Abstract
Dysfunctions of gene transcription and translation in the nociceptive pathways play the critical role in development and maintenance of chronic pain. Circular RNAs (circRNAs) are emerging as new players in regulation of gene expression, but whether and how circRNAs are involved in chronic pain remain elusive. We showed here that complete Freund's adjuvant-induced chronic inflammation pain significantly increased circRNA-Filip1l (filamin A interacting protein 1-like) expression in spinal neurons of mice. Blockage of this increase attenuated complete Freund's adjuvant-induced nociceptive behaviors, and overexpression of spinal circRNA-Filip1l in naive mice mimicked the nociceptive behaviors as evidenced by decreased thermal and mechanical nociceptive threshold. Furthermore, we found that mature circRNA-Filip1l expression was negatively regulated by miRNA-1224 via binding and splicing of precursor of circRNA-Filip1l (pre-circRNA-Filip1l) in the Argonaute-2 (Ago2)-dependent manner. Increase of spinal circRNA-Filip1l expression resulted from the decrease of miRNA-1224 expression under chronic inflammation pain state. miRNA-1224 knockdown or Ago2 overexpression induced nociceptive behaviors in naive mice, which was prevented by the knockdown of spinal circRNA-Filip1l. Finally, we demonstrated that a ubiquitin protein ligase E3 component n-recognin 5 (Ubr5), validated as a target of circRNA-Filip1l, plays a pivotal role in regulation of nociception by spinal circRNA-Filip1l. These data suggest that miRNA-1224-mediated and Ago2-dependent modulation of spinal circRNA-Filip1l expression regulates nociception via targeting Ubr5, revealing a novel epigenetic mechanism of interaction between miRNA and circRNA in chronic inflammation pain.
SIGNIFICANCE STATEMENT circRNAs are emerging as new players in regulation of gene expression. Here, we found that the increase of circRNA-Filip1l mediated by miRNA-1224 in an Ago2-dependent way in the spinal cord is involved in regulation of nociception via targeting Ubr5. Our study reveals a novel epigenetic mechanism of interaction between miRNA and circRNA in chronic inflammation pain.
Keywords: chronic inflammation pain, circRNA-Filipi1l, miRNA-1224, spinal, Ubr5
Introduction
Emerging evidence has shown that malfunctions in regulation of gene expression mediated by epigenetic mechanisms play the critical role in development and maintenance of chronic pain induced by diverse causes (Imai et al., 2013; Ji et al., 2016; Pan et al., 2016; Jiang et al., 2017). The existing body of research suggests that the epigenetic regulation of gene expression by the widespread noncoding RNAs (ncRNAs), including miRNA and long ncRNA, is involved in the process of inflammation or never injury-induced chronic pain (Zhao et al., 2013; Park et al., 2014; Jiang et al., 2016). However, the study of pain-related circular RNAs (circRNAs, a kind of ncRNAs) is still in its infancy.
circRNAs, a large class of circularized RNAs in different species ranging from human and mouse to Drosophila and Caenorhabditis elegans, are characterized by a high stable structure and high tissue-specific expression (You et al., 2015; Chen and Schuman, 2016). Their expression is associated with physiological and pathological processes, such as metabolism, cancer (Hansen et al., 2013a), atherosclerosis (Holdt et al., 2016), and myogenesis (Legnini et al., 2017). However, how they are causally linked to disease development remains elusive. Recent studies reveal that several circRNAs are specifically enriched in brain (Memczak et al., 2013; You et al., 2015; Chen and Schuman, 2016) or spinal cord (Zhou et al., 2017). Interestingly, these circRNAs are differentially expressed in various brain regions or in neuronal subcellular fraction, and notably involved in brain development, neuronal differentiation, and synaptic plasticity (Rybak-Wolf et al., 2015). These characteristics imply their potential involvement in the pathogenesis of a variety of CNS diseases. Accumulating evidence indicates that their aberrant expression or functional consequences contribute to the initiation, development, maintenance of various neurological disorders, such as epilepsy, Parkinson's disease (PD) (Kumar et al., 2018), Alzheimer's disease (AD) (Shao and Chen, 2016), and pain (Cao et al., 2017; Zhou et al., 2017). The expression profiling shows that spared nerve injury leads to 68 upregulation and 120 downregulation of circRNAs in rat spinal cord; furthermore, in vitro luciferase assay shows that circ-0006928 regulates chronic pain by targeting miRNA-184 (Zhou et al., 2017). Despite the fact in favor of circRNA's relevant potential therapeutic tool for CNS-related diseases, the role of circRNAs in the aberrant gene expression has not been explored in chronic pain. circRNA-Filip1l, named as circ-0000691 in circbase data (Memczak et al., 2013), is first found in mouse cerebella tissue (Glazar et al., 2014), and its expression is further confirmed in mammalian brain (Rybak-Wolf et al., 2015). Our circRNA profiling showed that circRNA-Filip1l was significantly increased in the spinal cord of complete Freund's adjuvant (CFA)-induced chronic inflammatory pain mice. However, it is unclear whether and how circRNA-Filip1l participates in the process of chronic pain.
Recently, a strong link between miRNA dysregulation and chronic pain has been established (Descalzi et al., 2015). Manipulation of miRNA expression in pain pathways from primary afferent nociceptors, DRG, spinal cord, and brain associated with pain perception prevents or reverses persistent inflammatory, neuropathic, and cancer pain behavior by post-transcription in cytoplasma (Park et al., 2014; Jiang et al., 2016; Gandla et al., 2017; Zhang et al., 2017). Growing findings suggest that the majority of miRNAs exist in both nucleus and cytoplasma, and some are preferentially enriched in the nucleus (Roberts, 2014; Rasko and Wong, 2017). Furthermore, the assembled Ago2-miRNA complexes are required for modulation of splicing or transcription of mRNA or circRNA through miRNA binding in nucleus. miRNA-671 directs the cleavage of a circular antisense transcript of cerebellar degeneration-related protein 1 (CDR1) in an Ago2-dependent manner in nucleus, resulting in the downregulation of circular antisense, suggesting a crucial function of miRNA-mediated AGO2 cleavage in the modulation of circRNA expression (Hansen et al., 2011). miRNA-1224 is relatively conserved in mammal cells and is abundantly expressed in CNS tissues, such as brain cerebral (Hunsberger et al., 2012), hippocampus, and the marginal division (Shu et al., 2013). In HEK293T cell, upregulation of miRNA-1224 with mimics silences the expression of LRRK2 and α-synuclein associated with PD (Sibley et al., 2012), supporting the potential regulatory role of miRNA-1224 in CNS diseases-related genes. However, it is still unknown whether miRNA-1224 is involved in chronic pain.
In the current work, we found the increase of circRNA-Filip1l and decrease of miRNA-1224 in mouse spinal cords in a CFA-induced inflammation pain model. Moreover, miRNA-1224 is predictively bound to the splice junction of precursor-circRNA-Filip1l (pre-circRNA-Filip1l). Thus, we hypothesized that the circRNA-Filip1l cleaved by miRNA-1224 in an Ago2-dependent manner contributes to the development and maintenance of chronic inflammatory pain.
Materials and Methods
Animals, pain model, and behavior testing.
All animal procedures were approved by the animal care committee of Xuzhou Medical University (Xuzhou, China). All efforts were made to minimize animal suffering and to reduce the number of animals used. Mice were housed at 23 ± 3°C with humidity ranges between 25% and 45%, and maintained on a 12:12 light/dark cycle (06:00 to 18:00 h) with access to food and water ad libitum. Adult male Shanghai populations of Kunming mice (20–25 g) were used in this study. The animals were randomized to either a control or an experimental group. Chronic inflammatory pain was induced by subcutaneous administration of CFA (40 μl; F5881; Sigma-Aldrich) into the plantar surface of the left hindpaw. A 0.9% saline solution was used as a control for CFA. Unilateral sciatic nerve chronic constrictive injury (CCI) model was performed as described previously (Pan et al., 2014). Mice were anesthetized with inhalation anesthesia by isoflurane in O2. Under the anesthesia condition, blunt dissection was made into the skin overlying the area between the gluteus and biceps femoris muscles, and the common left sciatic nerve of the hindpaw was exposed at the mid-thigh level; ∼7 mm of nerve was freed, proximal to the sciatic trifurcation, and three loose ligatures (∼1 mm space) of 7–0 silk thread were placed across the sciatic nerve, until a brief twitch was observed. Sham-operative groups underwent identical procedures but no ligation of the respective nerve. After surgery, all mice were maintained in a warm electric blanket with stable temperature until they recovered from anesthesia.
Paw withdrawal latency to a thermal stimulus and paw withdrawal thresholds to a mechanical stimulus were used to measure hyperalgesia and allodynia as described previously (Pan et al., 2017). Before nociceptive behavior testing, mice were acclimatized to the environment for 1 h. Thermal hyperalgesia was assessed with an analgesia meter (IITC Model 336 Analgesia Meter, Series 8; IITC Life Science) by focusing a beam of light on the plantar surface of the hindpaw to generate heat. The time required for the stimulus to elicit withdraw of the hindpaw was recorded. The radiant heat intensity was adjusted to obtain basal paw withdrawal latency of 11–14 s. An automatic 20 s cutoff was used to prevent tissue damage. Thermal stimuli were delivered three times to each hindpaw at 5 min intervals. Mechanical allodynia was assessed using von Frey filaments (Stoelting), starting with a 0.16 g and ending with a 6.0 g filament. The filaments were presented five times, respectively, at 5 min intervals, in ascending order of strength, perpendicular to the plantar surface with sufficient force to cause slight bending against the paw. A brisk withdrawal or flinching of the paw was considered a positive response. All behavioral tests were performed in a double-blind trial fashion in this study.
Locomotor function.
Three reflex tests were performed as follows. To test the grasping reflex, climbing tests were performed according to previously described procedures (Zhang et al., 2014). A 0.5-mm-diameter metal wire mesh with a 5-mm-wide grid was placed vertically 30 cm above the table. Each mouse started at the bottom of the mesh with its head facing downward. After the mouse was released, the time required for it to climb all the way to the top was recorded. A maximum time of 60 s was applied for animals that could not successfully complete this task. Two sessions were performed for each mouse with a 30 min interval, and the shorter time was recorded. To test the placing reflex (Tao et al., 2003), we held the mouse with the hind limbs slightly lower than the forelimbs and brought the dorsal surfaces of the hindpaws into contact with the edge of a table. The experimenter recorded whether the hindpaws were placed on the table surface reflexively. To test the righting reflex (Tao et al., 2003), we placed the mouse on its back on a flat surface; the experimenter noted whether it immediately assumed the normal upright position. Scores for placing, grasping, and righting reflexes were based on the counts of each normal reflex exhibited in five trials.
Spinal tissue collection.
Mice were anesthetized with 10% chloral hydrate, and the spinal cord within the lumbar segments (L3–L5) was removed rapidly. The dorsal spinal cord ipsilateral to CFA was separated and snap-frozen in liquid nitrogen, and stored at −80°C.
circRNA microarray.
Total RNA from each of 6 samples was quantified using the NanoDrop ND-2000 (Thermo Scientific). The sample preparation and microarray hybridization were performed according to Arraystar Mouse circRNA's standard protocols (Arraystar). Briefly, total RNA from each sample was first treated with Rnase R (Epicenter) to obtain circRNA through removing linear RNAs. Then, each sample was amplified and transcribed into fluorescent cRNA using a random priming method (Super RNA Labeling Kit; Arraystar). The labeled cRNAs were purified by RNeasy Mini Kit (QIAGEN). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-2000 (Thermo Scientific); 1 μg of each labeled cRNA was fragmented by adding 5 μl 10 × blocking agent and 1 μl of 25 × fragmentation buffer, then heated the mixture at 60°C for 30 min; finally, 25 μl 2 × hybridization buffer was added to dilute the labeled cRNA; 50 μl of hybridization solution was dispensed into the gasket slide and assembled to the circRNA expression microarray slide (6 × 7K, Arraystar). The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven. After having washed the slides, the arrays were scanned by the Axon GenePix 4000B microarray scanner. Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Quantile normalization and subsequent data processing were performed using the R software package. After quantile normalization of the raw data, low-intensity filtering was performed, and the circRNAs that at least 1 of 3 samples have flag “expressed” (>2 times background SD) were retained for further analyses. Differentially expressed circRNAs with statistical significance between two groups were identified through Volcano Plot filtering. The statistical significance of the difference was conveniently estimated by t test. circRNAs having fold changes ≥ 2, and p values ≤ 0.05 were selected as the significantly differentially expressed.
RNA, circRNA, miRNA, and qRT-PCR.
Total RNA was isolated with a Trizol reagent (15596–026; Invitrogen) to generate cDNA templates by reverse transcription reactions with oligo(dT) for Ago2, and Filip1l, or with random primers for circRNA-Filip1l, and pre-circRNA-Filip1l, and reverse transcriptase M-MLV (2641A; Takara) at 42°C for 60 min. cDNA products were used as templates to detect gene mRNA (Ago2: forward: 5′-CGTCCTTCCCACTACCACG-3′, reverse: 5′-CCAGAGGTATGGCTTCCTTCA-3′;Ubr5: forward: 5′-TGAGGTTTCTACGATCTGTG GC-3′, reverse: 5′-AAACACACGTTTGCATTTTCCA-3′; Filip1l: forward: 5′-CACAGGGTAAACTAGCCCTTG-3′; reverse: 5′-TGGCGATTTTGACTGTCCTCA-3′ and pre-circRNA-Filip1l: forward: 5′-CTCTGGTCACCTGGTGGGAT-3′, reverse: 5′-TGGGTAGAGGCAATTTG GCA-3′) or circRNA expression (circRNA-Filip1l: forward: 5′-AGGCCTCGGGATCCCAC CTC-3′, reverse: 5′-TCCAGTCCGCCGAGGGCGC-3′;circRNA-014740: forward: 5′-AGACA TTGATGACTGCTTATGC-3′, reverse: 5′-CATAGCCCTGGTCACAACT-3′; circRNA-16648: forward: 5′-TTGGAGCTGCTGGCCCATCC-3′, reverse: 5′-GCATTGTTGGTCCAACCGGGTCT-3′; circRNA-005786: forward: 5′-CTTGGCCTCTTCCTCCTTTT-3′, reverse: 5′-TGG GCCTCAGGAAGTAGAGA-3′) via qRT-PCR with SYBR Premix ExTaqII (RR820A; Takara Bio) according to the manufacturer's instructions. miRNA was reversely transcribed at 16°C for 30 min, and 37°C for 30 min using specific primer 1224RT (5′-TTAACTGGATACGAAGGGTCCGAACACCGGTCGTATCCAGTTAActccacc-3′). qRT-PCR was performed using primer pairs 1224 forward: 5′-TGCGGGTGAGGACTGGGGAG-3′ and 1224 reverse: 5′-TACGAAGGGTCCGAACAC-3′. RNase R treatment was performed as follows: 5 μg of total RNA was diluted in 20 μl of water with 4 U RNase R/μg unless differently stated and 2 μl of enzyme buffer (Epicenter), then incubated 15 min at 37°C and purified by phenol chloroform extraction. Reactions were performed in triplicate. Gapdh (GF, 5′-GGTGAAGGTCGGTGTGAACG-3′; GR, 5′-CTCGCTCCTGGAAGATGGTG-3′) was used as an internal control of Ago2, Ubr5, Filip1l, and pre-circRNA-Filip1l. U6 snRNA (6F, 5′-CTCGCTTCG GCAGCACATATACT-3′; 6R, 5′-ACGCTTCACGAATTTGCGTGTC-3′) was used as an internal control of miRNA-1224 and circRNA-Filip1l. The expression levels of the target genes were quantified relative to Gapdh or U6 snRNA expression (cycle threshold [Ct]) using the 2−ΔΔCT methods. Any value among triplicates that had a marked difference (≥1.00) compared with the average of the other two was omitted.
Spinal neuron culture.
The primary culture of spinal neurons was performed as described previously (Hugel and Schlichter, 2000). Briefly, after decapitation of 3- to 4-d-old mice under deep anesthesia, a laminectomy was performed, and the third dorsal of the spinal cord was cut with a razor blade. The tissue fragments were digested enzymatically for 45 min at 37°C with papain (20 U/ml, Sigma-Aldrich) in oxygenated divalent-free Earle's balanced salt solution (Invitrogen). The enzymatic digestion was stopped by adding 3 ml Earle's balanced salt solution containing BSA (1 mg/ml; Sigma-Aldrich), trypsin inhibitor (10 mg/ml; Sigma-Aldrich), and DNase (0.01%; Sigma-Aldrich), and a mechanical dissociation was performed with a 1 ml plastic pipette. The homogenate was deposited on top of 4 ml of a solution of composition similar to that described above, except that the concentration of BSA was increased to 10 mg/ml. After centrifugation (5 min at 500 rpm), the supernatant was removed and replaced with 5 ml of culture medium, the composition of which was as follows: MEM-α (Invitrogen), FCS (5% v/v; Invitrogen), heat-inactivated horse serum (5% v/v; Invitrogen), penicillin and streptomycin (50 IU/ml for each; Invitrogen), transferrin (10 mg/ml; Sigma-Aldrich), insulin (5 mg/ml; Sigma-Aldrich), putrescine (100 nm; Sigma-Aldrich), and progesterone (20 nm; Sigma-Aldrich). After trituration with a fire-polished Pasteur pipette, the cells were plated on 35 mm collagen-coated plastic culture dishes in the central compartment, which was delimited by a small (internal diameter 15 mm) circular glass ring. This ring was glued onto the bottom of the dish with paraffin wax and could easily be removed before electrophysiological experiments. Cultures were maintained in a water-saturated atmosphere (95% air, 5% CO2) at 37°C until use (10–15 d). Two days after the cells were seeded, cytosine arabinoside (10 μm) was added to the culture medium for 24 h to reduce glial proliferation.
Spinal astrocytes and microglia cultures.
The isolation of spinal astrocytes and microglia cells was performed as described previously (Monif et al., 2016) with few modifications. The third dorsal of spinal cord from 3- to 4-d-old mice was cut, the tissue fragments were digested enzymatically for 45 min at 37°C with trypsin (0.25%, Invitrogen), and stopped by adding equal volume DMEM with 10% FBS, then centrifuged at 1000 × g for 30 s, removed the supernatant, washed three times with DMEM with 10% FBS, and centrifuged at 1000 × g for 30 s each time. The mixed cells were suspended by the use of DMEM with 20% FBS and filtered with 200 mesh sieve. The suspended cells were plated into 75 cm2 plate. After 30 min, we transferred the medium to the 75 cm2 flask, cultured at 37°C for 7–10 d. To harvest the astrocytes, the flasks of mixed glial cells were shaken at 220 rpm at 37°C overnight, discarded the supernatant, the left was the astrocytes, then added DMEM with 10% FBS, and continued to culture at 37°C until the cellular astrocyte density required. To harvest microglial cells, the flasks of mixed glial cells were shaken at 150 rpm at 37°C for 4 h, we collected the supernatant containing the microglial cells to the new plates and continued to perform the culture at 37°C, until the density of microglial cells required
Cellular fraction and RNA isolation.
PARIS Kit (Invitrogen, #AM1921) was used to separately isolate nuclear and cytoplasmic RNA from cultured mouse spinal neurons, following the manufacturer instructions.
Immunofluorescence and FISH.
The procedure was performed as described in a previous study (Pan et al., 2014). In brief, spinal cords were rapidly dissected from perfused mice and fixed with 4% PFA, then cryoprotected in 30% sucrose. For FISH in cultured cells, digoxin-labeled circRNA-Filip1l probe (Dig-Filip1l, 5′-Dig-CGCCGG GGAGGTGGGATCCCGA-Dig-3′) or miRNA-1224 probe (Dig-1224, 5′-Dig-CTCC ACCTCCCCAGTCCTCAC-Dig-3′) was hybridized to spinal slices as instructed in the FISH kit (Guangzhou Exon), and incubated with then fluorescent-conjugated secondary anti-digoxin, and then after PBS wash 3 times FISH sections were incubated with NeuN antibody (MAB377, Millipore), finally after PBS wash 3 times incubated with fluorescent-conjugated secondary antibody (Alexa-594, Cell Signaling Technology). After the sections were rinsed in 0.01 m PBS, coverslips were applied.
Northern blot.
Northern blot was performed as described previously (Legnini et al., 2017) with modified. Briefly, 10 μg RNA was denatured with 1 volume of glyoxal loading dye (Ambion) at 50°C for 30 min and loaded on 1.2% agarose gel. Electrophoresis was performed for 2.5 h at 60 V. RNA was transferred on Hybond N+ membrane (GE Healthcare) by capillarity overnight in 10× SSC. Transferred RNA was cross-linked with UV at 1200 × 100 mJ/cm2, and the membrane was washed in 50 mm Tris, pH 8.0, at 45°C for 20 min. Prehybridization and hybridization were performed in Northern Max buffer (Ambion) at 68°C for 30 min and overnight, respectively; 500 ng of DIG-labeled probe in 10 ml was used for hybridization. The membrane was then washed with 2× SSC 0.1% SDS twice 30 min, then once 30 min and once 1 h with 0.2× SSC 0.1% SDS at hybridization temperature. The membrane was the processed for DIG detection (hybridization with anti-DIG antibody, washing, and luminescence detection) with the DIG luminescence detection kit (Roche Diagnostics), according to the manufacturer's instructions. DIG-labeled probes were produced by in vitro transcription with DIG-RNA labeling kit (11175025910, Roche Diagnostics) of PCR templates produced with the primers DFilip1lF (5′-AGGCCTCGGGATCCCACCTC-3′) and DT7-Filip1lR (5′-TAATACGACTCACTATAGGTCCAGTCCGCCGAGGGCGC-3′), used with mouse cDNAs. circRNA transcription with T7 RNA polymerase (Promega) was performed at 37°C for 2 h and then purified with Micro Bio-Spin 30 Chromatography Column (732–6223, Bio-Rad).
Synthetic anti-circRNA and circRNA mimics.
The anti-circRNA-Filip1l mimics and linear circRNA-Filip1l were obtained by in vitro transcription from a PCR-generated template, respectively, with anti-circRNA-Filip1l primer pair (DFilip1lF and DT7-Filip1lR) and linear circRNA-Filip1l primer pair (DT7-Filip1lF, 5′-TAATACGACTCACTATAGGGCTCCCCGGCGCGGCGGG-3′/Filip1lR, 5′-GAG GTGGGATCCCGAGGCCT-3′) using mouse cDNA as PCR template in the presence of T7 polymerase (Promega) following the manufacturer's instructions, and purified with Micro Bio-Spin 30 Chromatography Column (732–6223, Bio-Rad) after DNase treatment. circRNA-Filip1l was synthesized using linear circRNA-Filip1l as described previously (Legnini et al., 2017). A phosphate group was then attached to the 5′-OH using ATP and T4 polynucleotide kinase (BioLaboratories); the linear transcript carrying no 5′-phosphate and 3′-OH ends was subjected to ethanol precipitation in the presence of 10 mg of glycogen (Roche Diagnostics). Ligase reaction was performed in a final volume of 100 μl, incubating the linear transcript at 95°C for 2 min followed by 5 min at 75°C in the presence of 10% DMSO. T4 RNA ligase (BioLaboratories), 1×T4 RNA ligase buffer, 10 mm ATP, and RNase inhibitor were then added, and the reaction was performed at 16°C for 16 h. The circularized RNA product was separated and purified from the linear transcript by PAGE. A total of 30 ng of both circular and linear transcripts were subjected to RNase R treatment followed by qRT-PCR to assess the circularity of the gel-purified RNA molecules.
Plasmid construction.
All constructs were produced by the use of standard molecular methods and confirmed by DNA sequencing. To construct circRNA-Filip1l, miRNA-1224, and Ago2 overexpression vectors (OE): one insert prepared by PCR using primer pairs [circRNA-Filip1l OE: PWFilpF, 5′-ACGCTCGAGAGTGGCCCACTAGGCACTC-3′ (Xhol)/PWFilpR, 5′-GGCGTTTAAACAAACAATAAGTCTGGGAGAG-3′ (PmeI); miRNA-1224 OE: PW1224F, 5′-CGGGATCCGAGCCCATATCTCCTACTGG-3′ (BamhI)/PW1224R, 5′-AATACGCGTTCGACACAGGCGTTCTTGAG-3′ (MluI); and Ago2 OE: PW-Ago2F, 5′-AATGGATCCATGTACTCGGGAGCCGGCCC-3′ (BamHI)/PW-Ago2R, 5′-AATACGCGTTCAAGCAAAGTACATGGTGCGC-3′ (MluI)] and PWPXLvector were digested by corresponding double restriction endonucleases (NEB), and then ligated with T4 ligase. To construct circRNA-Filip1l overexpression vectors in DNA3.1 plasmid: one insert from PCR with PCR pair [5′-ACGAAGCTTAGCCTGAGTTTGCCATCTTG-3′ (HindIII) and 5′-ACGCTCGAGTCAAAGAAACTAACGGCAAC-3′ (Xhol)] and the digested cDNA3.1 vector, and then ligated with T4 ligase. To construct circRNA-Filip1l, miRNA-1224, and Ubr5 knockdown vector (KD), LV-anti-Filip1lF [5′-P-CGCGCCCACCTCCCTCCCCGGCGCGGCGGGCGAGACGGGCCGGTGG-3′ (MluI)] and LV-anti-Filip1lR [5′-P-CGCCACCGGCCCGTCTCGCCCGCCGCGCCGGGGAGGGAGGTGGG-3′ (ClaI)], or PLV-1224F [5′-P-CGCGGTGAGGACACCGAGGTGGAGtagcGTGAGGACACCGAGGTGGAG-3′ (MluI)] and PLV-1224R [5′-P-CGCTCCACCTCGGTGTCCTCACgctaCTCCACCTCGGTGTCCTCAC-3′(ClaI)] or PLV-Ubr5F [5′-P-CGCGGAATGTACTGGAGCAGGCTACTATTCGAAAATAGTAGCCTGCTCCAGTACATTC-3′ (MluI)] and PLV-Ubr5R [5′-P-CGGAATGTACTGGAGCAGGCTACTATTTTCGAATAGTAGCCTGCTCCAGTACATTCCG-3′ (ClaI)] was annealed and ligated to the digested PLVTHM vector, respectively.
Lentivirus production and verification.
The constructed core plasmid (16 μg) and two envelope plasmids, PSPAX2 (12 μg) and PMD2G (4.8 μg), were cotransfected into HEK293T cells in a 6-well plate according to the manufacturer's instructions of Lipofectamine 2000 (11668-027, Invitrogen). The supernatant was collected at 48 h after transfection and concentrated by using a Centricon Plus-70 filter unit (UFC910096, Millipore). Lentivirus with titers 108 TU/ml was used in the experiment. The assays of lentivirus in vitro and in vivo infection were performed according to a previous study (Pan et al., 2014). Briefly, 20 μl lentivirus and 1.5 μl polybrene (1.4 μg/μl; H9268, Sigma-Aldrich) were added in a 24-well plate containing 1 × 105 HEK293T cells and DMEM without FBS; after 24 h, the transfection medium was replaced with 500 μl fresh complete medium containing 10% FBS; cells were collected at 48 h after culture.
For in vivo verification of lentivirus, daily intrathecal injections of lentivirus or vector (1 μl) were performed for 2 consecutive days in naive or pain mice and then collected samples day 3 after the first injection (otherwise, see the specified injection time points detailed in corresponding figure legend).
siRNA, mimics, inhibitor, and lentivirus delivery.
Injections were performed by holding the mouse firmly by the pelvic girdle and inserting a 30-gauge needle attached to a 25 μl microsyringe between L5 and L6 vertebrae. Proper insertion of the needle into the subarachnoid space was verified by a slight flick of the tail after a sudden advancement of the needle. Injections of 5 μl of 20 μm siRNAs, mimics, and inhibitor for circRNA-Filip1l (5′-GCGCCGGGGAGGUGGAGCACGAGC-3′), Ago2 (sense: 5′-GCGCCGGGGAGGCGGAGCCACGAGCTT-3′, antisense: 5′-G CTCGTGGCTCCGCCTCCCCGGCGCTT-3′), Ubr5 (sense: 5′-GAAUGUACUGGAGCAGGCUACUATT-3′, antisense: 5′-UAGUAGCCUGCUCCAGUACAUUCT T-3′), or 1 μl Lentivirus were performed daily for 3 d in a double-blind trial fashion. Knockdown via Ago2-siRNA, ubr5-siRNA, and PLV-Ubr5 was confirmed with qRT-PCR from samples of the ipsilateral dorsal spinal cord taken 72 h after the last injection. Animals receiving intrathecal injections of scrambled siRNA or an empty vector were used as control groups.
Construction of reporter vector.
The defined region of Ubr5 promoter was amplified from mouse genomic DNA using primer pairs (G6-U5F, 5′-ACGCTCGAGCAGGCTGCGAGACGGAGAAAC-3′ and G6-U5R, 5′-AATAAGCTTCAGCGGGTGGACCACGAAAT-3′), and cloned into pGL6 plasmid (Beyotime) via XhoI and HindIII digestion. Empty pGL6 vector was used as control plasmid. To construct the psiCK-wt-Filip1l or psiCK-mut-Filip1l reporter vector, psiCK-wtF [5′-P-TCGAGTCCTAACGCGTACGCTCGTGCTCCACCTCCCCGGCGCGGCGGGCGAGACGGGC-3′ (Xhol)] (underline indicates the reverse complementary fragment in pre-circRNA-Filip1l to miRNA-1224) and psiCK-wtR [5′-P-GGCCGCCCGTCTCGCCCGCCGCGCCGGGGAGGTGGAGCACGAGCGTACGCGTTAGGAC-3′ (NotI)], or psiCK-mutF [5′-P-TCGAGTCCTAACGCGTACGCTCGTGCTAATCAGTTCCGGCGCGGCGGGCGAGACGGGC-3′ (Xhol)] and psiCK-mutR [5′-P-GGCCGCCCGTCTCGCCCGCCGCGCCGGAACTGATTAGCACGAGCGTACGCGTTAGGAC-3′ (NotI)], were annealed and ligated to the digested psiCHECK2 vector, respectively.
Single-cell RT-PCR.
Single-cell RT-PCR for spinal neurons was performed as described previously with few modifications (Jiang et al., 2016). Briefly, the contents of dissociated spinal neurons from CFA mice were harvested into patch pipettes with tip, placed gently into reaction tubes with Dnase I at 37°C for 30 min, and heated to 80°C for 5 min to remove genomic DNA. Reverse transcriptase (SuperScript III Platinum; Invitrogen) and specific reverse outer primer or 1224-RT was added, the sample was incubated at 50°C for 50 min, and the reaction was terminated at 70°C for 15 min. The cDNA products were used in gene-specific nested PCR. The first-round PCR was performed with the outer primer pair (outF and outR) in the FastStart universal SYBR Green master kit (Roche Diagnostics). PCR conditions were as follows: 1 cycle of 3 min at 94°C; 5 cycles of 15 s at 95°C and 5 min at 56°C, 30 s at 72°C; then 20 cycles of 15 s at 95°C and 15 s at 60°C, 30 s at 72°C, and 1 cycle of 10 min at 72°C. The second round of PCR was performed using 0.5 μl of the first PCR product as the template and with inner PCR primers (inF and inR). The amplification: 1 cycle of 3 min at 94°C; 35 cycles of 15 s at 95°C and 15 s at 60°C, 30 s at 72°C, and 1 cycle of 3 min at 72°C. A negative control was obtained from pipettes that were submerged in the bath solution only. Gapdh was used as the reference gene. The primers are shown as follows: circRNA-Filip1l: outF/inF, 5′-ACTGGAGAGGCCTCGGGATC-3′; outR, 5′-CGCCGAGGGCGCACCACC-3′; inR, 5′-CGCACCACCGGCCCGTGGC-3′. miRNA-1224: RT, and outF/outR same as 1224RT, and 1224F/1224R above, respectively; inF, 5′-GGGTGAGGACTGGGGAG-3′; inR, 5′-AAGGGTCCGAACACCGG-3′. Ago2: outF, 5′-AGTTTGACTTCTACCTGTGCA-3′; outR, 5′-TGTGTCCTGGTGGACCT GGA-3′. inF/inR same as Ago2 F/R above. Ubr5: outF, 5′-AGAAGCAATTGCCG TGACAAT-3′, outR, 5′-TGCTTGCCTGATCTGATGAC-3′. inF, 5′-TGAGGTTTCTACGATCTGTGGC-3′; inR, 5′-AAACACACGTTTGCATTTTCCA-3′. NeuN: outF, 5′-AGACAGACAACCAGCAACTC-3′; outR, 5′-CTGTTCCTACCACAGGGTTTAG-3′. inF, 5′-ACGATCGTAGAGGGACG-3′; inR, 5′-TTGGCATATGGGTTCCCAGG-3′. Gapdh: outF, 5′-AGGTTCATCAGGTAAACTCAG-3′; outR, 5′-ACCAGTAGACTCCACGACAT-3′. inF, ACCAGGGCTGCCATTTGCA; inR, 5′-CTCGCTCCTGGA AGATGGTG-3′.
Luciferase reporter assay.
HEK293T cells were cultured in DMEM with10% FBS. HEK293T cells were seeded at 1 × 105 cells per 24-well plate. Identification of target was performed by transfecting reporter plasmids (50 ng) and DNA3.1-Filip1l (80 ng) or Lenti-Ago2 vector (80 ng), or miRNA-1224 mimics (80 ng) or inhibitor (50 ng) into HEK293T cells using Lipofectamine 2000 (11668–027, Invitrogen) in a 24-well plate. Cell lysates were prepared and subjected to luciferase assays using the Double luciferase reporter kit (E1910, Promega) at 48 h after transfection according to the manufacturer's instruction. pRL-TK plasmid was used as an internal control (Promega).
RNA-binding protein immunoprecipitation (RPIP).
Immunoprecipitations were performed using Magna RIPRNA-Binding Protein Immunoprecipitation Kit (17–700, Millipore). Briefly, spinal cord was harvested and placed in ice-cold PBS, then homogenized and centrifuged 1500 rpm for 5 min at 4°C, and we obtained the supernatant. We added 50 μl of magnetic beads and ∼5 μg of the antibody of Ago2 (ab186733, Abcam) in each tube, incubated with rotation for 30 min at room temperature. Then, we resuspended the mixture by RIP immunoprecipitation buffer and added the tissue supernatant, incubated with rotation for overnight at 4°C and pulled down the RNA on the magnetic rack. Finally, we digested the protein with proteinase K and extracted RNA for qRT-PCR.
RNA-RNA in vivo precipitation (RRIP).
According to Su et al. (2015), with modification, biotin-labeled miRNA-1224 probe (Bio-1224, 5′-CT CCACCTCCCCAGTCCTCAC-Bio-3′) was used to perform the RRIP experiment assay. Spinal cord was harvested 24 h after intrathecal injection of Bio-1224 (5 μl, 20 μm) and fixed by 2.5% formaldehyde for 10 min, lysed, and sonicated. After centrifugation, 50 μl of the supernatant was retained as input, and the remaining part was incubated with Dynabeads M-280 Streptavidin (11205D, Thermo Fisher Scientific) mixture overnight at 4°C. The next day, a beads-probes-RNAs mixture was washed and incubated with 200 μl lysis buffer and proteinase K to reverse the formaldehyde crosslinking. Finally, the mixture was added with TRizol for RNA extraction and detection.
Western blot analysis.
Proteins (20–50 μg/sample) were separated by 10% SDS-PAGE, transferred onto nitrocellulose membranes, and incubated simultaneously at 4°C overnight in the corresponding antibodies against the following: Ago2 (1:500, ab186733; Abcam), Ubr5 (1:1000, 65344; Cell Signaling Technology), or control tubulin β polyclonal antibody (1:5000; AP0064; Bioworld). The membranes were then washed twice in TBS with Tween 20 at room temperature for 10 min, incubated with HRP-labeled goat anti-rabbit IgG (1:1000; A0208; Beyotime) at room temperature for 1 h, and washed twice again in TBS with Tween 20 at room temperature for 10 min. The immune complexes were detected with a nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate assay kit (72091; Sigma-Aldrich). Band analyses were performed in ImageJ software, with the intensities of the target signals normalized to those of β-actin for statistical analyses.
Statistical analysis.
All data were presented as mean ± SEM. The data were statistically analyzed with a one-way or two-way ANOVA or paired or unpaired Student's t test. When ANOVA showed a significant difference, pairwise comparisons between means were tested by the post hoc Tukey method. Statistical analyses were performed with Prism (GraphPad 5.00). p < 0.05 was considered statistically significance in all analyses.
Results
Profiling of spinal circRNAs in CFA-induced chronic inflammatory pain
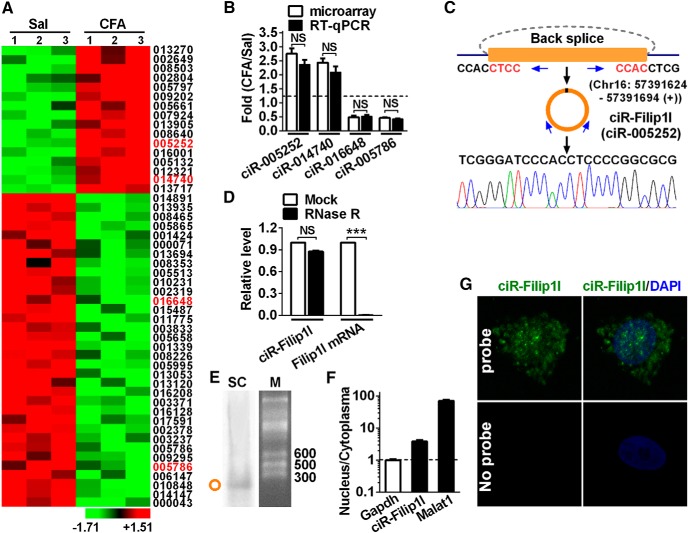
The abundance of circRNAs in the CNS suggests its potential roles in dynamic regulation of structure and function of the CNS (Shao and Chen, 2016). To identify spinal circRNAs involved in chronic inflammation pain, we analyzed the circRNA expression profiling from ipsilateral dorsal spinal cord of chronic inflammation pain mice and control mice. Sixteen upregulated and 34 downregulated circRNAs with >2.0-fold difference were obtained from 1099 candidates (Fig. 1A). circRNA-005252 with most significant difference, and other three randomly selected circRNAs, including 1 upregulated and 2 downregulated in microarray analysis, were further confirmed by qRT-PCR (Fig. 1B). circRNA-005252 (i.e., circRNA-Filip1l or mmu_circ_0000691 in cirbase database) (Memczak et al., 2013) is upregulated by 2.8-fold. The further analysis showed that circRNA-Filip1l was located on Chr16: 57391624–57391694 (+) and back spliced by intron 1 of Filip1l; the distinct product of the expected size was amplified using outward-facing primers and confirmed by Sanger sequencing (Fig. 1C). We then investigated the stability and localization of circRNA-Filip1l in spinal cells. Resistance to digestion with RNase R exonuclease showed that circRNA-Filip1l was resistant to RNase R digestion, whereas linear Filip1l mRNA was easily degraded, further confirming that circRNA-Filip1l species was circular in form (Fig. 1D). Northern blot assay verified the size of circRNA-Filip1l expected in spinal RNA of adult mouse (Fig. 1E). Furthermore, to clarify the distribution of circRNA-Filip1l in nucleus and cytoplasma, we separated the nucleus RNA and cytoplasmic RNA from in vitro cultured spinal neurons to detect the content of circRNA-Filip1l. qRT-PCR showed that the circRNA-Filip1l was localized in both nucleus and cytoplasma fraction (Fig. 1F). FISH further confirmed its localization in nucleus and cytoplasma of spinal neurons (Fig. 1G). These findings suggest that circRNA-Filip1l is an abundant and stable circular noncoding RNA expressed in spinal cord of mice.
Figure 1.
Profiling of circRNAs and circRNA-Filip1l expression in mouse spinal cord. A, The expression profiling of differential circRNAs with twofold or more was generated from circRNA microarray. Spinal cord was collected 3 d after CFA or saline (Sal) injection. n = 3 per group. The number indicates the identification of circRNA in Arraystar Mouse circRNA's Microarray. B, Four differential circRNAs were subjected to qRT-PCR verification. n = 4 per group. There was no significance (NS) versus the corresponding microarray groups by two-tailed paired Student's t test. C, The genomic loci of circRNA-Filip1l (ciR-Filip1l) is shown. The junction of circRNA-Filip1l was amplified by the use of back-to-back primers and then sequenced by Sanger sequencing. Arrows indicate divergent primers binding to the genome region of circRNA-Filip1l. D, Total RNAs were digested with RNase R followed by qRT-PCR detection of circRNA-Filip1l expression. Filip1l mRNA was detected as the RNase R-sensitive control. n = 4 per group. ***p < 0.001 versus the corresponding mock groups (two-tailed paired Student's t test). E, Northern blot for circRNA-Filip1l in spinal cord of mice. SC, Spinal cord; M, RNA marker. F, Distribution of circRNA-Filip1l in the nucleus and cytoplasma of spinal neuron cultured in vitro. Gapdh, Coding RNA control; Malat1, noncoding RNA control. Their levels were normalized to Gapdh. Spinal nucleus and cytoplasma RNA were collected, respectively, from 48 h cultured mouse spinal neurons in vitro. n = 4 per group. G, circRNA-Filip1l fluorescence in situ hybridization (FISH) in the spinal neuron cultured in vitro. DAPI, Nucleus staining dyes.
Expression patterns of spinal circRNA-Filip1l underlying chronic pain
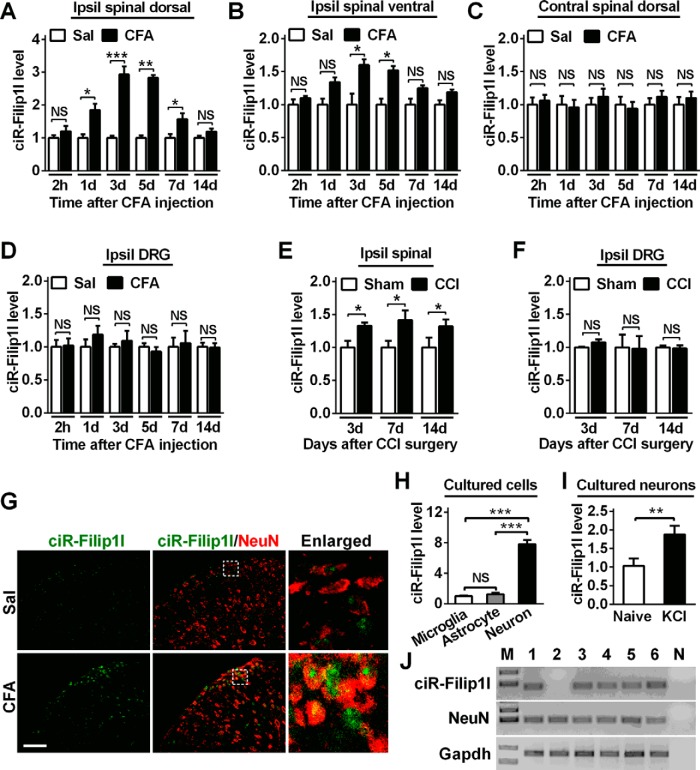
To uncover a temporal expression pattern of circRNA-Filip1l in spinal cord in chronic inflammation pain mice, we detected expression level of circRNA-Filip1l in the spinal dorsal or ventral horn of mice from hour 2 to day 14 after subcutaneous injection of CFA. qRT-PCR results showed that circRNA-Filip1l expression in the spinal dorsal horn was not altered in the acute phase (2 h) after CFA injection; however, it was increased by 75% day 1, reached a peak (180%) day 3, diminished day 7, and then returned to almost basal level day 14 after CFA injection (Fig. 2A). The circRNA-Filip1l level in the ventral horn of spinal cord was increased merely by 60% day 3 and 53% day 7 but had no change at the other time points after CFA injection (Fig. 2B). Furthermore, we found that the expression of circRNA-Filip1l in the contralateral spinal dorsal horn (Fig. 2C) and ipsilateral DRG (Fig. 2D) was not altered from hour 2 to day 14 after CFA injection. These results suggest that ipsilateral spinal dorsal horn is a major region contributing to the increase of circRNA-Filip1l in chronic inflammation pain. This increase trend of spinal circRNA-Filip1l expression related to chronic pain also was found in ipsilateral spinal dorsal horn of CCI mice (another chronic pain model) on days 3, 7, and 14 after surgery (Fig. 2E) but not observed in the ipsilateral DRG of CCI mice (Fig. 2F). These data indicate the possible involvement of spinal circRNA-Filip1l, not only in chronic inflammatory pain, but also in chronic neuropathic pain. Then, we characterized the differential spatial patterns of expression of spinal circRNA-Filip1l by combining FISH and cell-type-specific immunofluorescence staining in vivo. We observed that circRNA-Filip1l was colocalized with NeuN, a neuronal maker, in spinal cord (Fig. 2G). To further measure the expression level of circRNA-Filip1l in spinal neurons and non-neurons, we analyzed its content in the cultured spinal neurons and glial cells, including astrocytes and microglial cells. As shown in Figure 2H, circRNA-Filip1l in neurons was 7.8-fold as that in microglial cells and 6.2-fold as that in astrocyte cells, indicating that circRNA-Filip1l is mainly expressed in spinal neuron cells. Because the cultured neurons can be depolarized with high-concentration KCl to mimic sensitized in vivo neurons by nociceptive response (Yang et al., 2015), we treated the cultured spinal neurons with 50 mm KCl for 12 h and found that circRNA-Filip1l was significantly increased by this treatment (Fig. 2I), supporting a consistent change trend of circRNA-Filip1l expression between the spinal neurons of CFA-treated mice and KCl-treated spinal neurons in vitro. In addition, single-cell RT-PCR showed that 5 of 6 spinal neurons from CFA mice expressed circRNA-Filip1l (Fig. 2J), further confirming that the majority of spinal neurons express circRNA-Filip1l. Collectively, these results suggest that spinal circRNA-Filip1l is increased under chronic pain conditions.
Figure 2.
Chronic inflammatory pain induces the increase of spinal circRNA-Filip1l. A, B, Chronic inflammatory pain time-dependently led to the increase of circRNA-Filip1l in the ipsilateral (Ipsil) spinal dorsal horn (A) and to the slight increase of Ipsil spinal ventral horn (B) of mice. n = 5 per group. *p < 0.05; **p < 0.01; ***p < 0.001 versus the corresponding Sal groups (two-tailed paired Student's t test). C, D, The injection of CFA did not change the expression of circRNA-Filip1l in the contralateral (Contral) spinal dorsal horn (C) and the Ipsil DRG (D) of mice. n = 5 per group. There was no significance versus the corresponding Sal groups (two-tailed paired Student's t test). E, F, Neuropathic pain induced by chronic constriction injury altered the content of circRNA-Filip1l on days 3, 7, and 14 in spinal cord (E), but not in DRG (F) after surgery. n = 5 per group. *p < 0.05 versus the related Sham groups (two-tailed paired Student's t test). G, Combined circRNA-Filip1l FISH (green) and NeuN (a neuronal marker, red) immunofluorescence staining in spinal cord at day 3 after CFA or saline injections. Scale bar, 25 μm. H, The relative level of circRNA-Filip1l was analyzed by RT-PCR, respectively, in spinal neurons, and astrocytes and microglial cells cultured in vitro. n = 6 per group. ***p < 0.001 versus the related microglial groups (two-tailed paired Student's t test). I, circRNA-Filip1l was increased after treatment with KCl (50 mm) for 12 h in cultured spinal neurons. n = 6 per group. **p < 0.01 versus the related naive groups (two-tailed paired Student's t test). J, Single-cell RT-PCR shows the colocalization of circRNA-Filip1l with NeuN. Nos. 1–6 indicate six different neurons; no. 7 (N) is a negative control. The spinal neurons were isolated from day 2 after CFA-injected mice at 4 weeks of age.
Regulation of nociception by spinal circRNA-Filip1l
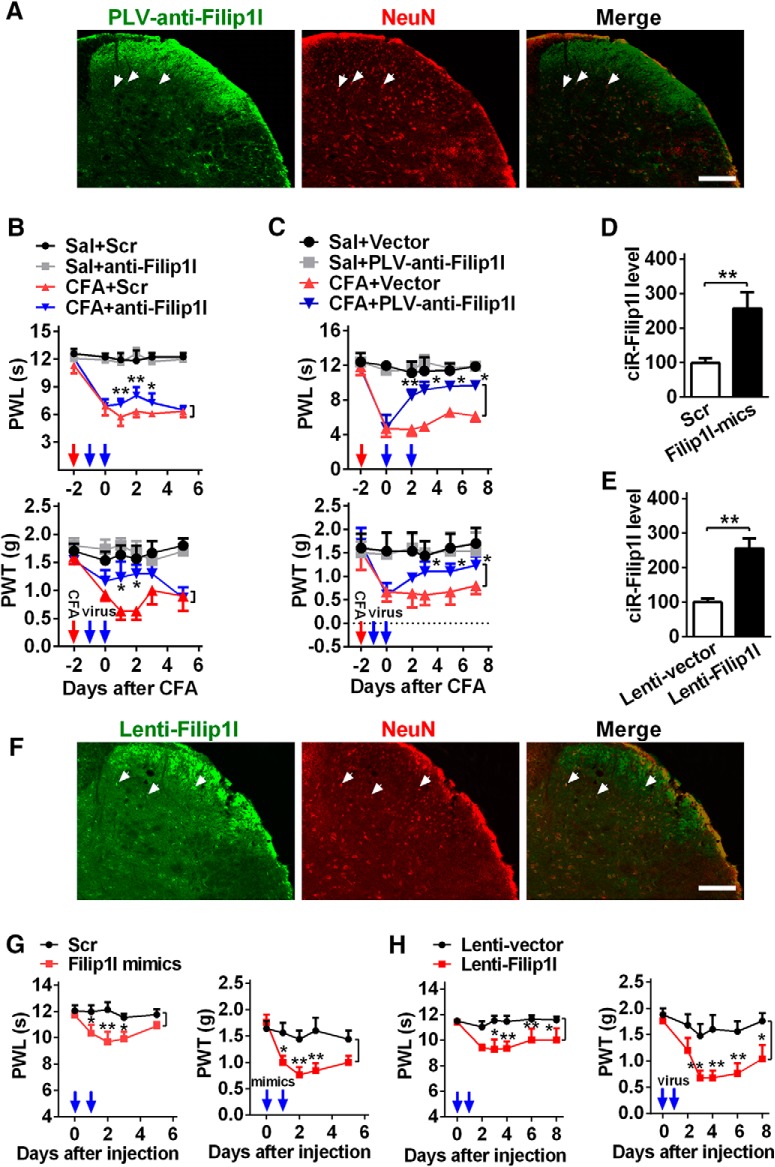
To further evaluate the therapeutic potential of spinal circRNA-Filip1l blockade in the relief of nociception response, we used the linear specific antisense of circRNA-Filip1l, including one exogenously synthesized anti-Filip1l by the use of in vitro T7 transcription, an antisense RNA of cirRNA-Filip1l that can prevent it from binding to its target gene; and another endogenous PLV-anti-Filip1l with GFP label in vivo expressed by lentivirus to block spinal circRNA-Filip1l in CFA mice. The analysis of GFP fluorescence intensity showed that PLV-anti-Filip1l mainly expressed in spinal neurons of naive mice day 3 after 2 consecutive days of intrathecal injection (Fig. 3A). Before the measure of nociceptive behavior, to exclude the possibility that the observed effects were affected by locomotor impairment, we observed the locomotor function in mice by testing their grasping reflex, placing reflex, and righting reflex. Results showed that blocking circRNA-Filip1l with anti-Filip1l or PLV-anti-Filip1l did not influence the locomotor function of the mice (Table 1). Thermal and mechanical nociceptive responses were attenuated at hour 24 after intrathecal injections of anti-Filip1l, but not scrambled control in CFA mice; these alleviative effects lasted at least 2 d (Fig. 3B). The antinociceptive effects of PLV-anti-Filip1l were also observed day 2 after intrathecal injection of PLV-anti-Filip1l in CFA mice (Fig. 3C); the effects had been maintained during the entire period of observation. To further explore the role of spinal circRNA-Filip1l in nociceptive regulation, we then examined whether circRNA-Filip1l overexpression mimics the nociception-like behavior in naive mice. To this end, two manipulating tools were used — exogenous circRNA-Filip1l mimics synthesized through cyclizing the linear circRNA-Filip1l from in vitro T7 transcription as described previously (Legnini et al., 2017), and endogenous Lenti-Filip1l lentivirus — to overexpress the circRNA-Filip1l in spinal cord of mice. Validation experiments showed that spinal circRNA-Filip1l expression in naive mice was upregulated by 157% day 2 after treatment with circRNA-Filip1l mimics and by 155% day 3 after Lenti-Filip1l treatment, respectively (Fig. 3D,E). Similarly, Lenti-Filip1l with GFP label mainly expressed in spinal neurons of naive mice day 3 after 2 consecutive days of intrathecal injection (Fig. 3F). Locomotor impairment was not found after overexpression of spinal circRNA-Filip1l by the use of circRNA-Filip1l mimics or Lenti-Filip1l (Table 1). However, the intrathecal injections of circRNA-Filip1l mimics or Lenti-Filip1l for 2 or 3 consecutive days, but not scrambled control or empty vector, significantly produced a nociception-like behavior as evidenced by a decrease threshold of mechanical or thermal stimulation (Fig. 3G,H). Together, these findings establish that spinal circRNA-Filip1l plays an essential role in physical and pathological nociceptive regulation.
Figure 3.
circRNA-Filip1l contributes to nociceptive behavior. A, The infection of PLV-anti-Filip1l with GFP reporter in spinal cord of naive mice on day 3 after 2 consecutive days of intrathecal injection (day 5 after the first injection). NeuN, a neuron marker. Scale bar, 25 μm. B, C, Inhibition of circRNA-Filip1l alleviated the thermal hyperalgesia and mechanical allodynia after 2 consecutive days of intrathecal injections of anti-Filip1l (B) or PLV-anti-Filip1l (C) in CFA mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Red arrow indicates CFA or saline injections. Blue arrows indicate anti-Filip1l or scrambled (Scr) and PLV-anti-Filip1l or vector injections. The anti-Filip1l sequence was antisense strand of full-length of circRNA beginning from junction. D, E, Intrathecal injections of circRNA-Filip1l mimics (D) or Lenti-Filip1l (E) for 2 consecutive days increased the spinal circRNA-Filip1l content in naive mice. n = 5 per group. Two-tailed paired Student's t test: **p < 0.01. F, The infection of Lenti-Filip1l with GFP reporter in spinal cord of naive mice on day 3 after 2 consecutive days of intrathecal injection. Scale bar, 25 μm. G, H, Overexpression of circRNA-Filip1l induced the generation of pain-like behavior after 2 consecutive days of intrathecal injections of circRNA-Filip1l mimics (G) or Lenti-Filip1l (H). n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Blue arrows indicate circRNA-Filip1l mimics or Scr and Lenti-Filip1l or Lenti-vector injections.
Table 1.
Test was performed 48 h after intrathecally injection of mimics, anti-RNA, and Scr or 72 h after intrathecal injection of lentivirus for 2 consecutive days in micea
| Treatment groups | Locomotor function test |
||
|---|---|---|---|
| Placing | Grasping | Righting | |
| Saline (5 μl) | 5 (0) | 5 (0) | 5 (0) |
| Negative control siRNA (Scr) | 5 (0) | 5 (0) | 5 (0) |
| PLVTHM empty vector (vector) | 5 (0) | 5 (0) | 5 (0) |
| Pwpxl empty vector (Lenti-vector) | 5 (0) | 5 (0) | 5 (0) |
| Sal + Scr | 5 (0) | 5 (0) | 5 (0) |
| Sal + anti-Filip1l | 5 (0) | 5 (0) | 5 (0) |
| CFA + Scr | 5 (0) | 5 (0) | 5 (0) |
| CFA + anti-Filip1l | 5 (0) | 5 (0) | 5 (0) |
| Sal + Vector | 5 (0) | 5 (0) | 5 (0) |
| Sal + PLV-anti-Filip1l | 5 (0) | 5 (0) | 5 (0) |
| CFA + Vector | 5 (0) | 5 (0) | 5 (0) |
| CFA + PLV-anti-Filip1l | 5 (0) | 5 (0) | 5 (0) |
| circRNA-Filip1l mimics | 5 (0) | 5 (0) | 5 (0) |
| Lenti-circRNA-Filip1l | 5 (0) | 5 (0) | 5 (0) |
| Sal + miRNA-1224 mimics | 5 (0) | 5 (0) | 5 (0) |
| CFA + miRNA-1224 mimics | 5 (0) | 5 (0) | 5 (0) |
| Sal + Lenti-miRNA-1224 | 5 (0) | 5 (0) | 5 (0) |
| CFA + Lenti-miRNA-1224 | 5 (0) | 5 (0) | 5 (0) |
| miRNA-1224 inhibitor | 5 (0) | 5 (0) | 5 (0) |
| PLV-miRNA-1224 | 5 (0) | 5 (0) | 5 (0) |
aData are mean (SEM). n = 5 mice per group; five trials. No significance; one-way ANOVA (response time vs the treated groups) followed by post hoc Tukey test.
miRNA-1224 is an upstream regulator of circRNA-Filip1l
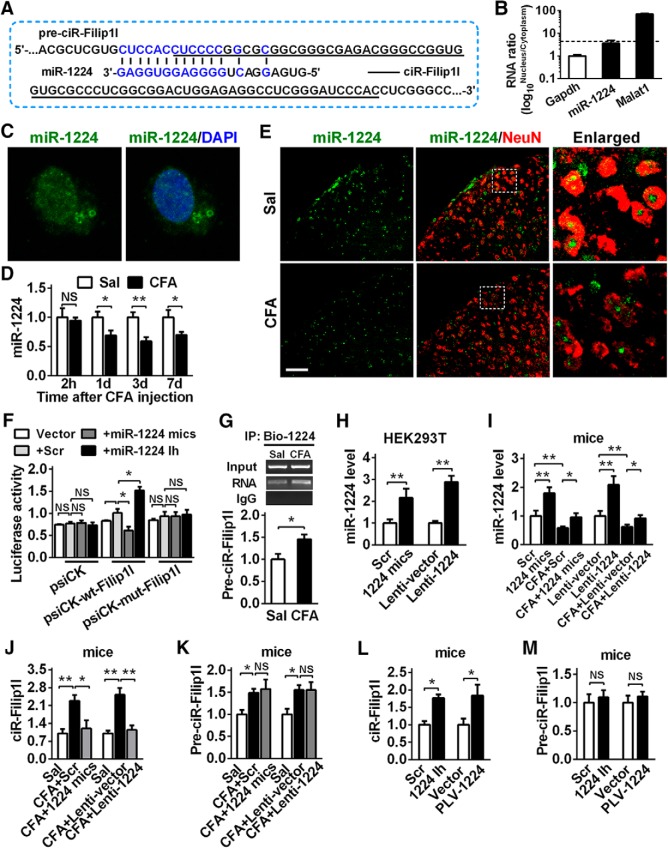
How is spinal circRNA-Filip1l upregulated under chronic inflammatory pain conditions? Data from a previous study suggest that miRNAs modulate the circRNAs expression via binding to the circRNA in cell nucleus (Hansen et al., 2011). Through search in mirbase database using sequence of circRNA-Filip1l or its precursor (pre-circRNA-Filip1l), we predicted possible miRNAs with the binding sites to circRNA-Filip1l or pre-circRNA-Filip1l. We found that the 14 nt fragment in miRNA-1224 was complementary to pre-circRNA-Filip1l region spanning its 5′ junction flank (Fig. 4A). Due to the location of pre-circRNA-Filip1l in spinal nucleus fraction, we supposed that miRNA-1224, as an upstream regulator, may be involved in the modulation of circRNA-Filip1l expression via the mediation of its cleavage. To confirm the existence of miRNA-1224 in spinal nucleus, we detected miRNA-1224 content in both nucleus and cytoplasma of spinal neuron cultured. miRNA-1224 was mainly located in nucleus, and its level in nucleus was 3.8-fold higher than that in cytoplasma (Fig. 4B). FISH further confirmed the preferential localization of miRNA-1224 in nucleus of spinal neurons (Fig. 4C). Furthermore, spinal miRNA-1224 expression was time-dependently decreased from hour 2 to day 7 after CFA injection (Fig. 4D). Costaining confirmed that miRNA-1224 was spatially localized in the spinal neurons and markedly increased under CFA-induced inflammatory pain conditions (Fig. 4E).
Figure 4.
miRNA-1224 is an upstream negative regulator of circRNA-Filip1l. A, Schematic presentation of miRNA-1224 (miR-1224) binding to the fragment of pre-circRNA-Filip1l (pre-ciR-Filip1l). Underline indicates circRNA-Filip1l. Blue represents the reverse complementary of miR-1224 to pre-ciR-Filip1l. B, Distribution of miRNA-1224 in the nucleus and cytoplasma of spinal neuron cultured in vitro. n = 4 per group. Spinal nucleus and cytoplasma RNA were separated from spinal neurons cultured in vitro for 48 h. C, miRNA-1224 FISH in the spinal neuron cultured in vitro. D, CFA induced the time-dependent decrease of spinal miRNA-1224. n = 5 per group. *p < 0.05; **p < 0.01 versus the corresponding Sal groups (two-tailed paired Student's t test). E, Combining FISH of miRNA-1224 and immunofluorescence staining (IF) of NeuN. Scale bar, 25 μm. F, The validation of miR-1224 negatively regulating circRNA-Filip1l by luciferase reporter assay in vitro. A fragment of pre-circRNA-Filip1l containing the bound region by miRNA-1224 was inserted into psiCHECK reporter vectors (psiCK-wt-pre-Filip1l). A mutation was generated via altering the sequence bound by miRNA-1224 as indicated (psiCK-mut-pre-Filip1l). The wild and mutation reporters were cotransfected into the HEK293T with miRNA-1224 mimics or inhibitor or the scrambled. n = 4 per group. Two-way ANOVA (effect vs plasmid × treated interaction) followed by post hoc Tukey test: *p < 0.05. G, Test for the binding capacity of miRNA-1224 to pre-circRNA-Filip1l in vivo. Spinal cord was harvested at hour 24 after intrathecal injection of Bio-miRNA-1224 (Bio-1224) probes, fixed by formaldehyde, and the bound pre-circRNA-Filip1l was pulled down by Dynabeads M-280 streptavidin. n = 4 per group. *p < 0.05 versus Sal group (two-tailed paired Student's t test). H, In vitro transfection of miRNA-1224 mimics or Lenti-1224 enhanced the miRNA-1224 level in HEK293T at hour 48 after treatment. n = 4 per group. **p < 0.01 versus the corresponding groups (two-tailed paired Student's t test). I, Intrathecal injections of miRNA-1224 mimics or Lenti-1224 for 2 consecutive days increased the miRNA-1224 content in spinal cord of naive mice. n = 4 per group. One-way ANOVA (expression vs the treated groups) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. J, K, Intrathecal injections of miRNA-1224 mimics (J) or Lenti-1224 (K) for 2 consecutive days inhibited the expression of spinal circRNA-Filip1l but did not change the spinal pre-circRNA-Filip1l level day 2 after last injection in CFA mice. n = 5 per group. One-way ANOVA (expression vs the treated groups) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. L, M, Intrathecal injections of miRNA-1224 inhibitor (L) or PLV-1224-sponge (M) for 2 consecutive days increased the expression of spinal circRNA-Filip1l but did not change the spinal pre-circRNA-Filip1l level day 3 after last injection in naive mice. n = 5 per group. *p < 0.05 versus the corresponding control groups (two-tailed paired Student's t test).
To experimentally validate the in silicon prediction of miRNA-1224 regulating circRNA-Filip1le expression, we cloned a bound fragment of pre-circRNA-Filip1l by miRNA-1224 into psiCHECK reporter vector and detected the effect of miRNA-1224 on the activities of the reporter in HEK293T cells. Cotransfection of miRNA-1224 mimics with the reporter psiCK-wt-pre-Filip1l decreased luciferase activities by 39% compared with mutated psiCK-mut-pre-Filip1l vector. Contrarily, miRNA-1224 inhibitor elevated luciferase activities by 52% in psiCK-wt-pre-Filip1l, but not in psiCK-mut-pre-Filip1l (Fig. 4F). These data indicate that miRNA-1224 in vitro negatively regulates the expression of circRNA-Filip1l. To seek to determine whether miRNA-1224 regulates circRNA-Filip1l expression in vivo via binding to pre-circRNA-Filip1l, we first examined the binding capacity of miRNA-1224 to pre-circRNA-Filip1l. We intrathecally injected bio-labeled miRNA-1224 probes into control and CFA mice, and then tested the content of pre-circRNA-Filip1l pulldown hour 24 after injection. qRT-PCR showed that pre-circRNA-Filip1l was pulled down by miRNA-1224 probes and was increased in spinal cord of CFA mice, compared with the saline group (Fig. 4G), confirming the binding ability of miRNA-1224 to the pre-circRNA-Filip1l in vivo. Next, we further determined whether miRNA-1224 can regulate circRNA-Filip1l expression by targeting pre-circRNA-Filip1l. Here, two tools, including miRNA-1224 mimics and Lenti-1224, were synthesized or constructed as the methods described previously (Pan et al., 2014) to upregulate the expression of miRNA-1224. Their work efficiencies were validated in vitro and in vivo. As shown in Figure 5A, HEK-293 cells transfected with miRNA-1224 mimics or Lenti-1224, but not scrambled or empty vector, displayed the increased miRNA-1224 by 118% or 189%, respectively (Fig. 4H). The expression of miRNA-1224 was upregulated by 79% at hour 48 after 2 consecutive days of miRNA-1224 mimics injection, or by 110% at hour 72 after 2 consecutive days of Lenti-1224 injection in naive mice and CFA mice (Fig. 4I). Furthermore, we found that overexpressing miRNA-1224 with miRNA-1224 mimics or Lenti-1224 in spinal cord of CFA mice blocked the CFA-evoked increase in spinal circRNA-Filip1l expression compared with the scrambled or Lenti-vector group (Fig. 4J); however, it did not change the increase in spinal pre-circRNA-Filip1l of CFA mice (Fig. 4K). In contrast, blocking miRNA-1224 with its inhibitor or PLV-1224 led to the increase of circRNA-Filip1l level (Fig. 4L) but unaltered the pre-circRNA-Filip1l level in spinal cord of naive mice (Fig. 4M). Together, these in vitro and in vivo results indicate that miRNA-1224 regulates expression of circRNA-Filip1l, but not pre-circRNA-Filip1l.
Figure 5.
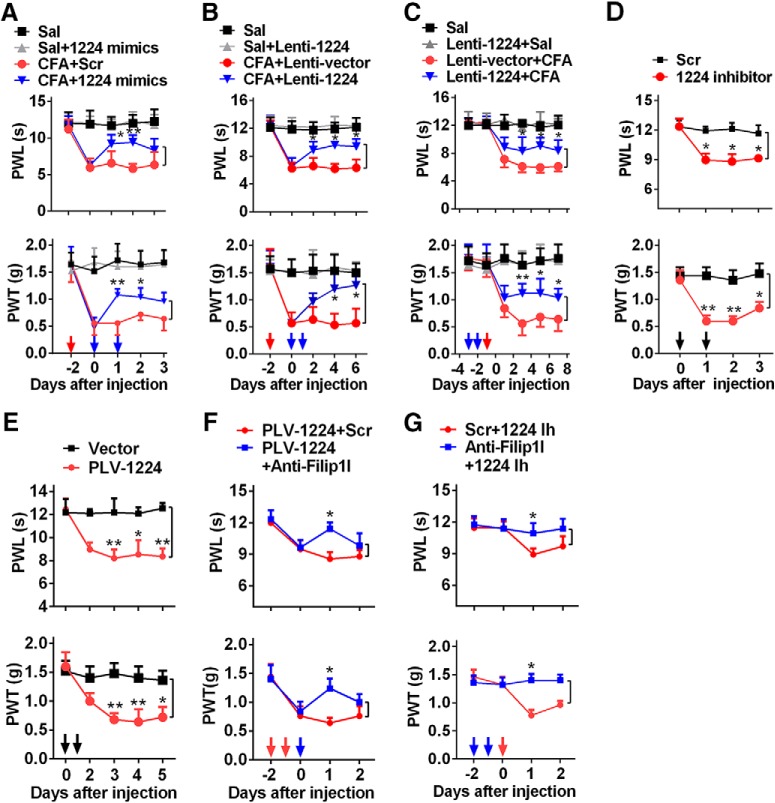
miRNA-1224 modulates the nociceptive behavior through the mediation of circRNA-Filip1l. A, B, Intrathecal injections of miRNA-1224 mimics (1224 mimics) (A) or Lenti-1224 (B) for 2 consecutive days reversed CFA-induced thermal hyperalgesia and mechanical allodynia during the maintenance period. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Red arrow indicates CFA or Sal injections. Blue arrows indicate miRNA-1224 mimics or Scr and Lenti-1224 or Lenti-vector injections. C, Intrathecal preinjection of Lenti-1224 for 2 consecutive days prevented the CFA-induced pain hypersensitivity during the development period. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Blue arrows indicate Lenti-1224 or Lenti-vector injections. Red arrow indicates CFA or Sal injections. D, E, Intrathecal injections of miRNA-1224 inhibitor (1224 inhibitor or 1224 Ih) (D) or PLV-1224 (E) for 2 consecutive days produced pain-like behavior in naive mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Black arrows indicate miRNA-1224 inhibitor or Scr and PLV-1224 or vector injections. F, Intrathecal injection of anti-Filip1l significantly inhibited or prevented the pain hypersensitivity induced by PLV-1224 in naive mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05. Red arrows indicate PLV-1224 injection. Blue arrow indicates anti-Filip1l or Scr injection. G, Pretreatment with anti-Filip1l significantly inhibited the pain hypersensitivity induced by miRNA-1224 inhibitor in naive mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05. Red arrow indicates miRNA-1224 inhibitor injection. Blue arrows indicate anti-Filip1l or Scr injections.
miRNA-1224 regulates nociception by mediation of circRNA-Filip1l
Our results indicate that miRNA-1224 modulates spinal circRNA-Filip1l expression under chronic inflammatory pain conditions. Therefore, spinal miRNA-1224 may also participate in the process of nociception regulation. Before pain behavior evaluation, we examined the effect of miRNA-1224 regulation tools on locomotor function. The reflex tests showed that miRNA-1224 overexpression with miRNA-1224 mimics or Lenti-1224 did not affect the locomotor function (Table 1). However, CFA-induced thermal and mechanical nociceptive responses were attenuated at hour 48 after the intrathecal injection of miRNA-1224 mimics (Fig. 5A) or at hour 72 after intrathecal injection of Lenti-1224 (Fig. 5B). Antinociceptive effect was undetected after the injection of scramble (Fig. 5A) or empty vector (Fig. 5B). We also observed that pretreatment with Lenti-1224, not empty vector (intrathecal injection of Lenti-1224 or empty vector for 2 consecutive days before CFA injection) significantly prevented CFA-induced nociceptive responses (Fig. 5C). Next, we tested whether knockdown of spinal miRNA-1224 in naive mice can induce the nociception-like behavior. The reflex tests confirmed no impairment of locomotor function after downregulation of spinal miRNA-1224 expression via intrathecally injecting miRNA-1224 inhibitor or PLV-1224 or their negative controls for 2 consecutive days in naive mice (Table 1), whereas the same treatment with miRNA-1224 inhibitor (Fig. 5D) or PLV-1224 (Fig. 5E), but not scramble or empty vector, significantly produced nociceptive responses as evidenced by a decrease of thermal and mechanical pain threshold (Fig. 5D,E). These findings suggest that spinal miRNA-1224 is involved in the process of nociceptive response. Finally, we checked whether miRNA-1224 regulates pain behavior through the mediation of circRNA-Filip1l. Naive mice were pretreated or post-treated with miRNA-1224 inhibitor or lentivirus to knockdown their spinal miRNA-1224 before or after intrathecal injection of anti-Filip1l, respectively, and then their nociceptive responses were measured. We observed that knockdown of circRNA-Filip1l significantly inhibited or reversed nociceptive responses induced by the block of miRNA-1224 with lentivirus (Fig. 5F) or inhibitor (Fig. 5G), suggesting that circRNA-Filip1l mediates the regulation of nociception by miRNA-1224. Together, these findings indicate that spinal miRNA-1224 regulates nociception via negatively targeting circR-Filip1l.
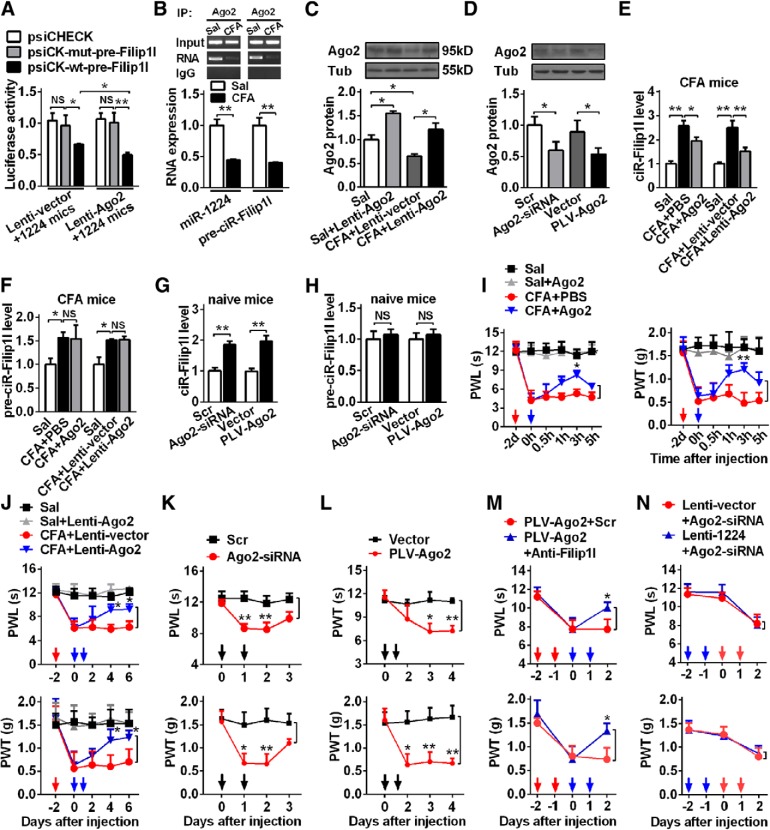
Ago2-mediated cleavage of pre-circRNA-Filip1l bound by miRNA-1224
To date, the modulatory mechanism of circRNA expression is poorly understood. In a recent study, it has been demonstrated that miRNA-671 directs cleavage of a circular antisense transcript of CDR1 in an Ago2-slicer-dependent manner (Hansen et al., 2011). Therefore, we hypothesized that miRNA-1224 mediates the cleavage of pre-circRNA-Filip1l in an Ago2-dependent manner to regulate the content of mature circRNA-Filip1l. To test the point, first, we cotransfected Ago2 overexpression plasmid (Lenti-Ago2) and miRNA-1224 mimics with wild reporter psiCK-wt-pre-Filip1l or mutation reporter psiCK-mut-pre-Filip1l in HEK293T cells. The transfection of miRNA-1224 mimics reduced the luciferase activities in psiCK-wt-pre-Filip1l group, compared with psiCK-mut-pre-Filip1l group; the overexpression of Ago2 further decreased the luciferase activities (Fig. 6A), suggesting that the overexpression of Ago2 inhibits the activity of pre-circRNA-Filip1l expression. We further examined whether AGO2 can combine to pre-cirRNA-Filip1l and miRNA-1224. We used AGO2 antibody to pull down spinal pre-circRNA-Filip1l and miRNA-1224. RPIP showed that both pre-circRNA-Filip1l and miRNA-1224 were pulled down by AGO2 antibody, and the harvested amounts were decreased in CFA mice compared with saline group (Fig. 6B). Next, we investigated whether Ago2 is involved in regulation of circRNA-Filip1l and pre-circRNA-Filip1l expression. Due to the reduced level of spinal AGO2 protein (Fig. 6C) day 3 after CFA injection, Lenti-Ago2 or AGO2 protein was intrathecally injected into CFA mice to overexpress AGO2; as well as PLV-Ago2 or Ago2-siRNA into naive mice to knockdown Ago2 expression; then the effect of Ago2 on expression of circRNA-Filip1l and pre-circRNA-Filip1l was evaluated. The tools of manipulating Ago2 were first validated. Ago2 protein was increased by 54% day 2 after intrathecal injection of Lenti-Ago2 in the saline group mice; the decreased Ago2 expression was reversed on day 3 after the same treatment in CFA mice (Fig. 6C), respectively. Ago2 expression was reduced by 41.2% or 39.3% on day 2 after intrathecal injection of Ago2-siRNA or on day 3 after PLV-Ago2 injection in naive mice, respectively (Fig. 6D). As expected, compared with the PBS or vector control group, the intrathecal injections of AGO2 protein or Lenti-Ago2 abolished the increase of spinal circRNA-Filip1l (Fig. 6E) but did not change the pre-circRNA-Filip1l level (Fig. 6F) in CFA mice. Intrathecal injections of Ago2-siRNA or PLV-Ago2 significantly elevated the expression of spinal circRNA-Filip1l (Fig. 6G), but not pre-circRNA-Filip1l level in naive mice (Fig. 6H). These results suggest that AGO2 affects the expression of circRNA-Filip1l, but not pre-circRNA-Filip1l. Finally, we wanted to know whether mice receiving the intrathecal injections of manipulation tools of Ago2 display behavioral changes in nociceptive thresholds. We observed that the injections of AGO2 protein (Fig. 6I) or Lenti-Ago2 (Fig. 6J), but not of PBS (Fig. 6I) or Lenti-vector (Fig. 6J), significantly blunted the thermal and mechanical nociception, respectively. On the contrary, injections of Ago2-siRNA (Fig. 6K) or PLV-Ago2 (Fig. 6L), but not of scramble (Fig. 6K) or vector (Fig. 6L), produced nociceptive responses. To further examine whether AGO2 regulates nociception via the mediation of circRNA-Filip1l or miRNA-1224, we evaluated the effect of blocking circRNA-Filip1l on nociception induced by knockdown of Ago2. As expected, blockage of circRNA-Filip1l by intrathecal preinjection of anti-Filip1l prevented nociceptive responses induced by Ago2 downregulation in naive mice (Fig. 6M). However, overexpression of spinal miRNA-1224 with Lenti-1224 did not prevent nociception induced by Ago2-siRNA in naive mice (Fig. 6N), indicating that Ago2 regulates nociceptive responses via circRNA-Filip1l, but not miRNA-1224. Collectively, these findings suggest that Ago2 is involved in regulation of physical and pathological nociception via miRNA-1224-dependent cleavage in circRNA-Filip1l.
Figure 6.
Ago2 mediates circRNA-Filip1l expression by cleavage of the pre-circRNA-Filip1l and regulates the nociceptive behavior. A, Luciferase activities of reporter plasmid after cotransfection of miRNA-1224 mimics, Ago2 overexpression plasmid (Lenti-Ago2) or empty vector (Lenti-vector), and psiCK-mut-pre-Filip1l or psiCK-wt-pre-Filip1l into HEK293T. n = 4 per group. Two-way ANOVA (effect vs plasmid × treated interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. B, Coimmunoprecipitation of AGO2 and pre-circRNA-Filip1l or miRNA-1224. The pre-circRNA-Filip1l, miRNA-1224, and Ago2 complex was pulled down using anti-Ago2 antibody for spinal tissues 3 d after CFA injection. n = 4 per group. **p < 0.01 versus the corresponding Sal groups (two-tailed paired Student's t test). C, D, The expression change of Ago2 protein after intrathecal injection of Lenti-Ago2 in naive mice or CFA-induced pain mice (C), and knockdown of Ago2 with Ago2-siRNA and PLV-Ago2 in naive mice (D). n = 4 per group. *p < 0.05, expression versus the treated groups (one-way ANOVA followed by post hoc Tukey test). Tub, β-Tubulin. E, F, Overexpression of Ago2 decreased the spinal circRNA-Filip1l content (E), but not pre-circRNA-Filip1l content (F) in CFA mice. Spinal cord was collected, respectively, at hour 3 after intrathecal injection of Ago2 protein and day 2 after Lenti-Ago2 intrathecal injection. n = 5 per group. One-way ANOVA (expression vs the treated groups) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. G, H, Knockdown of Ago2 increased the spinal circRNA-Filip1l expression (G), not changed pre-circRNA-Filip1l level (H) day 2 after 2 consecutive days of Ago2-siRNA or PLV-Ago2 injections in naive mice. n = 5 per group. **p < 0.01, versus the corresponding control groups (two-tailed paired Student's t test). I, J, Overexpression of Ago2 alleviated the thermal hyperalgesia and mechanical allodynia induced by intrathecal injection of Ago2 (I) or Lenti-Ago2 for 2 consecutive days (J) in CFA mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Red arrow indicates CFA injection. Blue arrows indicate Ago2 or PBS (left) and Lenti-Ago2 or Lenti-vector (right) injections. K, L, Knockdown of Ago2 induced the thermal and mechanical hypersensitivity after 2 consecutive days of intrathecal injections of Ago2-siRNA (K) or PLV-Ago2 (L) in naive mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Black arrows indicate Ago2-siRNA or Scr and PLV-Ago2 or vector injections. M, Inhibiting circRNA-Filip1l with anti-Filip1l prevented the thermal and mechanical hypersensitivity induced by knockdown of Ago2 after 2 consecutive days of intrathecal injection of PLV-Ago2 in naive mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05. Red arrows indicate PLV-Ago2 injection. Blue arrows indicate Anti-Filip1l or Scr injections. N, Overexpression of miRNA-1224 did not change the pain hypersensitivity induced by knockdown of Ago2 with Ago2-siRNA for 2 consecutive days of intrathecal injection in CFA mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test. Blue arrows indicate Lenti-1224 or Lenti-vector injections. Red arrows indicate Ago2-siRNA injection.
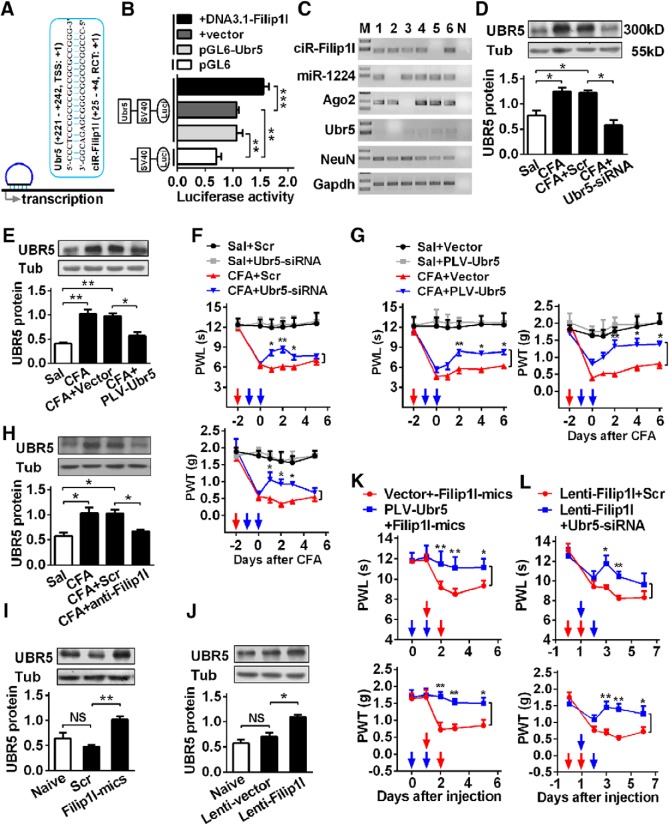
circRNA-Filip1l regulates nociception via targeting Ubr5
Many recent studies (Li et al., 2015; Long et al., 2017) have shown that nucleus circRNAs contribute to gene transcription via recruiting the RNA polymerase II. Therefore, we wanted to know what are the downstream of circRNA-Filip1l underlying regulation of nociception. First, we performed an in silicon target prediction using linear sequence of circRNA-Filip1l in BLAST program of NCBI. A total of 42 genes were predicted as the potential targets of circRNA-Filip1l. Some (e.g., Rn28s1, LOC105242388) are the ribosomal RNA or noncoding RNA; others are the unreported pain-related genes, such as Atxn2, Pwwp2a, Plxdc2, and Ubr5. Among them, ubiquitin protein ligase E3 component n-recognin 5 (Ubr5) can regulate the neuronal plasticity through activating the NMDA receptor in CNS and is implicated in the pathologic process of central neural system diseases, such as depression and epilepsy through ubiquitination of modification (Kato et al., 2012; Christensen et al., 2013). In Ubr5 gene, the specific region near transcription start site (TSS) of Ubr5 (221–242, TSS as 1) was found to be bound by circRNA-Filip1l (25–4, 5′ junction as 1) (Fig. 7A). Therefore, we chose to evaluate the potential role of Ubr5 as a target of circRNA-Filip1l. We hypothesized that circRNA-Filip1l facilitates the transcription of Ubr5 through its binding to Ubr5 TSS and recruits RNA polymerase. Second, to experimentally validate the in silicon predictions, according to our previous method (Pan et al., 2016), we cloned a 440 bp Ubr5 fragment containing the bound region by circRNA-Filip1l into pGL6 reporter vector and tested the effects of circRNA-Filip1l on the activities of the Ubr5 transcription in HEK293T cells. Expectedly, only pGL6-Ubr5 produced the relative strong luciferase activities compared with the empty vector (Fig. 7B), indicating that the cloned region contains the regulatory element and can drive Ubr5 expression. The cotransfection of pGL6-Ubr5 with circRNA-Filip1l overexpression plasmid enhanced the luciferase activities by 55.7% compared with cotransfection of pGL6-Ubr5 with empty vector (Fig. 7B). Third, we assessed whether Ubr5 coexpresses with other regulators: single-cell RT-PCR showed that 5 of 6 spinal neurons expressed Ubr5, that 4 of these 5 cells coexpressed with circRNA-Filip1l, 5 of them with miRNA-1224, and 4 of them with Ago2 (Fig. 7C), suggesting that they are able to coexpress in the spinal neurons. Thus, we further examined whether circRNA-Filip1l is involved in the regulation of Ubr5 expression in vivo. Spinal UBR5 was significantly increased by 67% day 3 after CFA injection (Fig. 7D,E), suggesting its possible regulatory role in the chronic inflammation pain. Furthermore, the increase of Ubr5 expression was efficiently reversed to the almost basal level day 2 after intrathecal injection of Ubr5-siRNA (Fig. 7D) day 3 after intrathecal injection of PLV-Ubr5 (Lentivirus Ubr5-shRNA) in CFA mice (Fig. 7E). It is apparent from our data that Ubr5-siRNA-or PLV-Ubr5-, but not scramble- or empty vector-injected, mice exhibited the antinociceptive effects in CFA-induced chronic inflammatory pain model (Fig. 7F,G). These findings indicate the involvement of Ubr5 in the process of chronic pain.
Figure 7.
circRNA-Filip1l regulates nociceptive response via positively targeting Ubr5. A, Schematic presentation of circRNA-Filip1l binding to near region of Ubr5 transcription start site (TSS). B, Validation of circRNA-Filip1l targeting Ubr5 by the use of luciferase reporter. The activities of the pGL6-Ubr5 encompassing TSS of Ubr5 region bound by circRNA-Filip1l were detected at hour 24 after cotransfection of pGL5 or pGL6-Ubr5 with DNA3.1-Filip1l by firefly luciferase reporter assays in HEK293T cells. The pGL6 plasmid (empty vector) was used as the negative control. pGL6-Ubr5, plasmid with Ubr5 region bound by circRNA-Filip1l; DNA3.1-Filip1l, plasmid of circRNA-Filip1l overexpression. Values of luciferase activities for each plasmid were normalized for transfection efficiency by cotransfection with pRL-TK plasmid. n = 4 per group. Two-way ANOVA (effect vs plasmid × treated interaction) followed by post hoc Tukey test: **p < 0.01; ***p < 0.001. C, Single-cell RT-PCR shows the coexpression of circRNA-Filip1l with miRNA-1224, Ago2, and Ubr5 in the spinal neurons of mice. No. 7 is a negative control. D, E, Knockdown of Ubr5 reversed the increase of spinal UBR5 protein 24 h after intrathecal injection of Ubr5-siRNA (D) or day 2 after 2 consecutive days of intrathecal injection of PLV-Ubr5 (E) in CFA mice. n = 5 per group. One-way ANOVA (expression vs the treated groups) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. F, Intrathecal injection of Ubr5-siRNA for 2 consecutive days alleviated the hypersensitivity to thermal or mechanical stimulus in CFA mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Red arrows indicate CFA or Sal injections. Blue arrows indicate Ubr5-siRNA or Scr injections. G, Intrathecal injection of PLV-Ubr5 for 2 consecutive days inhibited the pain sensitivity in CFA mice. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Red arrows indicate CFA or Sal injections. Blue arrows indicate PLV-Ubr5 or vector injections. H, Inhibiting circRNA-Filip1l via intrathecal injection of anti-Filip1l for 2 consecutive days reversed the increase of UBR5 protein in CFA mice. n = 5 per group. One-way ANOVA (expression vs the treated groups) followed by post hoc Tukey test: *p < 0.05. I, J, Upregulating circRNA-Filip1l via intrathecal injections of circRNA-Filip1l mimics (I) or Lenti-Filip1l (J) for 2 consecutive days increased the expression of UBR5 protein in naive mice. n = 5 per group. One-way ANOVA (expression vs the treated groups) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. K, Intrathecal preinjection of PLV-Ubr5 for 2 consecutive days prevented the thermal hyperalgesia and mechanical allodynia induced by circRNA-Filip1l during the development period. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Blue arrows indicate PLV-Ubr5 or vector injections. Red arrows indicate circRNA-Filip1l mimics or Scr injections. L, Intrathecal postinjection of Ubr5-siRNA for 2 consecutive days inhibited the pain hypersensitivity induced by Lenti-Filip1l during the development period. n = 6 per group. Two-way ANOVA (effect vs group × time interaction) followed by post hoc Tukey test: *p < 0.05; **p < 0.01. Red arrows indicate Lenti-Filip1l or Lenti-vector injections. Blue arrows indicate Ubr5-siRNA or Scr injections.
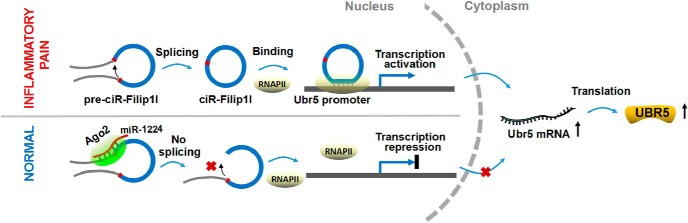
Next, we determined whether there is a regulatory role of circRNA-Filip1l in Ubr5 expression in vivo. The increase of Ubr5 was abolished by intrathecal injections of anti-Filip1l, but not the scrambled control in CFA mice (Fig. 7H), whereas overexpressing circRNA-Filip1l with circRNA-Filip1l mimics (Fig. 7I) or Lenti-Filip1l reduced (Fig. 7J) the level of UBR5 protein, compared with scrambled or empty vector in naive mice, suggesting that Ubr5 is a positive regulatory target of circRNA-Filip1l. Finally, to explore the role of Ubr5 in mediating nociceptive regulation by circRNA-Filip1l, we pretreated or post-treated animals with Ubr5-siRNA before or after overexpressing spinal circRNA-Filip1l, and then measured the nociceptive responses. Behavioral results showed that the knockdown of Ubr5 with intrathecal pretreatment with PLV-Ubr5 significantly prevented thermal and mechanical nociceptive responses induced by overexpression of circRNA-Filip1l through intrathecal injection of its mimics (Fig. 7K). Moreover, downregulation of Ubr5 with PLV-Ubr5 post-treatment significantly alleviated thermal and mechanical nociceptive responses induced by Lenti-Filip1l (Fig. 7L). These findings indicate a direct mediatory role of Ubr5 in regulation of nociception by circRNA-Filip1l. Together, these results suggest that spinal circRNA-Filip1l regulates nociception via positively targeting Ubr5. In conclusion, these data indicate that miRNA-1224 and Ago2 are involved in the regulation of circRNA-Filip1l-mediated Ubr5 expression (Fig. 8).
Figure 8.
The schematic of miRNA-1224 splicing circRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. RNAPII, RNA polymerase II.
Discussion
Chronic pain is one of the most intractable human complaints and is caused by inflammation, lesion, or dysfunction of the nervous system (Clark, 2016; Ji et al., 2018; Jing et al., 2018). The expression of aberrant pain-related gene in spinal neuronal or glial cells is the most prominent contributor in various nociceptive pathways underlying chronic pain (Ji et al., 2016; Jiang et al., 2017; Tsuda, 2018). Therefore, the unraveling of the genetic basis and its regulatory mechanisms will improve our insight into chronic pain, and provides potential targets for developing novel therapeutic strategies. Various studies have refined our understanding of miRNAs or long-strand ncRNA in the pathway of different pain models, and shown that noncoding RNAs have regulatory functions in the process of nociceptive signal. The current study identified an essential role of circRNA-Filip1l as a mediator of chronic inflammation pain by directly targeting Ubr5 at the spinal level. We further found that miRNA-1224 was an upstream regulator of circRNA-Filip1l in the Ago2-dependent manner. Our results are, for the first time, to functionally demonstrate that circRNA is an important player in the induction and maintenance of chronic pain.
circRNA, a noncoding RNA as the novel regulatory mechanism of gene expression, attracts widespread attention on its vital roles in biological processes and human diseases. A growing body of evidence suggests that circRNAs are enriched in the nervous system, such as different brain regions, primary neurons, and isolated synapses (Shao and Chen, 2016), a number of circRNAs are highly conserved among species, and their expressions are changed during neuronal differentiation (Rybak-Wolf et al., 2015). However, it is only beginning to be understood how circRNAs are involved in physiological and pathological processes of the nervous system. Several studies have gained an insight into the function of circRNAs in neurological disorders or diseases, such as PD (Kumar et al., 2018), AD (Lu and Xu, 2016; Zhao et al., 2016), amyotrophic lateral sclerosis, and spinal muscular atrophy (Scotti and Swanson, 2016). In 2017, circRNA was first reported to be associated with brain function: circRNA-Cdr1as-knock-out mice display the impaired sensorimotor gating due to the dysfunction of excitatory synaptic transmission in brain (Piwecka et al., 2017). circRNA serves as a critical player in CNS diseases. Recent work has focused on the relationship between circRNA and pain. Zhou et al. (2017) and Cao et al. (2017) found that spared nerve injury or chronic constriction injury-induced neuropathic pain causes the expression alteration of 188 or 469 spinal circRNAs, respectively. Despite that spinal circRNA profiles are changed in chronic neuropathic pain, it remains unknown whether circRNAs regulate the nociception behavior. The results of this study showed that CFA-induced chronic inflammation pain increased the content of circRNA-Filip1l in spinal cord of mice; knockdown of circRNA-Filip1l alleviated the nociceptive behavior, supporting the idea that circRNA-Filip1l regulates the nociceptive response.
miRNAs are ∼21 nt in length and well-studied noncoding RNA species in terms of their biogenesis and functions. Increasing evidence over the past few years from different pain models has linked miRNA to nociceptive pathways, including membrane receptors (Park et al., 2014; Jiang et al., 2016), ion channels (Pan et al., 2016; Peng et al., 2017), transcription factors (Willemen et al., 2012), translation factors (Sun et al., 2012), and other cellular signals (Zhou et al., 2016), from primary afferent nociceptors, DRG, spinal cord, and brain areas. Previous reports on nucleus miRNAs uncover their important functions in the RNA splice or transcription (Liao et al., 2010). For example, nucleus miR-671 can mediate the cleavage of circRNA-Cdr1as in mouse brain (Hansen et al., 2011). Although a growing number of molecular and functional data support the involvement of cytoplasma miRNA in the process of chronic pain, little is known about whether nucleus miRNA is related to chronic pain. In our work, the abundant expression of miRNA-1224 was identified in the mouse spinal nucleus. Indeed, miRNA-1224 has been discovered to differentially express in the liver of fatty liver disease (Dolganiuc et al., 2009), and in different tissues of bladder cancer (Dudziec et al., 2011), inflammation, and acute injured kidney (Niu et al., 2011; Bellinger et al., 2014; Roy et al., 2017). miRNA-1224 can be also detected in hippocampus and the marginal division of the neostriatum in rats (Shu et al., 2013). But up to now, far too little attention has been paid to the function of miRNA-1224 in diseases. We provide the first evidence that spinal nucleus miRNA-1224 is implicated in the modulation of chronic inflammatory pain. Furthermore, we reveal a novel mechanism of miRNA-122 in the pain process through splicing circRNA-Filip1l in an Ago2-dependent manner.
Indeed, it is still challenging to understand how circRNA expression itself is regulated in pathological processes. Several reports show that antisense oligonucleotide or miRNA are associated with the splice of circRNA and change their expression level (Havens et al., 2013; Jeck and Sharpless, 2014). As miRNA-671 almost fully binds to circRNA-Cdr1as to form the miRNA-671-circRNA-Cdr1as complex in cell nucleus, Ago2 slices circRNA-Cdr1as after recognizing the complex (Piwecka et al., 2017). In the present study, we found that AGO2 recognized and sliced the complex formed by miRNA-1224 and pre-circRNA-Filip1l, resulting in the reduction of mature circRNA-Filip1l in the spinal nucleus. Our data suggest that miRNA-mediated AGO2 cleavage for pre-circRNA may play a crucial role in the modulation of circRNA biogenesis, at least in spinal nucleus. Therefore, combined with previous reports, circRNA may be regulated through two ways: AGO2 cleaved circRNA or their precursors after miRNAs binding. Our findings will allow for a new optional strategy in prevention and treatment of pain or other CNS diseases via affecting circRNA biogenesis by miRNA.
Ago2, termed as Eif2c2, is a member of the Ago family (1, 2, 3, and 4) characterized with a high conservation among species and broad expression in different tissues (Ye et al., 2015). Interestingly, Ago2 is the only one with catalytic activity among family members and efficiently silences the expression of small RNAs. Distinguishing from the other members, mice with Ago2 knock-out are lethal (Shekar et al., 2011). Extensive studies have shown that AGO2 participates in miRNA generation (Schaefer et al., 2010), miRNA-mediated mRNA degradation (Meister et al., 2004; Cifuentes et al., 2010), translation repression (Friend et al., 2012), and hetero-chromatinization (Moshkovich et al., 2011) in Dicer (an endoribonuclease)-independent means. Knockdown of Ago2 leads to the decrease of global miRNA (Morita et al., 2007). For example, Ago2, not 1, 3, and 4, is downregulated in brain lysates from encephalomyelitis mice, the expression level of several miRNAs, including let-7a-5p, let-7e-5p, let-7f-5p, 106b-5p, 144-3p, and 188a-5p, display a significant reduction (Lewkowicz et al., 2015), suggesting that Ago2 is involved in the etiology of CNS diseases (Savas et al., 2008). Deficiency of Ago2 in D2R neurons relieves self-administration of cocaine due to the declined content of miRNAs in the mouse striatum (Schaefer et al., 2010), revealing a vital role of Ago2 in the treatment of CNS diseases (Carrick et al., 2016). Here, we found that nociceptive behavior caused the decrease of spinal Ago2 level, and overexpressing Ago2 significantly attenuated the pain hypersensitivity. It can therefore be concluded that Ago2 is an important player in nociceptive response.
Despite a large number of circRNAs having been found, how circRNA regulates gene expression has been a major problem for a long time. The relatively well-studied mechanism is that circRNA adsorbs miRNAs (a process known as miRNA sponge) and thereby decreases the binding of miRNAs to their target mRNAs (Hansen et al., 2013b). As circRNA-HIPK3 adsorbs 9 different miRNAs, the knockdown of circRNA-HIPK3 inhibits cell growth via enhancing the level of miRNAs binding to the target mRNAs (Zheng et al., 2016). miRNA-7 is involved in the formation of dendritic spine density (Choi et al., 2015; Zhang et al., 2015), Cdr1as is known as the sponge for miRNA-7 and regulates the stability or transport of miRNA-7 in neuronal cells; hence, the function-loss-circRNA-Cdr1as impairs the sensorimotor gating and synaptic transmission of mouse brain tissues, including cerebellum, cortex, hippocampus, and olfactory bulb, by enhancing the number of freely available miRNA-7 (Piwecka et al., 2017). Although, relatively to act as sponge of miRNAs, only few circRNAs are studied in the function of gene transcription, it is now well established from various studies that circRNAs can bind to not only proteins, such as AGO and RNA polymerase II (RNAPII), but also to linear RNAs and DNAs in cellular nucleus. circRNA from Fmn gene lavishly presents in nucleus and promotes the transcription of Fmn gene by enhancing the binding capacity of PoII to Fmn promoter (Chao et al., 1998; Zhang et al., 2013). Similarly, the complex formed by circRNA-EI and U1 snRNP can also recruit PoII to the promoter of circRNA-EI parental gene in nucleus, initiating the transcription process (Li et al., 2015). Interestingly, circRNA-Mble regulates the biogenesis of parent linear Mble RNA (Ashwal-Fluss et al., 2014). Consistently, our findings showed that circRNA-Filip1l enhanced the transcription level of Ubr5 via binding to the near region in TSS. But further research should be undertaken to investigate whether the binding sites recruit the RNA polymerase. Our observations may provide an approach to obtain the possible downstream targets of nucleus circRNAs. The findings of the current study support the previous reports: lncRNAs, such as RoX2, TERC, and HOTAIR, are enriched in the mammalian nucleus and bind to gene bodies and GA-rich DNA regions to control gene transcription via recruiting RNA polymerase (Chu et al., 2011; Bonasio and Shiekhattar, 2014). Consequently, we speculate that circRNA or lncRNA may share the regulatory mechanism of gene expression through transcriptional interference (Bonasio and Shiekhattar, 2014; Fatica and Bozzoni, 2014).
UBR5 is an HECT (homologous to E6-associated protein at the carboxy terminus) E3 Ub ligase recognizing n-degrons and has a catalytic ability of directly recognizing and ligating ubiquitin to degrading proteins. It is very well deliberated that Ubr5 participates in CNS-related diseases, such as neuroinflammation, cognitive disorders, and depression (Gudjonsson et al., 2012; Rutz et al., 2015). Depression increases Ubr5 expression in the lateral habenula tissues of rats; the administration of escitalopram, a selective serotonin reuptake inhibitor, for 4 weeks alleviates the depression behavior by decreasing the level of Ubr5 expression (Christensen et al., 2013). Ubr5 mutation clinically impairs the cognitive capability in AD (Hu et al., 2011), suggesting that Ubr5 has an essential role in CNS diseases. In our work, Ubr5 was significantly increased in spinal cord of chronic inflammatory pain model, the knockdown of Ubr5 attenuated the pain behavior, and therefore, Ubr5 could serve as an important regulator in the process chronic pain.
In conclusion, we demonstrate that spinal miRNA-1224-mediated splice of circRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. These findings shed light on new circRNA mechanism underlying nociceptive information processing and may provide the rationale for the future development of potential targeted interventions via circRNA modulating pain-related gene expression.
Footnotes
The work was supported by National Natural Science Foundation of China Grants 81671096 and 81271231 to Z.P., Grants 31771161 and 81720108013 to J.-L.C., and Grant 31500855 to L.-J.Z., Natural Science Foundation of Jiangsu Education Department Key Project 15KJA320004 to Z.P., the Project Funded by the Qing Lan Project, the Six Talent Summit Project, and the 333 High-level Personnel Training Project.
The authors declare no competing financial interests.
References
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Bellinger MA, Bean JS, Rader MA, Heinz-Taheny KM, Nunes JS, Haas JV, Michael LF, Rekhter MD (2014) Concordant changes of plasma and kidney microRNA in the early stages of acute kidney injury: time course in a mouse model of bilateral renal ischemia-reperfusion. PLoS One 9:e93297. 10.1371/journal.pone.0093297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R (2014) Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48:433–455. 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Deng W, Li Y, Qin B, Zhang L, Yu S, Xie P, Xiao Z, Yu T (2017) Chronic constriction injury of sciatic nerve changes circular RNA expression in rat spinal dorsal horn. J Pain Res 10:1687–1696. 10.2147/JPR.S139592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrick WT, Burks B, Cairns MJ, Kocerha J (2016) Noncoding RNA regulation of dopamine signaling in diseases of the central nervous system. Front Mol Biosci 3:69. 10.3389/fmolb.2016.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CW, Chan DC, Kuo A, Leder P (1998) The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 4:614–628. 10.1007/BF03401761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Schuman E (2016) Circular RNAs in brain and other tissues: a functional enigma. Trends Neurosci 39:597–604. 10.1016/j.tins.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Choi SY, Pang K, Kim JY, Ryu JR, Kang H, Liu Z, Kim WK, Sun W, Kim H, Han K (2015) Post-transcriptional regulation of SHANK3 expression by microRNAs related to multiple neuropsychiatric disorders. Mol Brain 8:74. 10.1186/s13041-015-0165-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T, Jensen L, Bouzinova EV, Wiborg O (2013) Molecular profiling of the lateral habenula in a rat model of depression. PLoS One 8:e80666. 10.1371/journal.pone.0080666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ (2010) A novel miRNA processing pathway independent of dicer requires Argonaute2 catalytic activity. Science 328:1694–1698. 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JD. (2016) Preclinical pain research: can we do better? Anesthesiology 125:846–849. 10.1097/ALN.0000000000001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descalzi G, Ikegami D, Ushijima T, Nestler EJ, Zachariou V, Narita M (2015) Epigenetic mechanisms of chronic pain. Trends Neurosci 38:237–246. 10.1016/j.tins.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G (2009) MicroRNA expression profile in lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res 33:1704–1710. 10.1111/j.1530-0277.2009.01007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziec E, Miah S, Choudhry HM, Owen HC, Blizard S, Glover M, Hamdy FC, Catto JW (2011) Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin Cancer Res 17:1287–1296. 10.1158/1078-0432.CCR-10-2017 [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15:7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J (2012) A conserved PUF-ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol 19:176–183. 10.1038/nsmb.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandla J, Lomada SK, Lu J, Kuner R, Bali KK (2017) miR-34c–5p functions as pronociceptive microRNA in cancer pain by targeting Cav2.3 containing calcium channels. Pain 158:1765–1779. 10.1097/j.pain.0000000000000971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20:1666–1670. 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, Mailand N, Neumann B, Heriche JK, Shearer R, Saunders D, Bartek J, Lukas J, Lukas C (2012) TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150:697–709. 10.1016/j.cell.2012.06.039 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J (2011) miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30:4414–4422. 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Kjems J, Damgaard CK (2013a) Circular RNA and miR-7 in cancer. Cancer Res 73:5609–5612. 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013b) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Havens MA, Duelli DM, Hastings ML (2013) Targeting RNA splicing for disease therapy. Wiley Interdiscip Rev RNA 4:247–266. 10.1002/wrna.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gäbel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Pickering EH, Hall SK, Naik S, Liu YC, Soares H, Katz E, Paciga SA, Liu W, Aisen PS, Bales KR, Samad TA, John SL (2011) Genome-wide association study identifies multiple novel loci associated with disease progression in subjects with mild cognitive impairment. Transl Psychiatry 1:e54. 10.1038/tp.2011.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel S, Schlichter R (2000) Presynaptic P2X receptors facilitate inhibitory GABAergic transmission between cultured rat spinal cord dorsal horn neurons. J Neurosci 20:2121–2130. 10.1523/JNEUROSCI.20-06-02121.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsberger JG, Fessler EB, Wang Z, Elkahloun AG, Chuang DM (2012) Post-insult valproic acid-regulated microRNAs: potential targets for cerebral ischemia. Am J Transl Res 4:316–332. [PMC free article] [PubMed] [Google Scholar]
- Imai S, Ikegami D, Yamashita A, Shimizu T, Narita M, Niikura K, Furuya M, Kobayashi Y, Miyashita K, Okutsu D, Kato A, Nakamura A, Araki A, Omi K, Nakamura M, James Okano H, Okano H, Ando T, Takeshima H, et al. (2013) Epigenetic transcriptional activation of monocyte chemotactic protein 3 contributes to long-lasting neuropathic pain. Brain 136:828–843. 10.1093/brain/aws330 [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BC, Cao DL, Zhang X, Zhang ZJ, He LN, Li CH, Zhang WW, Wu XB, Berta T, Ji RR, Gao YJ (2016) CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J Clin Invest 126:745–761. 10.1172/JCI81950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BC, He LN, Wu XB, Shi H, Zhang WW, Zhang ZJ, Cao DL, Li CH, Gu J, Gao YJ (2017) Promoted interaction of C/EBPalpha with demethylated Cxcr3 gene promoter contributes to neuropathic pain in mice. J Neurosci 37:685–700. 10.1523/JNEUROSCI.2262-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Chamessian A, Zhang YQ (2016) Pain regulation by non-neuronal cells and inflammation. Science 354:572–577. 10.1126/science.aaf8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Nackley A, Huh Y, Terrando N, Maixner W (2018) Neuroinflammation and central sensitization in chronic and widespread pain. Anesthesiology 129:343–366. 10.1097/ALN.0000000000002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing PB, Cao DL, Li SS, Zhu M, Bai XQ, Wu XB, Gao YJ (2018) Chemokine receptor CXCR3 in the spinal cord contributes to chronic itch in mice. Neurosci Bull 34:54–63. 10.1007/s12264-017-0128-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Tamiya G, Koyama S, Nakamura T, Makino S, Arawaka S, Kawanami T, Tooyama I (2012) UBR5 gene mutation is associated with familial adult myoclonic epilepsy in a Japanese family. ISRN Neurol 2012:508308. 10.5402/2012/508308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Shamsuzzama, Jadiya P, Haque R, Shukla S, Nazir A (2018) Functional characterization of novel circular RNA molecule, circzip-2 and its synthesizing gene zip-2 in C. elegans model of Parkinson's disease. Mol Neurobiol 55:6914–6926. 10.1007/s12035-018-0903-5 [DOI] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22–37.e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewkowicz P, Cwiklinska H, Mycko MP, Cichalewska M, Domowicz M, Lewkowicz N, Jurewicz A, Selmaj KW (2015) Dysregulated RNA-induced silencing complex (RISC) assembly within CNS corresponds with abnormal miRNA expression during autoimmune demyelination. J Neurosci 35:7521–7537. 10.1523/JNEUROSCI.4794-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH (2010) Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 5:e10563. 10.1371/journal.pone.0010563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Long Y, Wang X, Youmans DT, Cech TR (2017) How do lncRNAs regulate transcription? Sci Adv 3:eaao2110. 10.1126/sciadv.aao2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Xu AD (2016) Mini review: circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front Genet 7:53. 10.3389/fgene.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15:185–197. 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Monif M, Reid CA, Powell KL, Drummond KJ, O'Brien TJ, Williams DA (2016) Interleukin-1beta has trophic effects in microglia and its release is mediated by P2X7R pore. J Neuroinflammation 13:173. 10.1186/s12974-016-0621-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I (2007) One argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89:687–696. 10.1016/j.ygeno.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP (2011) RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev 25:1686–1701. 10.1101/gad.16651211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Mo D, Qin L, Wang C, Li A, Zhao X, Wang X, Xiao S, Wang Q, Xie Y, He Z, Cong P, Chen Y (2011) Lipopolysaccharide-induced miR-1224 negatively regulates tumour necrosis factor-alpha gene expression by modulating Sp1. Immunology 133:8–20. 10.1111/j.1365-2567.2010.03374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Zhu LJ, Li YQ, Hao LY, Yin C, Yang JX, Guo Y, Zhang S, Hua L, Xue ZY, Zhang H, Cao JL (2014) Epigenetic modification of spinal miR-219 expression regulates chronic inflammation pain by targeting CaMKIIgamma. J Neurosci 34:9476–9483. 10.1523/JNEUROSCI.5346-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Zhang M, Ma T, Xue ZY, Li GF, Hao LY, Zhu LJ, Li YQ, Ding HL, Cao JL (2016) Hydroxymethylation of microRNA-365–3p regulates nociceptive behaviors via Kcnh2. J Neurosci 36:2769–2781. 10.1523/JNEUROSCI.3474-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Xue ZY, Li GF, Sun ML, Zhang M, Hao LY, Tang QQ, Zhu LJ, Cao JL (2017) DNA hydroxymethylation by ten-eleven translocation methylcytosine dioxygenase 1 and 3 regulates nociceptive sensitization in a chronic inflammatory pain model. Anesthesiology 127:147–163. 10.1097/ALN.0000000000001632 [DOI] [PubMed] [Google Scholar]
- Park CK, Xu ZZ, Berta T, Han Q, Chen G, Liu XJ, Ji RR (2014) Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron 82:47–54. 10.1016/j.neuron.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Li L, Zhang MD, Bengtsson Gonzales C, Parisien M, Belfer I, Usoskin D, Abdo H, Furlan A, Häring M, Lallemend F, Harkany T, Diatchenko L, Hökfelt T, Hjerling-Leffler J, Ernfors P (2017) miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science 356:1168–1171. 10.1126/science.aam7671 [DOI] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- Rasko JE, Wong JJ (2017) Nuclear microRNAs in normal hemopoiesis and cancer. J Hematol Oncol 10:8. 10.1186/s13045-016-0375-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC. (2014) The microRNA biology of the mammalian nucleus. Mol Ther Nucleic Acids 3:e188. 10.1038/mtna.2014.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Bantel H, Wandrer F, Schneider AT, Gautheron J, Vucur M, Tacke F, Trautwein C, Luedde T, Roderburg C (2017) miR-1224 inhibits cell proliferation in acute liver failure by targeting the antiapoptotic gene Nfib. J Hepatol 67:966–978. 10.1016/j.jhep.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Rutz S, Kayagaki N, Phung QT, Eidenschenk C, Noubade R, Wang X, Lesch J, Lu R, Newton K, Huang OW, Cochran AG, Vasser M, Fauber BP, DeVoss J, Webster J, Diehl L, Modrusan Z, Kirkpatrick DS, Lill JR, Ouyang W, et al. (2015) Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518:417–421. 10.1038/nature13979 [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, Shiekhattar R, Markey SP, Tanese N (2008) Huntington's disease protein contributes to RNA-mediated gene silencing through association with argonaute and P bodies. Proc Natl Acad Sci U S A 105:10820–10825. 10.1073/pnas.0800658105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Venø MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O'Carroll D, Greengard P (2010) Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med 207:1843–1851. 10.1084/jem.20100451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS (2016) RNA mis-splicing in disease. Nat Rev Genet 17:19–32. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Chen Y (2016) Roles of circular RNAs in neurologic disease. Front Mol Neurosci 9:25. 10.3389/fnmol.2016.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekar PC, Naim A, Sarathi DP, Kumar S (2011) Argonaute-2-null embryonic stem cells are retarded in self-renewal and differentiation. J Biosci 36:649–657. 10.1007/s12038-011-9094-1 [DOI] [PubMed] [Google Scholar]
- Shu SY, Qing D, Wang B, Zeng QY, Chen YC, Jin Y, Zeng CC, Bao R (2013) Comparison of microRNA expression in hippocampus and the marginal division (MrD) of the neostriatum in rats. J Biomed Sci 20:9. 10.1186/1423-0127-20-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CR, Seow Y, Curtis H, Weinberg MS, Wood MJ (2012) Silencing of Parkinson's disease-associated genes with artificial mirtron mimics of miR-1224. Nucleic Acids Res 40:9863–9875. 10.1093/nar/gks712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Wang H, Ge W, Yang M, Hou J, Chen T, Li N, Cao X (2015) An in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancer. Cancer Res 75:2875–2885. 10.1158/0008-5472.CAN-14-2218 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li XQ, Sahbaie P, Shi XY, Li WW, Liang DY, Clark JD (2012) miR-203 regulates nociceptive sensitization after incision by controlling phospholipase A2 activating protein expression. Anesthesiology 117:626–638. 10.1097/ALN.0b013e31826571aa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao YX, Rumbaugh G, Wang GD, Petralia RS, Zhao C, Kauer FW, Tao F, Zhuo M, Wenthold RJ, Raja SN, Huganir RL, Bredt DS, Johns RA (2003) Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J Neurosci 23:6703–6712. 10.1523/JNEUROSCI.23-17-06703.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M. (2018) Modulation of pain and itch by spinal glia. Neurosci Bull 34:178–185. 10.1007/s12264-017-0129-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemen HL, Huo XJ, Mao-Ying QL, Zijlstra J, Heijnen CJ, Kavelaars A (2012) MicroRNA-124 as a novel treatment for persistent hyperalgesia. J Neuroinflammation 9:143. 10.1186/1742-2094-9-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JX, Hua L, Li YQ, Jiang YY, Han D, Liu H, Tang QQ, Yang XN, Yin C, Hao LY, Yu L, Wu P, Shao CJ, Ding HL, Zhang YM, Cao JL (2015) Caveolin-1 in the anterior cingulate cortex modulates chronic neuropathic pain via regulation of NMDA receptor 2B subunit. J Neurosci 35:36–52. 10.1523/JNEUROSCI.1161-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Jin H, Qian Q (2015) Argonaute 2: a novel rising star in cancer research. J Cancer 6:877–882. 10.7150/jca.11735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun XY, Zhang LY (2015) MicroRNA-7/Shank3 axis involved in schizophrenia pathogenesis. J Clin Neurosci 22:1254–1257. 10.1016/j.jocn.2015.01.031 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chung SK, Chow BK (2014) The knockout of secretin in cerebellar Purkinje cells impairs mouse motor coordination and motor learning. Neuropsychopharmacology 39:1460–1468. 10.1038/npp.2013.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yang XN, Zang T, Luo J, Pan Z, Wang L, Liu H, Liu D, Li YQ, Zhang YD, Zhang H, Ding HL, Cao JL (2017) Astroglial MicroRNA-219–5p in the ventral tegmental area regulates nociception in rats. Anesthesiology 127:548–564. 10.1097/ALN.0000000000001720 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, Gao YJ, Hoffman PN, Cui H, Li M, Dong X, Tao YX (2013) A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci 16:1024–1031. 10.1038/nn.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ (2016) Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel) 7:E116. 10.3390/genes7120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xiong Q, Chen H, Yang C, Fan Y (2017) Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front Mol Neurosci 10:91. 10.3389/fnmol.2017.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yang L, Larson S, Basra S, Merwat S, Tan A, Croce C, Verne GN (2016) Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut 65:797–805. 10.1136/gutjnl-2013-306464 . [DOI] [PMC free article] [PubMed] [Google Scholar]