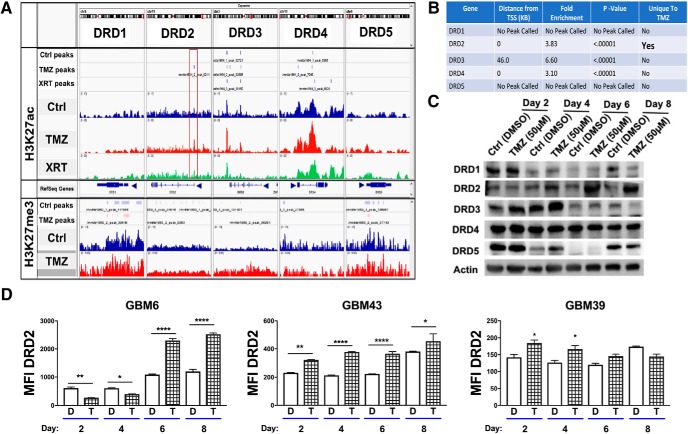
To begin our investigation of monoamines in GBM, we analyzed the epigenetic status of their receptors during therapeutic stress. We performed a genome-wide ChIP-Seq (chromatin immunoprecipitation with massively parallel DNA sequencing) analysis of PDX GBM43 cells for histone 3 lysine 27 (H3K27) acetylation (ac), a marker of open chromatin and active gene transcription, and H3K27 tri-methylation (H3K27me3), a maker of closed chromatin and transcriptional repression. Cells were treated with temozolomide (TMZ; 50 μm), equimolar vehicle control DMSO, or radiation (2Gy). ChIP-Seq was performed 96 h after treatment initiation. Examination of all dopamine, serotonin, and noradrenergic receptors showed that the DRD2 promotor alone possessed enrichment of an H3K27ac mark without any change in H3K27me3 (MACS2 peak score: 45.0, fold-enrichment: 4.023; p = 1.28 E−8; Fig. 1A,B and Fig. 1-1). Further, analysis of The Cancer Genome Atlas (TCGA) revealed that, of the five dopamine receptors, DRD2 mRNA is present at the highest levels (Fig. 1-2).
Figure 1.
Chemotherapeutic stress induces DRD2 expression in the patient-derived xenograft glioma lines. A, B, H3K27ac and H3K27me3 marks distribution around the transcription start sites of dopamine receptors following therapy. ChIP-Seq was performed for the open chromatin state H3K27 acetylation in PDX line GBM43 after 4 d of single exposure to TMZ (50 μm), or treated with one fractionated dose of radiotherapy (2 Gy). DMSO was used as a vehicle control. The red box in DRD2 highlights a key residue near the promoter that uniquely exhibited increased H3K27ac following TMZ treatment. (MACS2 peak score: 45.0, fold-enrichment: 4.023, p < 0.0001). Table summarizes statistical information for each of the five DRDs. Figure 1-1 also shows peak calling of all monoamine receptors. C, Western blot analysis of dopamine receptors expression in PDX cells treated with TMZ over 8 d. D, FACS analysis of DRD2 on different subtypes of PDX cells treated with vehicle control DMSO or TMZ (50 μm). Figure 1-2 includes analysis of all five DRDs demonstrating elevated DRD2 expression in human samples. Bars represent means from three independent experiments and error bars show the SD. Student t tests were performed for each day separately. *p < 0.05, **p < 0.01, ****p < 0.0001.
Extended Data Figure 1-1: ChIP analysis of H3K27ac and H3K27me for other monoamine receptors. Download Figure 1-1, XLSX file (9.3KB, xlsx)
Extended Data Figure 1-2: GBM tumors express DRDs. (A) TCGA data reveals elevated expression of DRD2 relative to the other DRDs. Dots represent mRNA levels from individual tumors. Mean levels were compared using one-way ANOVA. (B) DRD2 expression in patient samples varies based on subtype. (C) Expression of DRDs in PDX GBM cells was determined using FACS analysis. Bars represent the mean of three individual experiments. (D) PDX GBM cells were implanted intracranially into the right hemisphere of athymic nude mice. Dots represent the mean from each individual mouse. Download Figure 1-2, TIF file (6.9MB, tif)
To determine whether this alteration in histone status of the DRD2 promoter leads to increased protein expression, we analyzed protein levels of all DRDs in PDX GBM cells treated with either vehicle control DMSO or TMZ for 2, 4, 6, or 8 d by Western blot. We found that DRD2 was specifically increased after treatment (Fig. 1C) compared with the other dopamine receptors. We also performed FACS analysis of DRD2 expression in three different PDX cells lines over the same time course. In line with our Western blot data, we observed induction of DRD2 protein expression following TMZ treatment (Fig. 1D; GBM6: Day 2, adj p = 0.0018; Day 4, adj p = 0.0355; Day 6, adj p = 0.0001; Day 8, adj p = 0.0001; GBM39: Day 2, adj p = 0.0131; Day 4, adj p = 0.0193; Day 6, adj p = 0.2297; Day 8, adj p = 0.1585; GBM43: Day 2, adj p = 0.0087; Day 4, adj p = 0.0001; Day 6 adj p = 0.0001; Day 8, adj p = 0.0492). To confirm DRD2 expression was not caused by culture conditions, we quantified the expression of DRD2 in orthotopic xenografts from the brains of athymic nude (ν/ν) mice, as well as the presence of DRD2 mRNA in human patient samples (Fig. 1-1A). Our results corroborate several recent studies. First, GBM tumors in patients express DRD2; these levels are often elevated compared with healthy brain (Li et al., 2014). DRD2 is known to interact with EGFR signaling, a pathway frequently mutated or hyperactive in GBM (Winkler et al., 2004; Giannini et al., 2005; Mukherjee et al., 2009; Schunemann et al., 2010; Verhaak et al., 2010; Snuderl et al., 2011; Szerlip et al., 2012). Other groups have shown that DRD2 inhibition in conjunction with EGFR antagonists reduced GBM proliferation (O'Keeffe et al., 2009; Li et al., 2014). Based on these results, we selected DRD2 as our primary receptor for further exploration.
CD133+ glioma initiating cells respond to DRD2 activation with electrophysiological changes
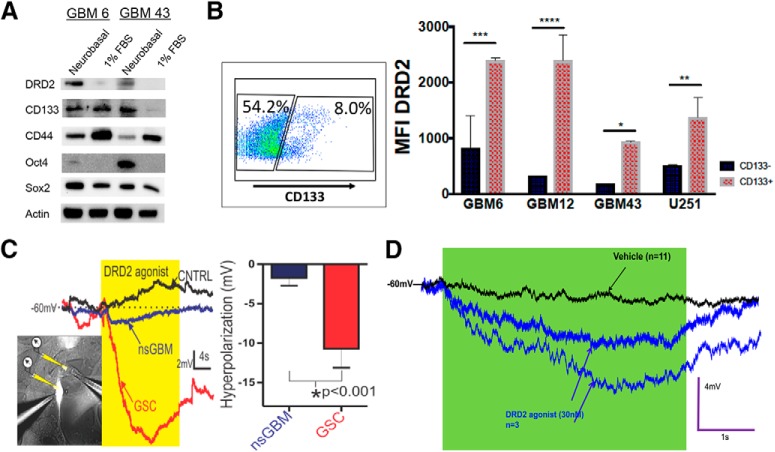
We next tested whether DRD2 expression correlates with certain cellular phenotypes. First, we examined how DRD2 protein expression is affected by alterations in culture conditions known to promote the GIC state. We examined expression of DRD2 in PDX cells growing either in differentiation condition media (DMEM with 1% FBS) or as spheres in supplemented neurobasal media. We found that cells growing as spheres in GIC maintenance media had elevated expression not only of GIC markers like CD133 and Sox2, but also heightened DRD2 (Fig. 2A), indicating that cellular state can influence this receptor's expression. We next analyzed whether DRD2 expression correlates with the expression of CD133, a widely used GIC marker. FACS analysis revealed that DRD2 expression is elevated in the CD133+ population (Fig. 2B; GBM6: adj p = 0.0001, GBM12: adj p = 0.0393, GBM39: adj p = 0.0199, GBM43: adj p = 0.00257).
Figure 2.
Glioma initiating cells preferentially express DRD2 and specifically respond to receptor activation with hyperpolarization. A, Two different subtypes of PDX lines freshly isolated from animals were cultured in tumor sphere maintenance media (neurobasal media containing EGF and FGF) or differentiation media (DMEM containing 1% FBS) and subjected to immunoblot analysis for the expression of DRD2 and various GIC markers. B, PDX cells were subjected to FACS analysis for GIC marker CD133 and DRD2. Using controls, we established gates for CD133+ and CD133- populations. We then quantified the mean fluorescence intensity (MFI) for DRD2 expression in each of these populations. C, Human glioma U251 cells expressing RFP under the control of the CD133 promoter were patch-clamped and electrophysiological recordings were taken during exposure to highly selective DRD2 agonist. Puffing DRD2 agonist (30 nm; yellow) onto fluorescently labeled GIC (brightly labeled cell, left; red membrane potential trace, middle) induces a more robust hyperpolarization of the membrane potential (mV) than non-GICs exposed to the same agonist (nsGBM; non-labeled cell, left; blue trace, middle), or versus puffing vehicle control (n = 4 each). Graph: DRD2 agonist hyperpolarizes GIC more strongly than non-stem GBM cells (right). D, Electrophysiological recordings of PDX cells confirms that only certain cells respond to the agonist with hyperpolarization. Bars represent means from three independent experiments and error bars show the SD. Student t tests were performed for each day separately. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Intrigued by this differential expression, we analyzed functional differences in DRD2 signaling in GICs and non-GICs within a heterogeneous cell population. One of DRD2's main effects is to alter the polarization of brain cells (Beaulieu and Gainetdinov, 2011). We therefore used electrophysiology to determine the effects of DRD2 activation on GBM cellular polarization. We have previously developed a GIC-reporter line in which expression of red fluorescent protein (RFP) is under the control of a GIC-specific CD133-promoter. This system enables real-time, faithful identification of tumor-initiating GICs in live cells (Lee et al., 2016). We treated our high-fidelity U251 GIC reporter cell line with the highly specific DRD2 agonist (3′-fluorobenzylspiperone maleate; Tocris Bioscience) at a dose of 30 nm, the estimated concentration of dopamine in CSF (Keefe et al., 1993; Floresco et al., 2003). Electrophysiological recordings revealed that RFP+CD133+ cells responded to DRD2 activation to a greater extent than RFP− non-GICs in the same population (Fig. 2C; p = 0.0010). Notably, this activation produced hyperpolarization's similar in amplitude and duration to neuronal responses (Beaulieu and Gainetdinov, 2011). We also confirmed that only certain PDX cells respond to the agonist (Fig. 2D). These results show that DRD2 receptors in GBM cells are functional and produce GIC-specific effects.
Activation of DRD2 increases GBM cell self-renewal and tumor engraftment capacity
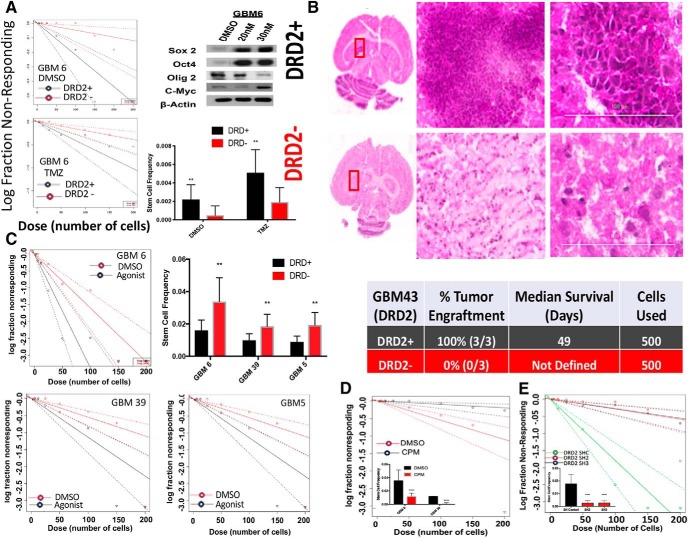
We next sought to understand the relationship between DRD2 expression and tumor cell phenotype during therapy, specifically self-renewal ability and tumor engraftment capacity. To that end, PDX GBM cells were treated with either DMSO or TMZ. Following 4 d of exposure, cells were sorted based on DRD2 expression and plated for limiting dilution neurosphere assays. We found that DRD2-positive (DRD+) cells exhibited approximately fourfold elevation in sphere formation capacity compared with DRD2-negative (DRD−) cells (Fig. 3A; DMSO pretreatment: p = 0.00709, TMZ pretreatment: p = 0.00616), suggesting that the DRD2+ population has enhanced self-renewal capacity. Exposure to physiological dose of TMZ also enhanced the self-renewal capacity in both DRD2+ compared with DRD2− population. Most importantly, post-TMZ therapy enhancement of self-renewal resulted in enhanced tumor engraftment capacity in the orthotopic xenograft model because 100% of the mice developed tumor when 500 DRD2+ cells were implanted in the brain of the immunocompromised nu/nu mice (Fig. 3B).
Figure 3.
Activation of DRD2 signaling increases the self-renewal capacity of GBM cells. A, PDX GBM cells treated with either DMSO or TMZ were sorted based on DRD2 expression and neurosphere assays were performed. Using the extreme limiting dilution assay algorithm, frequency of GICs was calculated (DMSO, DRD2 + 1/458, and DRD2- 1/2091, p < 0.001; TMZ, DRD+ 1/196, and DRD2 − 1/515, p < 0.001). B, In vivo engraftment of 500 cells sorted on DRD2 ± expression demonstrates DRD2+ cells engraft more efficiently. Figure 3-1 includes more images of engrafted tumors from other mice. PDX lines treated with dopamine agonist demonstrate elevation in key stem cell genes via Western blot. Figure 3-1 includes Western blots from other PDX lines demonstrating similar trends. C, PDX GBM cells were treated with either DRD2 agonist (30 nm) or equimolar vehicle control DMSO and plated for neurosphere assays. Using the extreme limiting dilution assay algorithm, frequency of GICs was calculated [GBM6: DMSO stem cells 1/62 and agonist stem cells 1/29.2 (p < 0.01); GBM39: DMSO stem cells 1/181.9 and agonist stem cells 1/87.8 (p < 0.05); GBM5: DMSO stem cells 1/118.9 and agonist stem cells 1/67.4 (p < 0.05)]. Figure 3-1 includes a nonresponding GBM12 neurosphere assay. D, Two PDX lines were treated with either CPM (1 μm) or equimolar DMSO and plated for neurosphere assays as above. Fraction Nonresponding graph is shown for GBM 39. Difference between groups was determined by score test of heterogeneity. Error bars represent upper and lower limit of 95% confidence interval computed for stem-cell frequency. E, Viral knock-out of DRD2 using two different SH-RNA constructs in GBM43 demonstrates reduced stem-cell frequency. Student t tests were performed for each comparison. **p < .001, ****p < 0.0001.
Extended Data Figure 3-1: Activation of DRD2 increases expression of GIC markers. (A) Neurosphere assay of PDX GBM12 treated with either DRD2 against (30nM) or equimolar DMSO. Extreme limiting dilution assays reveal no significant difference in stem cell frequency. (B)Mice injected with DRD2+ cells show efficient tumor engraftment compared to DRD2- cells. (C) PDX GBM cells treated with agonist or DMSO and Ki67 levels were determined by FACS. No difference in Ki67 was detected. (D) PDX cells treated with agonist or DMSO and the expression of GIC markers was examined by western blot after four days. Beta-actin was used as a loading control. (E) U251-CD133 RFP driven reporter line was treated with agonist for 24 hours and RFP (reporter) and CD133 (ligand) were assayed. Download Figure 3-1, TIF file (34.8MB, tif)
Based on our data, as well as recently reported data demonstrating that DRD2 signaling influences normal progenitor phenotype and differentiation (Ohtani et al., 2003; Baker et al., 2004; O'Keeffe et al., 2009; Winner et al., 2009), we next examined whether the observed self-renewal capacity of DRD2+ cells was indeed DRD2 dependent and whether tumor subtype influences such characteristics. Multiple subtypes of PDX cell lines were plated in the presence of either vehicle control DMSO or 30 nm DRD2 agonist and, after 8 d, neurospheres were counted. Our results demonstrate that, for three of the PDX lines tested, DRD2 activation increases sphere-forming capacity (Fig. 3C; GBM5: p = 0.00239, GBM6: p = 0.00362, GBM39: p = 0.0138). GBM12, however, did not respond to the agonist, thus indicating there may be some subtype-specific variability when responding to DRD2 signaling (Fig. 3-1 To confirm this increase in GIC, we analyzed the expression of several key GIC markers following treatment with the DRD2 agonist and found that cell lines that responded to the agonist by increasing sphere formation also showed increased expression of genes associated with stemness (Fig. 3-1. Importantly, treatment did not increase cell proliferation as measured by the Ki67 FACS, suggesting our result comes from cellular plasticity and not increased GIC proliferation (Fig. 3-1. In addition, inhibition of DRD2 by chlorpromazine (CPM; 1 μm), a selective DRD2 antagonist(Beaulieu and Gainetdinov, 2011) in agonist-responsive cell lines GBM6 and GBM39, markedly reduced sphere formation, as measured by neurosphere assays (Fig. 3D; GBM6: p = 1.42E−5, GBM: 39 p = 4.93E−10). Finally, SH-RNA mediated knock-down of DRD2 showed significant reduction in sphere forming capacity in GBM 43 across two different SH-RNA constructs (Fig. 3E; sh2 p = 1.19E−10, sh3 p = 4.53E−10).
GBM cells express TH and secrete dopamine
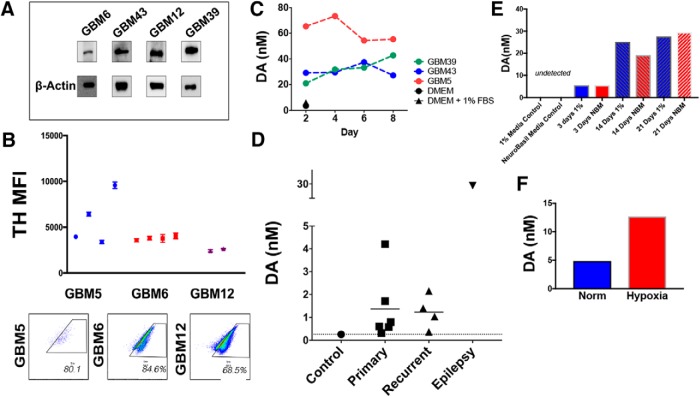
Our data indicate that DRD2 activation alters the phenotype of GBM cells. We next investigated the possibility that this DRD2 activation occurs via autocrine release of its natural ligand, dopamine. Production of monoamines like dopamine is usually a tightly regulated process limited to specific dopaminergic neuronal populations. Critically, all our experiments were performed in purified monocultures of cancer cells in the absence of dopaminergic neurons, raising the question of the mechanism of DRD2 activation. To investigate the possibility that tumor cells synthesize their own dopamine, we first examined expression of tyrosine hydroxylase (TH), the rate-limiting enzyme of dopamine synthesis, in PDX GBM cells (Fig. 4A). To confirm this expression occurs in vivo, we intracranially implanted PDX GBM cells into athymic nude mice; upon visible signs of tumor burden, mice were killed and whole brains were obtained. FACS analysis revealed that human leukocyte antigen (HLA+) tumor cells do in fact express TH in vivo (Fig. 4B). Examination of GlioVis data (Bowman et al., 2017) also demonstrated that patient GBM samples express TH and other enzymes necessary for dopamine synthesis, as well as transporters for vesicle loading and reuptake (Fig. 4-1.
Figure 4.
GBM cells synthesize and secrete dopamine. A, Western blot analysis of TH expression, the rate-limiting enzyme in dopamine synthesis. β-Actin was used as a loading control. B, PDX cells were implanted intracranially into athymic nude mice. Upon sickness from tumor burden, mice were killed and whole brains extracted. FACS analysis of HLA+ tumor cells demonstrates that GBM tumors continue to express TH in vivo. Each dot represents individual mice; FACS analyses were run in technical triplicates. Inset, Scatter plots are representative for FACS results in each cell line tested. C, Conditioned media from PDX GBM cells growing in purified monocultures were analyzed by HPLC-MS for dopamine. Unconditioned DMEM media with and without 1% FBS was used as a control. D, HPLC-MS was performed on GBM samples from patients undergoing resection. Cortical tissue from an epilepsy surgery was used as the control. E, PDX GBM6 cells were growing in either 1% FBS containing media or neurobasal media (NBM). Conditioned media were collected at various days and dopamine levels were calculated using HPLC. F, PDX GBM43 cells were cultured in the presence of either 20% or 1% oxygen (hypoxia) for 3 h. Conditioned media was collected, and dopamine levels determined. Dots represent values from each time point and/or sample tested. Figure 4-1 includes other genes involved in dopamine synthesis (VMAT2, DAT, DDC, TH) measured in patient tumor samples as well as HPLC performed on various PDX subtypes undergoing a TMZ treatment time course.
Extended Data Figure 4-1: (A) Patient tumors express mRNA for several genes involved in synthesis, secretion, and reuptake of dopamine. Data were taken from the Cancer Genome Atlas (TCGA). Expression was examined in astrocytomas (Grade 1, II, III, IV) and within GBM based on subtype (Classical, Mesenchymal, Proneural). (B) PDX lines were exposed to 50μm TMZ and collected after 2, 4, 6, and 8 days and subjected to HPLC. Results are shown normalized to equimolar DMSO controls. Download Figure 4-1, TIF file (12.4MB, tif)
To determine whether PDX monocultures were generating dopamine, we assayed dopamine levels using high performance liquid chromatography with mass spectrometry (HPLC-MS). Conditioned media of PDX monocultures were found to have dopamine levels of 20–65 nm, between 10 and 30 times greater than cell-free media (Fig. 4C). The levels of dopamine were relatively stable in samples taken over 8 d, suggesting that the PDX cells produce and maintain basal dopamine synthesis. Next, we quantified dopamine in human tumor samples. A portion of cortex removed from an epilepsy surgery was also analyzed to determine dopamine levels in noncancerous brain tissue. These assays revealed dopamine expression in all cases (Fig. 4D).
In light of our previous data indicating that dopamine signaling is associated with cell state, we next examined how cell state influences dopamine secretion. PDX GBM6 cells were cultured in either 1% FBS or neurobasal media. Conditioned media were collected after 3, 14, and 21 d and dopamine levels determined by HPLC-MS. We found that dopamine production was time dependent, but not clearly linked to culture conditions (Fig. 4E). Furthermore, it has been established that hypoxia induction can influence the ability of stem cells to differentiate and produce dopamine (Wang et al., 2013). We therefore analyzed dopamine levels from media in which PDX GBM cells were growing exposed to either 20% normoxia or 1% hypoxia for 3 h. HPLC-MS revealed that hypoxia increase dopamine levels (Fig. 4F), suggesting a similar mechanism.
Activation of DRD2 induces expression of HIF proteins
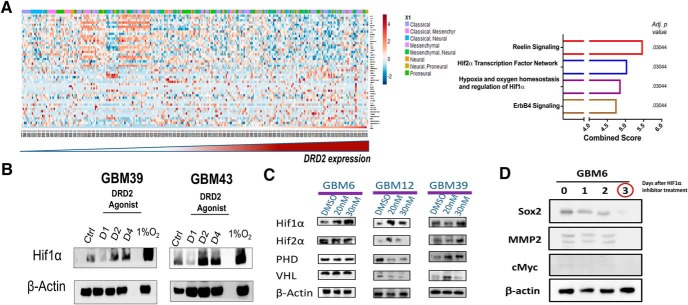
To determine how DRD2 signaling may underlie the observed changes in self-renewing capacity, we analyzed patient tumor samples from TCGA. Patient samples were stratified from lowest to highest expression of DRD2 and correlations were determined for 12,042 genes by Pearson correlation coefficient analysis; coefficients >0.5 or <−0.5 and false discovery rate (FDR) < 0.05 were used. Genes fitting these parameters were then analyzed using Enrichr pathway analysis program. Our analysis revealed that DRD2 levels are positively correlated with the expression of hypoxia inducible factors (HIF) 1α and HIF2α pathways, as well as with the Reelin and ErbB4 signaling networks (adj p = 0.03044; Fig. 5A).
Figure 5.
DRD2 signaling activates hypoxia inducible factors in normoxic cultures. A, Correlation between DRD2 and 12042 genes from TCGA was determined by Pearson correlation coefficients. Forty-nine genes with coefficients >0.5 or <−0.5 and FDR <0.05 were selected. Genes fitting these parameters were then analyzed using Enrichr. Top hits are shown in the graph, with combined score and adjusted p value. B, PDX GBM cells were treated with DRD2 agonist (30 nm) for 1, 2, or 4 d and then probed for expression of HIF1α by immunoblot analysis. DMSO-treated cells as vehicle-treated control and cells exposed to hypoxic conditions (1.5% O2) were used as a positive control for HIF expression, and β-actin was used as a loading control. C, Western blots were used to analyze the level of proteins in the VHL–PHD regulatory pathway after treatment with either DMSO or DRD2 agonist (20 or 30 nm). β-Actin was used as a loading control. D, PDX GBM6 were exposed to DRD2 agonist (30 nm) and treated with HIF inhibitor for 24, 48, and 72 h; SOX2, MMP2, and CMYC protein levels were assayed.
Hypoxia is well known to influence the phenotype of GBM cells, pushing them toward a more GIC phenotype (Evans et al., 2004; Soeda et al., 2009). Further, we have previously shown that GBM cells use HIF signaling to promote cellular plasticity following therapy (Lee et al., 2016). To investigate how DRD2 stimulation affects HIF signaling, PDX cells GBM39 and GBM43 were treated with agonist under normal and hypoxic conditions. Immunoblot analysis demonstrated a substantial, time-dependent increase in HIF1α levels in both cell lines following activation of DRD2 signaling, regardless of oxygen availability (Fig. 5B).
HIF protein levels are controlled both at the level of gene transcription and via hydroxylase-dependent degradation. To determine whether DRD2's activation of HIF was occurring at the level of gene transcription or degradation, we analyzed the levels of proline-hydroxylase domain (PHD) and von Hippel-Lindau (VHL), key regulators of HIF stabilization, following treatment with the DRD2 agonists by Western blot. These assays revealed cell-line-specific changes in the HIF pathway. In GBM12, the expected correlations were observed; elevated HIF levels matched decreased levels of both PHD and VHL. However, in GBM39, the increased HIF expression actually correlated with elevated PHD and VHL (Fig. 5C). Notably, these results indicate that increases in HIF levels following DRD2 activation occurs in both cell lines that are responsive and nonresponsive in neurosphere assays. This common shared HIF induction with differential functional outcomes suggests that shared activation of HIF leads to unique downstream effects.
We next sought to determine whether the increased GIC population following DRD2 activation was dependent on the previously identified increase in HIF1α in the clinically relevant PDX model. GBM6 treated with HIF1α inhibitor after exposure to DRD2 agonist show loss of Sox2, CMYC, and MMP2 expression (Fig. 5D).
Activation of DRD2 induces alterations in expression of HIF-responsive genes and functional changes in metabolic phenotype
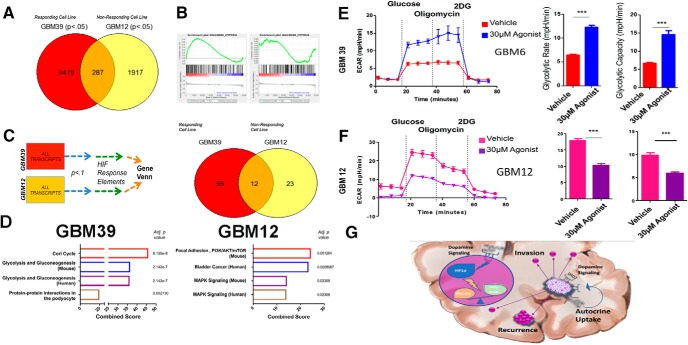
HIF signaling can have profound effects on cancer cells, ranging from shifts in gene expression to altered functional attributes. For a full transcriptome profile of the mechanisms governing DRD2's influence on GBM, we performed microarray analysis of gene expression in cells treated with either vehicle control DMSO or 30 nm DRD2 agonist for 4 d. We chose two cell lines with distinct responses to dopamine in sphere-forming assays; neurosphere-responsive GBM39 and nonresponsive GBM12. To determine the changes that were specific to each cell line, we used the online GeneVenn tool. Transcripts with p < 0.05 from each cell line, comparing agonist and DMSO controls, were identified and overlap was analyzed by GeneVenn. We found that 3479 increases in gene expression were specific to neurosphere-responsive GBM39, whereas 1917 increases were unique to neurosphere-nonresponsive GBM12. These two cell lines showed shared increases in only 287 genes (Fig. 6A). Enrichr analysis of these cell lines' dopamine-induced transcriptomes revealed activation of unique signaling pathways (Fig. 6B). Given our result indicating a connection between HIF signaling and DRD2, we next examined genes known to be regulated by HIF during canonical hypoxia responses. We therefore performed Gene set enrichment analysis (GSEA) of only the hypoxia-response gene sets. This analysis confirmed a subtype-dependent response to DRD2 activation. GBM39 cells did not activate canonical HIF targets following agonist, whereas GBM12 did (Fig. 6B). These results indicate a specific change in gene expression following the agonist and suggest that these differences may underlie the functional changes in sphere formation.
Figure 6.
DRD2 triggers alterations in gene expression and metabolic phenotype. A, Microarray analysis of gene expression was performed in PDX cells treated with DRD2 agonist after 4 d. GeneVenn analysis was performed on gene transcripts found to be significantly elevated in the two cell lines, revealing subtype-specific alterations in expression profiles. B, GSEA was performed on each cell line for canonical hypoxia response signaling. C, D, Microarray results were then limited to genes with confirmed hypoxia response elements. E, F, Seahorse analysis was performed to quantify the glycolytic rate in two PDX lines; (E) GBM39, which responds to DRD2 activation with increased sphere-formation, and (F) GBM12, which are unresponsive to DRD2 activation in neurosphere formation. Cells were treated with either DMSO or 30 nm DRD2 agonist and subjected to Seahorse analysis after 96 h. Figure 6-1 includes a responding PDX line, GBM5, as an assay control demonstrating similar effects. Figure 6-1 also includes FACS analysis of glucose uptake and fatty acid uptake in GBM12 and 39 as further validation of seahorse assays. G, Summary schematic of dopamine's role within the tumor microenvironment and at the single-cell level. Microarray was performed on biological triplicates. Bars represent means from three independent experiments and error bars represent SD. Student t tests were performed for each separate cell line. ***p < 0.001.
Extended Data Figure 6-1 (A) Alkylating chemotherapy alters the uptake of glucose and fatty acid. PDX GBM cells were treated with either DMSO or TMZ. Four days later, cells were exposed to fluorescently-labeled glucose and palmitate. Uptake was determined by FACS analysis. (B) Seahorse analysis was used to determine the glycolytic rate of PDX lines after 4 days of 30nm agonist, or equimolar DMSO treatment. Comparisons were made using student t-Tests. *p<.05, **p<.01, ***p<.001. Download Figure 6-1, TIF file (14.3MB, tif)
Although many genes are activated in response to HIF proteins, certain genes are known to be activated via direct binding of HIFs to gene promoters at conserved regions termed HIF-responsive elements (HRE). To determine whether the observed changes in gene expression occurred at the level of actual HIF binding, we collected all the gene transcripts that were significantly upregulated after treatment with the DRD2 agonist and known to have a promoter containing the HRE (previously confirmed by ChIP-Seq; Schödel et al., 2011). These sets of genes were then analyzed using the Enrichr platform. Once again, we found that the two cell lines showed drastically different responses to the agonist (Fig. 6C). Treated GBM39 cells showed increased levels of HIF-regulated genes, including glycolysis (p = 2.142E-7), whereas GBM12 showed increased activation of the PI3K/AKT pathway (p = 0.001264; Fig. 6D). These results suggest that the differential effects of DRD2-induced activation of HIF in classical GBM39 and proneural GBM12 depend on unique signaling mechanisms.
Changes in gene expression, although informative, do not guarantee a functional alteration in cellular behavior. We therefore sought to determine whether activation of DRD2 does in fact lead to GBM39-specific alterations in metabolism. First, we analyzed the uptake of glucose and fatty acid via FACS, using 2-NBDG, a established fluorescent analog of glucose, and our proprietary fatty-acid tagged quantum dots (Muroski et al., 2017). Only GBM39 showed altered fatty acid uptake following DRD2 activation. However, all cells treated with the agonist showed increases in glucose uptake (Fig. 6-1. To further characterize the effects of DRD2 signaling on glucose metabolism, we performed extracellular flux analysis. We discovered a differential effect predicted by each cell line's behavior in neurosphere assays. GBM39, of the classical subtype and responsive to the agonist in neurosphere assays, showed a marked increase in glycolytic rate as measured by glucose stimulated ECAR (p = 0.00010; Fig. 6E). In contrast, GBM12, the nonresponsive proneural subtype, actually showed decreases in glycolysis following DRD2 agonist treatment (Fig. 6F; p = 0.00011).
Discussion
In this study, we investigated how DRD2 activation contributes to the molecular and functional phenotype of GBM. We demonstrated that therapeutic stress induced by anti-glioma chemotherapy alters the epigenetic status of the DRD2 promoter and subsequently increases DRD2 protein expression. Notably, this expression was elevated in GICs and could be induced by GIC-promoting culture conditions. In classical GBM cells, DRD2 activation increased sphere forming capacity, an indicator of GIC state and tumorgenicity, as well as the expression of several key GIC markers. We further found that GBM cells themselves can generate dopamine in purified monocultures, and therefore exhibit autocrine/paracrine DRD2 activation capabilities. In all lines tested, activation of DRD2 induced the expression of HIF proteins, regardless of oxygen availability. Whereas the HIF induction by DRD2 activation was common to all cell lines tested, the downstream functional changes varied. Classical GBM39 cells exhibited increased uptake of glucose and glycolytic rate following DRD2 stimulation. In contrast, proneural GBM12 cells showed no change in sphere-forming capacity. However, activation of DRD2 did induce expression of canonical HIF-responsive genes and led to a reduction in their glycolytic rate. Cells exposed to low oxygen conditions typically activate HIF and subsequently upregulate glycolysis; the fact that proneural cells do not upregulate this metabolism, despite elevated HIF levels, suggests a unique HIF regulatory mechanism. These results indicate a key role for DRD2 signaling in influencing GBM cellular plasticity, at the level of both gene expression and functional attributes.
Induction of cellular plasticity, specifically in steering cells toward a treatment-resistant GIC state, is a key problem in GBM and contributes to tumor recurrence (Dahan et al., 2014; Olmez et al., 2015; Safa et al., 2015; Lee et al., 2016). Our data demonstrate that dopamine signaling plays a role in this cellular conversion. Functionally, exposure to DRD2 agonist was sufficient to increase sphere-forming capacity of PDX GBM cells. Given the concurrent increase in GIC marker expression as well as engraftment efficiency, this treatment appears to induce the conversion of GBM cells to a more stem-like state.
Our results indicate that activation of DRD2 induces unique downstream effects dependent on the genetic subtype of cell line used. Even more interestingly, these unique changes in gene expression and phenotypes occurred despite shared activation of a key signaling node; HIFs. GBM39, of the classical tumor subtype, responds to the agonist with increased sphere-formation, expression of key GIC markers, altered gene expression related to metabolism, and increased glycolytic rate. In contrast, GBM12, a proneural cell line, did not increase neurosphere formation or expression of GIC markers, but did show induction of HIFs and altered gene expression related to canonical HIF signaling. It remains an open question as to which other signaling mechanisms when activated in these two subtypes enable common signaling nodes to induce cell-line-specific changes in gene expression and functional phenotype. The unique genetic makeup of each cell line may provide an explanation of this phenomenon. Analysis of patient sample gene expression and orthotopic xenograft tumor protein levels showed that proneural tumors and GBM12 xenografts have somewhat elevated expression of DRD2 (Fig. 1-1) suggesting that this signaling axis may already be active. Further, GBM39 is of the classical subtype and carries the EGFRvIII mutation, whereas GBM12 expresses wild-type EGFR (Giannini et al., 2005). EGFR is connected to dopamine signaling in neural development (Höglinger et al., 2004); therefore, the fact that EGFR mutant cell lines respond with increased sphere-formation suggests the possibility that increased EGFR activity may play a role in the ability of DRD2 to alter cellular plasticity.
These differential responses to activation of the same receptor further highlight a key challenge facing the neuro-oncology community; intratumoral heterogeneity. GBM tumors have been shown to contain cells corresponding to each of the three subtypes within a single tumor (Sottoriva et al., 2013; Patel et al., 2014). This wide range of cell types and diverse molecular phenotypes currently limit the effective treatment of GBM. These data highlight the importance of this heterogeneity, by suggesting a mechanism for the same molecular signal to exert unique influences on different subpopulations of tumor cells. It is not hard to imagine that dopamine may generate multiple adaptive features, including elevated self-renewal capacity or maintenance of the GIC pool via increased glycolysis, and neo-angiogenesis in GBM cells.
These data illustrate that dopamine signaling exerts influence on global as well as HIF-dependent gene expression. Further, our data demonstrate that these changes in gene transcription have a functional effect, as dopamine signaling is capable of altering the metabolism of GBM cells. GBM39 showed a highly elevated glycolytic rate following agonist treatment, whereas GBM12 cells had slightly reduced rates of glycolysis. How this increase in glucose uptake and glycolytic rate relates to induction of the GIC state, however, remains an open question. Several groups have recently demonstrated that altered metabolic phenotypes influence the acquisition of a stem cell phenotype in cancer cells (Currie et al., 2013; Mao et al., 2013); it is not outside the realm of possibility that a similar effect is at play here. Further research will delineate the precise connection between elevated glucose uptake and glycolytic rate and altered cell state.
It should be noted that a recent report suggests that dopamine may actually inhibit growth of glioma. Lan et al. (2017) showed that dopamine reduced growth of U87 and U251 cells growing as subcutaneous xenografts. Although differences in cell lines (U87 vs PDX) as well as the subcutaneous placement of tumors may account for this difference, this result suggests a dose-dependent effect of dopamine. Critically, our experiments were performed with doses of DRD2 agonist between 20 and 30 nm, whereas Lan et al. (2017) performed experiments with doses of dopamine between 10 and 25μΜ. Dopamine receptor signaling is highly sensitive to concentration of ligands and antagonists, which complicates understanding its role in GBM (for review, see Caragher et al., 2018). It is possible that the influence of dopamine signaling is dose-dependent. Further, this work highlights the complex nature of dopamine's multiple receptors. Adding a pan-dopamine receptor agonist, as they did, will activate all five receptors, which have unique downstream signaling pathways.
Our data also show that GBM cells can synthesize and secrete dopamine, an ability assumed to be the sole province of neurons. However, there is mounting evidence that GBM cells have the ability to adopt certain neuronal functions. Osswald et al. (2015) demonstrated that GBM cells use GAP43, neuronal outgrowth cone protein, to form networks. Further, it has been shown that GBM cells synthesize and secrete glutamate, an excitatory neurotransmitter, and alter cortical activity (Buckingham et al., 2011). Developmental neurobiology also provides some clues as to how these cells can attain a neuron-specific behavior. Neural stem cells engrafted into the striatum of mice can spontaneously attain dopaminergic traits, suggesting that secreted factors in the brain can induce the formation of dopamine secreting cells (Yang et al., 2002). Given the highly similar expression profile of NSCs and Glioma initiating cells, a similar mechanism may induce the formation of a dopamine-producing population of glioma cells. Finally, a recent report using TCGA datasets indicated that high TH expression correlates with reduced median survival (Dolma et al., 2016). Critically, these data analyzed mRNA expression in GBM cells themselves, not in surrounding brain cells. Therefore, the survival difference relies on how TH functions in the tumor.
In short, GBM cells exhibit a variety of neuron-like properties, including the ability to synthesize and secrete dopamine. Our in vitro data indicate that the GBM cells are synthesizing dopamine rather than taking up dopamine from the surrounding dopamine neurons and storing it. Our results clearly fit within this emerging theory that GBM tumors may display neuron-specific behaviors. More research is needed to delineate the specific subpopulations of GBM tumors that secrete dopamine and the mechanisms governing this process. This result, combined with the evidence that DRD2 signaling promotes a shift to the GIC state, suggests that tumors use dopamine as a mechanism to control their microenvironment and promote a pro-growth, pro-stemness milieu.
It could be argued that even if GBM cells do use dopamine in an autocrine manner, they must also rely on ambient dopamine in the brain microenvironment to induce major signaling changes. It remains possible that non-tumor dopamine also influences GBM. It has been shown previously that proteins from the synaptic cleft of nearby neurons can influence glioma growth (Venkatesh et al., 2015). We therefore cannot rule out the possibility that our tumor sample data represent cells that have absorbed dopamine rather than produced it (Fig. 4). Indeed, TCGA indicates that GBM cells express the dopamine transporter (Fig. 4-1). In addition, it has been shown that patients with Parkinson's disease, characterized by the loss of dopaminergic neurons, have a lower incidence of high-grade gliomas (Diamandis et al., 2009). Further, studies of tumor localization indicate that GBM tumors preferentially localize to areas of dopamine innervation (Larjavaara et al., 2007). We do not exclude or reject the possibility that ambient dopamine influences tumor behavior, with two key caveats. First, given that only specific portions of the brain contain dopaminergic neurons (Ungerstedt, 1971) and that GBM tumor growth often disrupts the normal neural circuitry in their area, it cannot be guaranteed that dopamine from the microenvironment is widely available for every tumor. Second, the three-dimensional shape of the tumor is likely to limit perfusion of certain molecules from distal sources into the cleft. As such, ambient dopamine released from neurons may also influence signaling in GBM tumors.
Finally, it remains a lingering question the extent to which the effects of dopamine signaling observed here are GBM-specific or represent a recapitulation of the behavior of normal NSCs and brain progenitor cells. It has been demonstrated, for example, that NSCs in the subventricular zone respond to dopamine with increased proliferation (Baker et al., 2004; O'Keeffe et al., 2009; Winner et al., 2009). Activation of dopamine receptors on progenitors in the developing brain has also been shown to expand neuronal populations and neurite outgrowth (Yoon and Biak, 2013), and alter cell cycle status (Ohtani et al., 2003). Finally, dopamine signaling in oligodendrocyte precursors expands the pluripotent population (Kimoto et al., 2011). These studies suggest that GBM cells have hijacked an existing role for dopamine in brain development for their own gains. The connection of metabolism, however, was not identified in these studies or other relevant literature. Further research will undoubtedly expand on this interesting connection.
In sum, our data indicate that GBM cells respond to DRD2 activation at the level of both gene expression and functional phenotype and that specific inactivation of DRD2 itself is required to provide therapeutic benefit (Fig. 3 and Fig. 3-1). Further research will shed light on the dynamics of tumor-derived dopamine and mechanisms influencing key factors like rate of production and secretion, interaction with noncancerous cells in the environment, and the effects of tumor-derived dopamine on therapy resistance. Novel creation of specific dopamine receptor inhibitors may provide better clinical relevance toward treatment of tumor types that rely on dopamine signaling dynamics. Overall, this study highlights a novel mechanism by which GBM cultivates a supportive microenvironmental niche to colonize the CNS. It also suggests that GBM tumors possess a remarkable ability to hijack cellular mechanisms previously thought to be the exclusive provenance of neurons. Given the growing chorus of studies indicating that dopamine antagonists inhibit GBM growth (Shin et al., 2006; Karpel-Massler et al., 2015; Kang et al., 2017), these results provide critical context for the source of tumor-influencing dopamine and the mechanism underlying dopamine's role in gliomagenesis.
Footnotes
This work was supported by the National Institute of Neurological Disorders and Stroke Grant 1R01NS096376, the American Cancer Society Grant RSG-16-034-01-DDC (to A.U.A.), R01NS095642 (to C.D.J.) and National Cancer Institute Grant R35CA197725 (to M.S.L.) and P50CA221747 SPORE for Translational Approaches to Brain Cancer. We thank Meijing Wu for the statistical analysis for the preparation of this paper.
The authors declare no competing financial interests.
References
- Alifieris C, Trafalis DT (2015) Glioblastoma multiforme: pathogenesis and treatment. Pharmacol Ther 152:63–82. 10.1016/j.pharmthera.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Baker SA, Baker KA, Hagg T (2004) Dopaminergic nigrostriatal projections regulate neural precursor proliferation in the adult mouse subventricular zone. Eur J Neurosci 20:575–579. 10.1111/j.1460-9568.2004.03486.x [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Gainetdinov RR (2011) The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev 63:182–217. 10.1124/pr.110.002642 [DOI] [PubMed] [Google Scholar]
- Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M (2017) GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol 19:139–141. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham SC, Campbell SL, Haas BR, Montana V, Robel S, Ogunrinu T, Sontheimer H (2011) Glutamate release by primary brain tumors induces epileptic activity. Nat Med 17:1269–1274. 10.1038/nm.2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caragher SP, Hall RR, Ahsan R, Ahmed AU (2018) Monoamines in Glioblastoma: complex biology with therapeutic potential. Neuro Oncol 20:1014–1025. 10.1093/neuonc/nox210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H (2011) The brain tumor microenvironment. Glia 59:1169–1180. 10.1002/glia.21136 [DOI] [PubMed] [Google Scholar]
- Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr (2013) Cellular fatty acid metabolism and cancer. Cell Metab 18:153–161. 10.1016/j.cmet.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan P, Martinez Gala J, Delmas C, Monferran S, Malric L, Zentkowski D, Lubrano V, Toulas C, Cohen-Jonathan Moyal E, Lemarie A (2014) Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: possible involvement in radioresistance. Cell Death Dis 5:e1543. 10.1038/cddis.2014.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamandis P, Sacher AG, Tyers M, Dirks PB (2009) New drugs for brain tumors? Insights from chemical probing of neural stem cells. Med Hypotheses 72:683–687. 10.1016/j.mehy.2008.10.034 [DOI] [PubMed] [Google Scholar]
- Dolma S, Selvadurai HJ, Lan X, Lee L, Kushida M, Voisin V, Whetstone H, So M, Aviv T, Park N, Zhu X, Xu C, Head R, Rowland KJ, Bernstein M, Clarke ID, Bader G, Harrington L, Brumell JH, Tyers M, et al. (2016) Inhibition of dopamine receptor D4 impedes autophagic flux, proliferation, and survival of glioblastoma stem cells. Cancer Cell 29:859–873. 10.1016/j.ccell.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Judy KD, Dunphy I, Jenkins WT, Hwang WT, Nelson PT, Lustig RA, Jenkins K, Magarelli DP, Hahn SM, Collins RA, Grady MS, Koch CJ (2004) Hypoxia is important in the biology and aggression of human glial brain tumors. Clin Cancer Res 10:8177–8184. 10.1158/1078-0432.CCR-04-1081 [DOI] [PubMed] [Google Scholar]
- Floresco SB, West AR, Ash B, Moore H, Grace AA (2003) Afferent modulation of dopamine neuron firing differentially regulates tonic and phasic dopamine transmission. Nat Neurosci 6:968–973. 10.1038/nn1103 [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64:7011–7021. 10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed] [Google Scholar]
- Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, Schroeder MA, James CD (2005) Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol 7:164–176. 10.1215/S1152851704000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardee ME, Marciscano AE, Medina-Ramirez CM, Zagzag D, Narayana A, Lonning SM, Barcellos-Hoff MH (2012) Resistance of glioblastoma-initiating cells to radiation mediated by the tumor microenvironment can be abolished by inhibiting transforming growth factor-beta. Cancer Res 72:4119–4129. 10.1158/0008-5472.CAN-12-0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN (2009) The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 8:3274–3284. 10.4161/cc.8.20.9701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan A, Rich JN (2011) Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ 18:829–840. 10.1038/cdd.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, Spellman PT, Vandenberg SR, Berger MS, James CD (2009) Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol 11:477–487. 10.1215/15228517-2008-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglinger GU, Rizk P, Muriel MP, Duyckaerts C, Oertel WH, Caille I, Hirsch EC (2004) Dopamine depletion impairs precursor cell proliferation in Parkinson disease. Nat Neurosci 7:726–735. 10.1038/nn1265 [DOI] [PubMed] [Google Scholar]
- Huang M, Panos JJ, Kwon S, Oyamada Y, Rajagopal L, Meltzer HY (2014) Comparative effect of lurasidone and blonanserin on cortical glutamate, dopamine, and acetylcholine efflux: role of relative serotonin (5-HT)2A and DA D2 antagonism and 5-HT1A partial agonism. J Neurochem 128:938–949. 10.1111/jnc.12512 [DOI] [PubMed] [Google Scholar]
- Kang S, ong J, Lee JM, Moon HE, Jeon B, Choi J, Yoon NA, Paek SH, Roh EJ, Lee CJ, Kang SS (2017) Trifluoperazine, a well-known antipsychotic, inhibits glioblastoma invasion by binding to calmodulin and disinhibiting calcium release channel IP3R. Mol Cancer Ther 16:217–227. 10.1158/1535-7163.MCT-16-0169-T [DOI] [PubMed] [Google Scholar]
- Karpel-Massler G, Kast RE, Westhoff MA, Dwucet A, Welscher N, Nonnenmacher L, Hlavac M, Siegelin MD, Wirtz CR, Debatin KM, Halatsch ME (2015) Olanzapine inhibits proliferation, migration and anchorage-independent growth in human glioblastoma cell lines and enhances temozolomide's antiproliferative effect. J Neurooncol 122:21–33. 10.1007/s11060-014-1688-7 [DOI] [PubMed] [Google Scholar]
- Keefe KA, Zigmond MJ, Abercrombie ED (1993) In vivo regulation of extracellular dopamine in the neostriatum: influence of impulse activity and local excitatory amino acids. J Neural Transm Gen Sect 91:223–240. 10.1007/BF01245233 [DOI] [PubMed] [Google Scholar]
- Kimoto S, Okuda A, Toritsuka M, Yamauchi T, Makinodan M, Okuda H, Tatsumi K, Nakamura Y, Wanaka A, Kishimoto T (2011) Olanzapine stimulates proliferation but inhibits differentiation in rat oligodendrocyte precursor cell cultures. Prog Neuropsychopharmacol Biol Psychiatry 35:1950–1956. 10.1016/j.pnpbp.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Malenka RC (2014) Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology 76:351–359. 10.1016/j.neuropharm.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan YL, Wang X, Xing JS, Yu ZL, Lou JC, Ma XC, Zhang B (2017) Anti-cancer effects of dopamine in human glioma: involvement of mitochondrial apoptotic and anti-inflammatory pathways. Oncotarget 8:88488–88500. 10.18632/oncotarget.19691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larjavaara S, Mäntylä R, Salminen T, Haapasalo H, Raitanen J, Jääskeläinen J, Auvinen A (2007) Incidence of gliomas by anatomic location. Neuro Oncol 9:319–325. 10.1215/15228517-2007-016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Auffinger B, Guo D, Hasan T, Deheeger M, Tobias AL, Kim JY, Atashi F, Zhang L, Lesniak MS, James CD, Ahmed AU (2016) Dedifferentiation of glioma cells to glioma stem-like cells by therapeutic stress-induced HIF signaling in the recurrent GBM model. Mol Cancer Ther 15:3064–3076. 10.1158/1535-7163.MCT-15-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhu S, Kozono D, Ng K, Futalan D, Shen Y, Akers JC, Steed T, Kushwaha D, Schlabach M, Carter BS, Kwon CH, Furnari F, Cavenee W, Elledge S, Chen CC (2014) Genome-wide shRNA screen revealed integrated mitogenic signaling between dopamine receptor D2 (DRD2) and epidermal growth factor receptor (EGFR) in glioblastoma. Oncotarget 5:882–893. 10.18632/oncotarget.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, Wang M, Hu B, Cheng SY, Sobol RW, Nakano I (2013) Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A 110:8644–8649. 10.1073/pnas.1221478110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, Hatanpaa KJ, Mickey B, Madden C, Maher E, Boothman DA, Furnari F, Cavenee WK, Bachoo RM, Burma S (2009) EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res 69:4252–4259. 10.1158/0008-5472.CAN-08-4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroski ME, Miska J, Chang AL, Zhang P, Rashidi A, Moore H, Lopez-Rosas A, Han Y, Lesniak MS (2017) Fatty acid uptake in T cell subsets using a quantum dot fatty acid conjugate. Sci Rep 7:5790. 10.1038/s41598-017-05556-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Goto T, Waeber C, Bhide PG (2003) Dopamine modulates cell cycle in the lateral ganglionic eminence. J Neurosci 23:2840–2850. 10.1523/JNEUROSCI.23-07-02840.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keeffe GC, Tyers P, Aarsland D, Dalley JW, Barker RA, Caldwell MA (2009) Dopamine-induced proliferation of adult neural precursor cells in the mammalian subventricular zone is mediated through EGF. Proc Natl Acad Sci U S A 106:8754–8759. 10.1073/pnas.0803955106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmez I, Shen W, McDonald H, Ozpolat B (2015) Dedifferentiation of patient-derived glioblastoma multiforme cell lines results in a cancer stem cell-like state with mitogen-independent growth. J Cell Mol Med 19:1262–1272. 10.1111/jcmm.12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, Huang L, Ratliff M, Karimian Jazi K, Kurz FT, Schmenger T, Lemke D, Gömmel M, Pauli M, Liao Y, Häring P, et al. (2015) Brain tumour cells interconnect to a functional and resistant network. Nature 528:93–98. 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K (2015) Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis 2:152–163. 10.1016/j.gendis.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR (2011) High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117:e207–e217. 10.1182/blood-2010-10-314427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunemann DP, Grivicich I, Regner A, Leal LF, de Araújo DR, Jotz GP, Fedrigo CA, Simon D, da Rocha AB (2010) Glutamate promotes cell growth by EGFR signaling on U-87MG human glioblastoma cell line. Pathol Oncol Res 16:285–293. 10.1007/s12253-009-9223-4 [DOI] [PubMed] [Google Scholar]
- Shin SY, Choi BH, Ko J, Kim SH, Kim YS, Lee YH (2006) Clozapine, a neuroleptic agent, inhibits akt by counteracting Ca2+/calmodulin in PTEN-negative U-87MG human glioblastoma cells. Cell Signal 18:1876–1886. 10.1016/j.cellsig.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, Louis DN, Iafrate AJ (2011) Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20: 810–817. 10.1016/j.ccr.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, Kassam AB, Pollack IF, Park DM (2009) Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene 28:3949–3959. 10.1038/onc.2009.252 [DOI] [PubMed] [Google Scholar]
- Sottoriva A, I Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 110:4009–4014. 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, Di Meco F, Lieberman F, Zhu JJ, Stragliotto G, Tran D, Brem S, Hottinger A, Kirson ED, Lavy-Shahaf G, Weinberg U, et al. (2017) Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 318:2306–2316. 10.1001/jama.2017.18718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, Brennan CW (2012) Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A 109:3041–3046. 10.1073/pnas.1114033109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Aoyagi M, Ando N, Ogishima T, Wakimoto H, Yamamoto M, Ohno K (2013) Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy. J Neurosurg 119:1145–1155. 10.3171/2013.7.JNS122417 [DOI] [PubMed] [Google Scholar]
- Ungerstedt U. (1971) Stereotaxic mapping of the monoamine pathways in the rat brain. Acta Physiol Scand Suppl 367:1–48. [DOI] [PubMed] [Google Scholar]
- Venkatesh HS, Johung TB, Caretti V, Noll A, Tang Y, Nagaraja S, Gibson EM, Mount CW, Polepalli J, Mitra SS, Woo PJ, Malenka RC, Vogel H, Bredel M, Mallick P, Monje M (2015) Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell 161:803–816. 10.1016/j.cell.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh HS, Tam LT, Woo PJ, Lennon J, Nagaraja S, Gillespie SM, Ni J, Duveau DY, Morris PJ, Zhao JJ, Thomas CJ, Monje M (2017) Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 549:533–537. 10.1038/nature24014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, et al.; Network Cancer Genome Atlas Research (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, Gu Y, Lum KM, Masui K, Yang H, Rong X, Hong C, Turner KM, Liu F, Hon GC, Jenkins D, Martini M, Armando AM, Quehenberger O, Cloughesy TF, et al. (2016) An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell 30:683–693. 10.1016/j.ccell.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang J, Li H, Wang X, Zhu L, Fan M, Wang X (2013) Hypoxia promotes dopaminergic differentiation of mesenchymal stem cells and shows benefits for transplantation in a rat model of Parkinson's disease. PLoS One 8:e54296. 10.1371/journal.pone.0054296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F, Kozin SV, Tong RT, Chae SS, Booth MF, Garkavtsev I, Xu L, Hicklin DJ, Fukumura D, di Tomaso E, Munn LL, Jain RK (2004) Kinetics of vascular normalization by VEGFR2 blockade governs brain tumor response to radiation: role of oxygenation, angiopoietin-1, and matrix metalloproteinases. Cancer Cell 6:553–563. 10.1016/S1535-6108(04)00305-8 [DOI] [PubMed] [Google Scholar]
- Winner B, Desplats P, Hagl C, Klucken J, Aigner R, Ploetz S, Laemke J, Karl A, Aigner L, Masliah E, Buerger E, Winkler J (2009) Dopamine receptor activation promotes adult neurogenesis in an acute parkinson model. Exp Neurol 219:543–552. 10.1016/j.expneurol.2009.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Stull ND, Berk MA, Snyder EY, Iacovitti L (2002) Neural stem cells spontaneously express dopaminergic traits after transplantation into the intact or 6-hydroxydopamine-lesioned rat. Exp Neurol 177:50–60. 10.1006/exnr.2002.7989 [DOI] [PubMed] [Google Scholar]
- Yoon S, Baik JH (2013) Dopamine D2 receptor-mediated epidermal growth factor receptor transactivation through a disintegrin and metalloprotease regulates dopaminergic neuron development via extracellular signal-related kinase activation. J Biol Chem 288:28435–28446. 10.1074/jbc.M113.461202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1-1: ChIP analysis of H3K27ac and H3K27me for other monoamine receptors. Download Figure 1-1, XLSX file (9.3KB, xlsx)
Extended Data Figure 1-2: GBM tumors express DRDs. (A) TCGA data reveals elevated expression of DRD2 relative to the other DRDs. Dots represent mRNA levels from individual tumors. Mean levels were compared using one-way ANOVA. (B) DRD2 expression in patient samples varies based on subtype. (C) Expression of DRDs in PDX GBM cells was determined using FACS analysis. Bars represent the mean of three individual experiments. (D) PDX GBM cells were implanted intracranially into the right hemisphere of athymic nude mice. Dots represent the mean from each individual mouse. Download Figure 1-2, TIF file (6.9MB, tif)
Extended Data Figure 3-1: Activation of DRD2 increases expression of GIC markers. (A) Neurosphere assay of PDX GBM12 treated with either DRD2 against (30nM) or equimolar DMSO. Extreme limiting dilution assays reveal no significant difference in stem cell frequency. (B)Mice injected with DRD2+ cells show efficient tumor engraftment compared to DRD2- cells. (C) PDX GBM cells treated with agonist or DMSO and Ki67 levels were determined by FACS. No difference in Ki67 was detected. (D) PDX cells treated with agonist or DMSO and the expression of GIC markers was examined by western blot after four days. Beta-actin was used as a loading control. (E) U251-CD133 RFP driven reporter line was treated with agonist for 24 hours and RFP (reporter) and CD133 (ligand) were assayed. Download Figure 3-1, TIF file (34.8MB, tif)
Extended Data Figure 4-1: (A) Patient tumors express mRNA for several genes involved in synthesis, secretion, and reuptake of dopamine. Data were taken from the Cancer Genome Atlas (TCGA). Expression was examined in astrocytomas (Grade 1, II, III, IV) and within GBM based on subtype (Classical, Mesenchymal, Proneural). (B) PDX lines were exposed to 50μm TMZ and collected after 2, 4, 6, and 8 days and subjected to HPLC. Results are shown normalized to equimolar DMSO controls. Download Figure 4-1, TIF file (12.4MB, tif)
Extended Data Figure 6-1 (A) Alkylating chemotherapy alters the uptake of glucose and fatty acid. PDX GBM cells were treated with either DMSO or TMZ. Four days later, cells were exposed to fluorescently-labeled glucose and palmitate. Uptake was determined by FACS analysis. (B) Seahorse analysis was used to determine the glycolytic rate of PDX lines after 4 days of 30nm agonist, or equimolar DMSO treatment. Comparisons were made using student t-Tests. *p<.05, **p<.01, ***p<.001. Download Figure 6-1, TIF file (14.3MB, tif)