Abstract
Background
Obstetric complications have long been retrospectively associated with a wide range of short- and long-term health consequences, including neurodevelopmental alterations such as those observed in schizophrenia and other psychiatric disorders. However, prospective studies assessing fetal well-being during pregnancy tend to focus on perinatal complications as the final outcome of interest, while there is a scarcity of postnatal follow-up studies. In this study, the cerebroplacental ratio (CPR), a hemodynamic parameter reflecting fetal adaptation to hypoxic conditions, was analyzed in a sample of monozygotic monochorionic twins (60 subjects), part of them with prenatal complications, with regard to (i) epigenetic age acceleration, and (ii) DNA methylation at genes included in the polygenic risk score (PRS) for schizophrenia, and highly expressed in placental tissue.
Results
Decreased CPR measured during the third trimester was associated with epigenetic age deceleration (β = 0.21, t = 3.362, p = 0.002). Exploration of DNA methylation at placentally expressed genes of the PRS for schizophrenia revealed methylation at cg06793497 (EP300 gene) to be associated with CPR (β = 0.021, t = 4.385; p = 0.00008, FDR-adjusted p = 0.11). This association was reinforced by means of an intrapair analysis in monozygotic twins discordant for prenatal suffering (β = 0.027, t = 3.924, p = 0.001).
Conclusions
Prenatal adverse environment during the third trimester of pregnancy is associated with both (i) developmental immaturity in terms of epigenetic age, and (ii) decreased CpG-specific methylation in a gene involved in hypoxia response and schizophrenia genetic liability.
Electronic supplementary material
The online version of this article (10.1186/s13148-019-0674-5) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Obstetric complications, Prenatal stress, Hypoxia, EP300 gene, Epigenetic clock, Monozygotic twins, Schizophrenia
Main text
Background
Prenatal environment constitutes the first modulating agent the developing fetus encounters as it progresses through gestation. The tremendous impact of any environmental threat occurring during this period for both short- and long-term consequences is now widely accepted and well-known as the Developmental Origins of Health and Disease (DOHaD) hypothesis [1]. Also known as the theory of fetal programming, the embedding of early life and its ability to exert long-term effects in late-life is thought to rely on epigenetic mechanisms [2, 3].
Recently, several DNA methylation-based epigenetic clocks have been developed in order to predict chronological age with high accuracy [4, 5]; afterward, Knight and colleagues developed a new predictor specifically aimed to predict gestational age (GA) in perinatal samples [6]. Although epigenetic and chronological age robustly show high correlation across studies, the difference between both variables allows the estimation of the so-called age acceleration (i.e., when epigenetic age is higher than chronological age).
On the one hand, epigenetic age acceleration in adult subjects has been associated with cumulative lifetime stress, lifestyle, and all-cause mortality, among others, suggesting its utility as a better predictor for life expectancy than chronological age itself [7–9]. On the other hand, epigenetic GA deceleration (i.e., when chronological age is higher than epigenetic age), as measured in cord blood, has been described in newborns born to women with low socioeconomic status, Sjögren syndrome, insulin-treated gestational diabetes mellitus, and experiencing antenatal depressive symptoms [6, 10, 11]. Such findings suggest that newborns exposed to prenatal stressors are born in an immature state independently of their chronological GA. In this regard, boys—but not girls—who exhibited lower epigenetic GA at birth exhibited more internalizing problems, such as anxious-depressive symptoms or somatic complaints, at follow-up (mean age 3.7 years), suggesting they are born with a developmental disadvantage [11].
Nevertheless, there is a dearth of studies examining the putative relationship between ultrasound parameters acquired during pregnancy and epigenetic GA acceleration. In this regard, the cerebroplacental ratio (CPR) has been reported to be associated with adverse perinatal outcomes not only in growth-restricted fetuses, but also in low-risk population [12, 13]. Briefly, CPR is calculated by dividing the middle cerebral artery (MCA) pulsatility index (PI) by the umbilical artery (UA) PI [14]. The PI is a parameter reflecting vascular impedance or resistance, i.e., decreased blood flow. Specifically, fetal brain blood supply is known to increase in front of hypoxic stimuli thus decreasing PI in the MCA [15]; while placental insufficiency decreases umbilical blood flow hence increasing UA PI, and has been associated with both short- and long-term detrimental outcomes, including increased cardiovascular risk and deficits in cognition [16, 17]. Consequently, a decreased CPR reflects the combination of both alterations and is an indicator of fetal adaptation to adverse conditions [12].
Obstetric complications (OCs) constitute one of the risk factors more reliably associated with psychopathology, particularly with neurodevelopmental disorders; specifically, the putative association between OCs and schizophrenia has been debated since the 1970s [18–20]. In this regard, a recent umbrella review evaluating all published meta-analysis regarding risk factors and biomarkers for schizophrenia spectrum disorders revealed a history of OCs to significantly increase the risk for developing the disorder with an odds ratio of 2 [21]. Furthermore, exposure to severe OCs together with increased genetic vulnerability, as measured with the polygenic risk score (PRS) for schizophrenia, interact to increase the risk to suffer the disorder up to an odds ratio of 8.36 [22]. In the same study, authors further explored the putative relevance of placental expression of genes included in the PRS; following this approach, they reported (i) an enrichment of PRS genes expressed in placental tissue and (ii) differential expression of PRS genes in placentae from complicated pregnancies (specifically in pre-eclampsia and intrauterine growth restriction). Specifically, the described gene-environment interaction between exposure to OCs and the PRS for schizophrenia was driven by those genes highly expressed in placenta and/or dynamically regulated in complicated pregnancies [22]. Since CPR is a robust indicator of prenatal stress and a predictor of perinatal and long-term morbidity, DNA methylation analysis of genes included in the placental PRS for schizophrenia could shed light on the epigenetic mechanisms mediating the interaction between OCs and neurodevelopmental disorders.
Monozygotic twins have been instrumental for the elucidation of environmental and genetic risks in the etiology of complex traits and disorders. Actually, the differential role of the prenatal environment in shaping psychopathological proneness was first described thanks to monozygotic twin designs [23–25]; these pioneering studies focused on dermatoglyphic measures assessed at birth, which can be used as surrogate measures of altered neurodevelopment during the second trimester of pregnancy [26]. Furthermore, monozygotic twin pregnancies and, more specifically, monochorionic twin pregnancies—i.e., those in which both fetuses share the placenta—are at a higher risk of obstetric complications, the more prevalent being twin-to-twin transfusion syndrome (TTTS) and selective intrauterine growth restriction (sIUGR) [27–29]. Thus, the thorough and prospective ultrasound assessment of prenatal development through monochorionic twin pregnancies offers a quasi-experimental study design in which the genetic and environmental components of epigenetic variability can be dissected.
The objective of the current study was to investigate whether prenatal adverse environment (i) alters human development in terms of epigenetic age, and if (ii) it can get embedded through epigenetic mechanisms in genes previously identified as risk factors for schizophrenia acting during prenatal stages. We hypothesized that a higher exposure to prenatal adverse environment would be associated with (i) delayed development and (ii) DNA methylation at genes involved in the pathogenesis of schizophrenia. While CPR can have diverse effects on genome-wide DNA methylation, with potential relevance for a multitude of phenotypes, the present study a priori examined how CPR epigenetically regulates risk genes for schizophrenia, previously described to interact with the presence of OCs [22].
Results
GA estimation using Knight’s epigenetic clock
After exclusion of two twin pairs (see Methods section), the final sample size was 30 twin pairs. The mean GA at birth of our twin cohort (n = 30 twin pairs) was 35.3 weeks (range = 31.7–37.1) and the mean DNA methylation GA at birth was 35 weeks (range = 31.4–37.7). To validate the epigenetic clock predictor in our sample, DNA methylation-based GA was tested for correlation with chronological GA (r = 0.76, p = 1.68 × 10−12; Fig. 1). The average absolute difference between epigenetic GA and chronological GA—hereinafter referred as ΔGA—was 0.9 weeks (range = 0.03–4.02), i.e., 6.3 days. In agreement with previous studies, there was a significant negative correlation between ΔGA and chronological GA (r = − 0.47; p < 0.001).
Fig. 1.

Correlation between chronological GA and epigenetic GA. Chronological GA was calculated using first-trimester crown-rump length measurement of the larger twin, and epigenetic age was calculated based on DNA methylation-based Knight’s clock. Both GA estimations were significantly correlated (r = 0.76; p = 1.68 × 10−12)
Association between ΔGA and CPR
ΔGA was tested for associations with CPR measured during the third trimester (mean = 33.8 weeks, range = 28.3–36.4), a few days before childbirth (median = 6.5 days). CPR was significantly associated with ΔGA (β = 0.21, t = 3.362, p = 0.002) when adjusting for sex, birthweight, diagnostic of either TTTS or sIUGR, surgery time interval (when laser fetoscopy had been applied), and gestational age at ultrasound as covariates. The positive association between CPR and ΔGA remained significant after correction for cell type proportion (β = 0.21, t = 2.616, p = 0.01). Figure 2 shows the positive association between third trimester CPR and ΔGA.
Fig. 2.

Association between epigenetic age acceleration and cerebroplacental ratio measured during the third trimester. Epigenetic age delta (ΔGA) corresponds to estimated epigenetic age minus chronological age. Thus, ΔGA-positive values reflect epigenetic age acceleration while negative values point out the presence of epigenetic age deceleration. The cerebroplacental ratio (CPR) is calculated as the ratio between the MCAPI and UAPI. Both variables were significantly correlated when adjusting for sex, chronological gestational age, birth weight, and gestational age at ultrasound
Epigenetic exploration of placental PRS for schizophrenia with regard to CPR
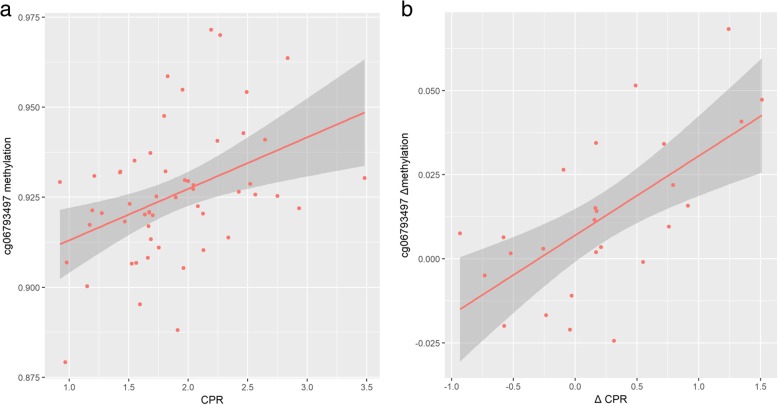
Following the approach developed by Ursini and collaborators (2018), association between CPR and DNA methylation was tested in all CpG sites included in the DNA methylation array located within genes of the PRS for schizophrenia expressed in placental tissue (placental PRS) [22]. There were 1400 CpG sites annotated to placental PRS genes out of 866,091 CpG sites included in the array. After FDR correction for multiple testing, methylation at one single CpG site, cg06793497, was significantly associated with CPR (β = 0.021, p = 0.00008, t = 4.385; qFDR adjusted = 0.11; Fig. 3a), such that increased cg06793497 methylation was associated with increased CPR. The top 10 CpG sites yielded by this approach are summarized in Table 1 (all q values > 0.75).
Fig. 3.
Schizophrenia PRS methylation exploration with regard to cerebroplacental ratio measured during the third trimester. a Methylation at cg06793497 (EP300 gene) was significantly associated with CPR in the whole sample (n = 54 twin subjects). b Intrapair methylation difference at cg06793497 was significantly associated with intrapair CPR difference (n = 26 twin pairs)
Table 1.
Top 10 CpG sites of the PRS methylomic exploration in association with CPR (1400 CpG sites tested)
| # | CpG probea | Genomic coordinatesb | Gene | PRS explorationc | Intrapaird | |||
|---|---|---|---|---|---|---|---|---|
| beta | p value | q value | beta | p value | ||||
| 1 | cg06793497 | 22: 41,542,898 | EP300 | 0.021 | 8.2E-05 | 0.115 | 0.027 | 0.001 |
| 2 | cg15620905 | 1: 44,024,150 | PTPRF | 0.040 | 0.002 | 0.804 | 0.008 | 0.563 |
| 3 | cg12252443 | 2: 198,364,630 | HSPD1 | -0.006 | 0.002 | 0.804 | − 0.007 | 0.055 |
| 4 | cg24936500 | 2: 233,499,637 | EFHD1 | 0.022 | 0.004 | 0.804 | 0.001 | 0.849 |
| 5 | cg00262246 | 3: 136,007,461 | PCCB | 0.012 | 0.004 | 0.804 | 0.012 | 0.029 |
| 6 | cg22495590 | 5: 138,161,059 | CTNNA1 | -0.020 | 0.004 | 0.804 | − 0.010 | 0.203 |
| 7 | cg14902598 | 5: 138,210,650 | CTNNA1 | 0.017 | 0.005 | 0.804 | 0.006 | 0.371 |
| 8 | cg01024069 | 14: 104,158,878 | KLC1 | -0.010 | 0.005 | 0.804 | − 0.010 | 0.033 |
| 9 | cg16362480 | 16: 30,077,084 | ALDOA | 0.031 | 0.005 | 0.804 | 0.024 | 0.126 |
| 10 | cg12955069 | 16: 58,593,852 | CNOT1; SNORA50 | -0.016 | 0.006 | 0.804 | − 0.021 | 0.026 |
a CpG site code according to the Illumina annotation
bGenomic coordinates correspond to hg19 built
cRefers to the statistics of each analysis in the first model encompassing 1400 CpG sites located in genes of the PRS for schizophrenia highly expressed in the placenta as described by Ursini et al.
dRefers to intrapair comparison of DNA methylation values and CPR. Thus, the n for these analyses was of 26 twin pairs
To further explore the association between cg06793497 methylation and CPR, it was analyzed in a monozygotic twin intrapair design. The intrapair twin design further allows controlling for chronological GA, sex, and timing of the Doppler ultrasound, since these variables are shared by co-twins of a pair. Four observations were removed from the analysis due to missingness for any of the variables in one of the co-twins of a pair. Thus, intrapair differences for these measures were calculated for all twin pairs of the sample were both measures were available for both twins of a pair (n = 27 twin pairs). Intrapair differences in cg06793497 methylation and CPR, measured during the third trimester, were significantly correlated (r = 0.64, p < 0.001; Fig. 3b). The association between both variables remained significant after adjusting for cell type count intrapair differences (β = 0.027, t = 3.924, p = 0.001). Intrapair exploration of the top 10 CpG sites (Table 1) revealed significant associations between CPR and DNA methylation at CpG probes cg00262246 (β = 0.012, p = 0.029), cg01024069 (β = − 0.01, p = 0.033), and cg12955069 (β = − 0.021, p = 0.026).
DNA methylation exploration of EP300 gene
To further explore the putative relevance of DNA methylation at other CpG sites located within the EP300 gene and its surrounding regions, DNA methylation at 27 CpG sites included in the array and annotated to this region was also explored with regard to CPR (see Table 2). All analyses were adjusted for cell sex, birthweight, gestational age at ultrasound, and cell type count. In addition to cg06793497, two additional CpG sites—cg12968540 and cg19011939—were significantly associated with CPR (p < 0.05); moreover, methylation at four additional CpG sites—cg04452260, cg24349919, cg11931284 and cg25888227—showed trend associations with CPR (p < 0.01). The intrapair approach was then applied for these newly identified six CpG sites revealing cg11931284 (β = 0.028, t = 2.985, p = 0.008) and cg19011939 (β = − 0.021, t = − 2.343, p = 0.03) to be significantly associated with CPR, when adjusting for cell types intrapair differences.
Table 2.
List of CpG sites included in the array located in the EP300 gene and its surrounding CpG island and antisense ncRNA (EP300-AS1)
| CpG probe | Genomic coordinates | beta | p value |
|---|---|---|---|
| cg00500400 | 41487283 | 0.001 | 0.67 |
| cg04452260 | 41487569 | 0.003 | 0.09 |
| cg09331127 | 41487734 | − 0.0006 | 0.79 |
| cg02046995 | 41487740 | 0.004 | 0.49 |
| cg24349919 | 41487761 | 0.005 | 0.09 |
| cg02107564 | 41488750 | − 0.005 | 0.48 |
| cg03427564 | 41489051 | 0.004 | 0.21 |
| cg03656483 | 41490340 | − 0.001 | 0.65 |
| cg00187244 | 41492007 | 0.011 | 0.23 |
| cg11931284 | 41492370 | 0.015 | 0.07 |
| cg13028324 | 41501680 | − 0.0005 | 0.89 |
| cg20730595 | 41513219 | − 0.001 | 0.85 |
| cg17439569 | 41513539 | − 0.005 | 0.16 |
| cg05997318 | 41542772 | − 0.014 | 0.12 |
| cg06793497 | 41542898 | 0.021 | 0.00008 |
| cg06329185 | 41544246 | − 0.0008 | 0.75 |
| cg07345240 | 41556691 | 0.002 | 0.73 |
| cg25299898 | 41563501 | 0.003 | 0.74 |
| cg12968540 | 41572924 | 0.009 | 0.008 |
| cg26901641 | 41573032 | − 0.006 | 0.12 |
| cg03950371 | 41573046 | − 0.003 | 0.48 |
| cg14455139 | 41573155 | − 0.0001 | 0.98 |
| cg05601844 | 41573176 | − 0.0005 | 0.89 |
| cg19011939 | 41591607 | − 0.01 | 0.01 |
| cg12917725 | 41592634 | 0.002 | 0.76 |
| cg25888227 | 41593581 | 0.004 | 0.07 |
| cg22037654 | 41593650 | 0.0001 | 0.94 |
DNA methylation at CpG sites highlighted in italics was significantly associated with CPR measured during the third trimester in an intrapair approach
Discussion
To the best of our knowledge, this is the first study analyzing the epigenetic age in association with adverse prenatal environment as measured by a hemodynamic ultrasound parameter. Firstly, we describe the significant association between CPR measured during the third trimester of pregnancy with epigenetic age acceleration. Specifically, subjects exhibiting decreased CPR—exposed to prenatal adverse conditions—were born with decelerated epigenetic age, i.e., prenatally stressed subjects were born immature adjusting for their gestational age at birth. Additionally, methylomic exploration of schizophrenia PRS genes known to be expressed in placenta revealed the association between CPR and EP300 gene CpG-specific methylation, at the cg06793497 probe, in our monochorionic twin sample.
Developmental deficits and developmental delays have been previously described in children who would later develop schizophrenia [30]; although such prodromal symptoms were in accordance with the neurodevelopmental hypothesis for schizophrenia, biological mechanisms mediating these effects remain largely unknown. Epigenetic immaturity in response to prenatal stress could be contributing to this developmental delay. Interestingly, epigenetic age deceleration has been previously described in association with maternal pathologies during pregnancy, such as maternal depression or Sjögren’s syndrome, suggesting it can be a robust biomarker of prenatal suffering [10, 11]. It is worth noting that CPR was measured a few days prior to childbirth; thus, it can be used as a surrogate marker of prenatal adaptation to adverse conditions experienced at the end of the pregnancy, i.e., as a marker of perinatal risk.
Integration of the schizophrenia PRS [31] with obstetric and placental information [22], allowed the identification of E1A binding protein p300 (EP300) gene CpG-specific methylation as a putative marker of exposure to prenatal stress. Interestingly, the EP300 gene encodes a histone acetyltransferase (HAT) involved in several cell pathways such as cell proliferation and differentiation. Mutations at EP300 gene have been described to cause Rubinstein-Taybi syndrome, a rare autosomal dominant neurodevelopmental disorder characterized by intellectual disability, psychomotor and language delay, and facial dysmorphisms [32]. Likewise, these symptoms, including developmental delay, learning problems, and cleft palate, characterize the 22q11.2 deletion syndrome, a well-defined congenital condition caused by the deletion of the 22q11.2 segment [33]. Notably, this syndrome is associated with a higher risk to develop schizophrenia, among other psychiatric conditions [34]; interestingly, EP300 gene is located on chromosome 22 at position 22q13.2.
Further exploration of differential DNA methylation in and around the EP300 gene revealed cg19011939 to be differentially methylated in association with prenatal adversity. While higher exposure to a prenatal adverse environment, as reflected by lower CPR during the third trimester, is associated with decreased methylation at cg06793497 in the hypoxia-responsive EP300 gene, there appears to be increased methylation at cg19011039 at EP300-AS1 gene. Thus, we speculate that higher exposure to prenatal stress might be associated with reciprocal patterns of EP300 and EP300-AS1 epigenetic regulation that could act synergistically, a hypothesis that may be explored in future studies [35].
Remarkably, EP300 has been identified as a co-activator of the hypoxia-inducible factor 1 alpha (HIF1A). In this regard, hypoxic conditions stimulate EP300 expression, which has a neuroprotective role [36]. Accordingly, genetic variability at EP300 gene has been associated with human adaptations to high altitude regions, e.g., the Tibet [37]. Likewise, pre- and peri-natal hypoxia have been associated with schizophrenia spectrum disorders, particularly by decreasing hippocampal volume [38, 39]; complementarily, a decreased or impaired response to hypoxia via neurotrophic factors has also been implicated in the etiology of schizophrenia [40]. Furthermore, DNA methylation at the IGF2BP1 gene, also involved in prenatal development [41], has been associated with both adult working memory and birthweight [42]; further highlighting the advantage of twin study designs to identify environmentally-driven epigenetic consequences of prenatal stress. Overall, these findings point to the existence of a GxE interaction between genetic vulnerability and exposure to prenatal hypoxia, as already highlighted by Ursini and collaborators [22]. In this framework, EP300 methylation could be one of the mediators of such interaction.
A number of limitations of the present study should be noted. First, the moderate sample size (n = 60 subjects, 30 twin pairs) limits the statistical power of the analysis; however, smaller sample sizes (n = 22 MZ twin pairs) have been described to be sufficient to identify methylation differences of 6% with > 80% power [43]. Moreover, a lenient significance threshold after correction multiple testing was used; however, previous epigenetic studies have described FDR values between 5 and 20% as markers of medium-confidence sites [44]. Another limitation regards the moderately small reported effect sizes (around 2%) questioning the biological relevance of our findings [45]; however, these findings are in agreement with a larger body of evidence regarding cord blood methylation after exposure to a number of prenatal stressors. Such small DNA methylation changes may act in conjunction with a myriad of other epigenetic signatures and biological processes in order to maintain homeostasis in the face of threats. Additionally, although epigenomic information was available from a methylomic array including more than 800,000 CpG sites distributed throughout the whole human genome, only 1400 CpG sites were analyzed; alternative approaches including the total of CpG sites included in the array would have yield different findings, probably pointing to genes involved in other neurodevelopmental disorders besides schizophrenia. Furthermore, while the set of genes analyzed in the current approach were described to be highly expressed in placental tissue [22], placentae were not available for this sample and cord blood was thus analyzed as the proxy tissue of choice with regard to exposure to prenatal adversity. Finally, MZ twin pregnancies are characterized by lower gestational ages at birth than singleton pregnancies; besides, obstetric scales commonly used in psychiatric studies include twin pregnancies as an obstetric complication. Thus, findings derived from the present design might not be generalizable to the general population.
Conclusions
Further studies are needed to test the time stability of the hereby identified methylation signature. It will be equally relevant to explore neurobehavioral correlates of EP300 methylation during early childhood along with its putative association with neurodevelopmental outcomes, including psychosis liability. Additionally, a longitudinal follow-up is required to test the role of postnatal environment in these phenotypes since both epigenetic age deceleration and CpG-specific differential methylation in association with CPR could return to basal levels after birth. Finally, genetic exploration of these subjects regarding schizophrenia PRS will be instrumental for the study of GxE interactions and genetic liability for an impaired hypoxia response during human development.
Methods
Study population
This was a prospective study including fetal pairs from monochorionic diamniotic twin pregnancies attended at Hospital Clínic de Barcelona (Spain) during a 2-year recruitment period. Monochorionic monoamniotic twin pregnancies were excluded from the present study to avoid putative confounding with regard to differential exposure to stress in both types of twin pregnancies. The study protocol was approved by the hospital ethics committee (HCB/2016/0046), and all patients provided written informed consent.
We included 32 monochorionic pregnancies (n = 64 samples). The sample was enriched for two monochorionic-specific severe obstetric complications: twin-to-twin transfusion syndrome (TTTS, n = 8) and selective intrauterine growth restriction (sIUGR, n = 9). All TTTS cases were treated upon detection by means of laser fetoscopy [46].
Maternal age and pre-pregnancy BMI were retrieved from hospital records. Gestational age was dated using first-trimester crown-rump length measurement of the larger twin [47].
Fetal ultrasound assessment
Ultrasound assessment was performed on a Voluson Expert 8 (General Electrical Medical Systems, Milwaukee, WI, USA) or a Siemens Sonoline Antares (Siemens Medical Systems, Erlangen, Germany) with 8- to 4-MHz or 6- to 4- MHz curved array probes, respectively. All fetuses underwent detailed ultrasound evaluation including fetal anatomy and Doppler measurements such as UAPI, MCAPI and ductus venosus PI. All Doppler evaluations were acquired at a normal fetal heart rate (FHR) in the absence of fetal body or respiratory movements and at an angle of insonation as close to 0° as possible (but always < 15°), and the mechanical and thermal indices were maintained below 1. CPR was calculated as the ratio between MCAPI and UAPI, according to previous studies [12].
DNA methylation
Umbilical vein cord blood samples were obtained from the clamped umbilical cord immediately after delivery of the fetus. All blood samples were collected in EDTA-treated tubes and processed within 1 h. Plasma was separated by centrifugation at 3000 rpm for 10 min at 4 °C, and stored at − 80 °C until further use. Genomic DNA was extracted from fetal cord blood using QIAamp DNA Mini Kit (Qiagen). DNA quality and quantity were assessed by NanoDrop One (Thermo Scientific). Genomic DNA was bisulfite converted using the Zymo EZ-96 DNA Methylation Kit (Zymo Research). Genome-wide DNA methylation levels were assessed over 850,000 CpG sites by means of the Infinium MethylationEPIC BeadChip Kit (Illumina Inc., CA, USA) according to the manufacturer’s protocol. Pre-processing and normalization were performed using the Bioconductor minfi package [48]. CpG probes containing common SNPs were discarded. All probes mapping to the X and Y chromosomes were also removed. Finally, cross-hybridizing probes as previously identified were excluded from further analysis [49]. All samples (n = 64) were run on the same plate.
Absence of maternal contamination was confirmed after retrieving DNA methylation values at 10 CpG sites previously described to identify sample contamination by maternal blood during sample collection [50]. None of the samples assayed exhibited DNA methylation values above the threshold at 5 or more of those CpG sites (see Additional file 1 for specific methylation values). Two samples (from the same twin-pair) were excluded from further analyses due to lack of monozygosity as assessed by 59 SNPs included in the array. One of the samples was removed from analysis due to insufficient DNA concentration, the co-twin sample was also excluded from further analysis.
Statistical analyses
All statistical analyses were conducted in R version 3.5.0 [51]. DNA methylation-based GA prediction was performed using the R code and statistical pipeline developed by Knight, based on the methylation profile of 148 CpG sites [6]; this predictor was developed using 15 Illumina DNA methylation datasets (n = 1434 neonates). Following Simpkin et al. recommendations, the Knight clock was preferred for our analysis as it was developed and tested in preterm infants datasets such as our monozygotic twin population, characterized by a mean gestational age at birth of 35.3 weeks [52]. The EPIC array lacks 6 of the CpG sites originally included in the Knight clock, these values were imputed manually as non-available. Interestingly, DNA methylation-based age estimation relying on EPIC array data has already been described to accurately predict age despite the lack of several CpG sites originally included in Horvath’s and Hannum’s clocks [53].
Gestational age acceleration (ΔGA) was calculated as the absolute difference between epigenetic GA and chronological GA. Since ΔGA was associated with chronological GA (r = − 0.47; p < 0.001), the latter was included as a covariate in all statistical models; this association has been already reported in prior studies exploring epigenetic-based GA estimations at birth [6, 10, 11].
Cell counts of CD4+ T cells, CD8+ T cells, B cells, NK cells, granulocytes, monocytes, and nucleated red blood cells (nRBCs) were estimated using the R code and statistical pipeline developed by Houseman [54].
A multiple linear regression model was built to analyze the correlation between ΔGA and CPR. Fetal sex, birthweight, diagnostic of either TTTS or sIUGR (binary variable), post-surgery interval (in TTTS cases where laser fetoscopy had been applied), and gestational age at ultrasound were included as independent variables in the model as they are known to influence either DNA methylation (from which ΔGA is calculated) or CPR. This analysis was conducted in the total MZ twin sample (n = 60).
DNA methylation at CpG sites annotated to the 43 genes of the Placental PRS1 as described by Ursini et al. [22] was retrieved to test their association with CPR. A second multiple linear regression model was then designed to explore putative effects of CPR upon methylation of PRS genes, testing 1,400 associations. The aforementioned confounding variables along with cell types proportions (CD4+ T cells, CD8+ T cells, B cells, NK cells, granulocytes, monocytes, and nRBCs) were included as covariates, as they are known to affect methylation values. False discovery rate (FDR) correction for multiple testing was applied, considering q values under 20% to be indicative of medium-confidence probes following prior studies [44].
A twin-based approach previously developed in our group [55] was also applied to refine the association between cg06793497 methylation and CPR. Briefly, intrapair differences for both variables of interest were computed for each twin pair; afterward, a regression model was fitted with an estimated intrapair cg06793497 methylation (Δmethylation) and intrapair CPR (ΔCPR). This last model was not adjusted for either sex or chronological gestational age since both variables are identical for both twins of a pair.
Additional file
DNA methylation values for CpG probes used to discard the presence of maternal contamination. (DOCX 29.6 kb)
Acknowledgements
We would like to thank Anna Valldeperas, Miriam Osorio, and Talita Micheletti for their technical assistance with the biological sample processing. We are indebted to the IDIBAPS Biobank, integrated in the Spanish National Biobank Network, for the sample and data procurement.
Funding
This work was supported by a NARSAD Distinguished Investigator Grant, awarded to Professor Lourdes Fañanás, the Biomedical Research Networking Center for Mental Health (CIBERSAM) and the Comissionat per a Universitats i Recerca del DIUE, Generalitat de Catalunya (grant number 2017SGR1577). This research was also partially funded by CERCA Programme/Generalitat de Catalunya “la Caixa” Foundation (LCF/PR/GN14/10270005), and AGAUR 2017 SGR grant number 1531. The organizations listed above had no further role in study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the paper for publication.
Availability of data and materials
The datasets used and/or analyzed during the current study together with the associated relevant meta-data are available from the corresponding author upon reasonable request.
Authors’ contributions
HPG contributed to the statistical analyses, writing/editing the manuscript, and prepared the figures. EE and FC contributed to the acquisition of the data, and writing/editing the manuscript. SM performed the methylation analysis and contributed to the statistical analysis. AZ contributed to the interpretation of the results and editing the manuscript. LF contributed to the conception and design of the work, acquisition of data, and writing/editing the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all participants after a detailed description of the study aims and design, as approved by the Bioethics Committee of the University of Barcelona. All procedures contributing to this work were performed in accordance with the Helsinki Declaration of 1975, as revised in 2008.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Graignic-Philippe R, Dayan J, Chokron S, Jacquet AY, Tordjman S. Effects of prenatal stress on fetal and child development: A critical literature review. Neurosci Biobehav Rev. 2014;43:137–162. doi: 10.1016/j.neubiorev.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 2.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014;5:5592. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palma-Gudiel H, Cirera F, Crispi F, Eixarch E, Fañanás L. The impact of prenatal insults on the human placental epigenome: A systematic review. Neurotoxicol Teratol Pergamon. 2018;66:80–93. doi: 10.1016/j.ntt.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda SV, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight AK, Craig JM, Theda C, Bækvad-Hansen M, Bybjerg-Grauholm J, Hansen CS, et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 2016;17:1–11. doi: 10.1186/s13059-016-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: Relevance of glucocorticoid signaling. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B, Marioni RE, Bressler J, Fornage M, Studenski S, Vandiver AR, et al. DNA methylation-based measures of biological age : meta-analysis predicting time to death. Aging (Albany NY). 2016;8:1–22. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Declerck K, Vanden Berghe W. Back to the future: epigenetic clock plasticity towards healthy aging. Mech Ageing Dev. 2018;174:18–29. doi: 10.1016/j.mad.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Girchenko P, Lahti J, Czamara D, Knight AK, Jones MJ, Suarez A, et al. Associations between maternal risk factors of adverse pregnancy and birth outcomes and the offspring epigenetic clock of gestational age at birth. Clinical Epigenetics. 2017;9:1–14. doi: 10.1186/s13148-017-0349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suarez A, Lahti J, Czamara D, Lahti-Pulkkinen M, Knight AK, Girchenko P, et al. The epigenetic clock at birth: associations with maternal antenatal depression and child psychiatric problems. J Am Acad Child Adolesc Psychiatry. 2018;57:321–328.e2. doi: 10.1016/j.jaac.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeVore GR. The importance of the cerebroplacental ratio in the evaluation of fetal well-being in SGA and AGA fetuses. Am J Obstet Gynecol. 2015;213:5–15. doi: 10.1016/j.ajog.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Morales-Roselló J, Khalil A, Morlando M, Papageorghiou A, Bhide A, Thilaganathan B. Changes in fetal Doppler indices as a marker of failure to reach growth potential at term. Ultrasound Obstet Gynecol. 2014;43:303–310. doi: 10.1002/uog.13319. [DOI] [PubMed] [Google Scholar]
- 14.Bahado-Singh RO, Kovanci E, Jeffres A, Oz U, Deren O, Copel J, et al. The Doppler cerebroplacental ratio and perinatal outcome in intrauterine growth restriction. Am J Obstet Gynecol. 1999;180:750–756. doi: 10.1016/S0002-9378(99)70283-8. [DOI] [PubMed] [Google Scholar]
- 15.Peeters LLH, Sheldon RE, Jones MD, Jr, Makowski EL, Meschia G. Blood flow to fetal organs as a function of arterial oxygen content. Am J Obstet Gynecol. 1979;135:637–646. doi: 10.1016/S0002-9378(16)32989-1. [DOI] [PubMed] [Google Scholar]
- 16.Mone F, Thompson A, Stewart MC, Ong S, Shields MD. Fetal umbilical artery Doppler pulsatility index as a predictor of cardiovascular risk factors in children—a long-term follow up study. J Matern Neonatal Med. 2014;27:1633–1636. doi: 10.3109/14767058.2013.871698. [DOI] [PubMed] [Google Scholar]
- 17.Mone Fionnuala, McConnell Barbara, Thompson Andrew, Segurado Ricardo, Hepper Peter, Stewart Moira C, Dornan James C, Ong Stephen, McAuliffe Fionnuala M, Shields Michael D. Fetal umbilical artery Doppler pulsatility index and childhood neurocognitive outcome at 12 years. BMJ Open. 2016;6(6):e008916. doi: 10.1136/bmjopen-2015-008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottesman II, Shields J. A critical review of recent adoption, twin, and family studies of schizophrenia: behavioral genetics perspectives. Schizophr Bull. 1976;2:360–401. doi: 10.1093/schbul/2.3.360. [DOI] [PubMed] [Google Scholar]
- 19.Cannon M, Ph D, Jones PB, Ph D, Murray RM, Sc D, et al. Obstetric complications and schizophrenia : historical and meta-analytic review. Am J Psychiatry. 2002;159:1080–1092. doi: 10.1176/appi.ajp.159.7.1080. [DOI] [PubMed] [Google Scholar]
- 20.Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, et al. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68:314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belbasis L, Köhler CA, Stefanis N, Stubbs B, Os J, Vieta E, et al. Risk factors and peripheral biomarkers for schizophrenia spectrum disorders: an umbrella review of meta-analyses. Acta Psychiatr Scand. 2017;137:88–97. doi: 10.1111/acps.12847. [DOI] [PubMed] [Google Scholar]
- 22.Ursini Gianluca, Punzi Giovanna, Chen Qiang, Marenco Stefano, Robinson Joshua F., Porcelli Annamaria, Hamilton Emily G., Mitjans Marina, Maddalena Giancarlo, Begemann Martin, Seidel Jan, Yanamori Hidenaga, Jaffe Andrew E., Berman Karen F., Egan Michael F., Straub Richard E., Colantuoni Carlo, Blasi Giuseppe, Hashimoto Ryota, Rujescu Dan, Ehrenreich Hannelore, Bertolino Alessandro, Weinberger Daniel R. Convergence of placenta biology and genetic risk for schizophrenia. Nature Medicine. 2018;24(6):792–801. doi: 10.1038/s41591-018-0021-y. [DOI] [PubMed] [Google Scholar]
- 23.Bracha HS, Torrey EF, Gottesman II, Bigelow LB, Cunniff C. Second-trimester markers of fetal size in schizophrenia: a study of monozygotic twins. Am J Psychiatry. 1992;149:1355–1361. doi: 10.1176/ajp.149.10.1355. [DOI] [PubMed] [Google Scholar]
- 24.Torrey EF, Taylor EH, Bracha HS, Bowler AE, McNeil TF, Rawlings RR, et al. Prenatal origin of schizophrenia in a subgroup of discordant monozygotic twins. Schizophr. Bull. US: National Institute of Mental Health; 1994. p. 423–432. [DOI] [PubMed]
- 25.Rosa A, Fañanas L, Bracha HS, Torrey EF, van Os J. Congenital dermatoglyphic malformations and psychosis: a twin study. Am J Psychiatry. 2000;157:1511–1513. doi: 10.1176/appi.ajp.157.9.1511. [DOI] [PubMed] [Google Scholar]
- 26.Fañanas L, van Os J, Hoyos C, McGrath J, Mellor CS, Murray R. Dermatoglyphic a-b ridge count as a possible marker for developmental disturbance in schizophrenia: replication in two samples. Schizophr Res. 1996;20:307–314. doi: 10.1016/0920-9964(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 27.Valsky DV, Eixarch E, Martinez JM, Crispi F, Gratacós E. Selective intrauterine growth restriction in monochorionic twins: pathophysiology, diagnostic approach and management dilemmas. Semin Fetal Neonatal Med. 2010;15:342–348. doi: 10.1016/j.siny.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Sebire NJ, Snijders RJM, Hughes K, Sepulveda W, Nicolaides KH. The hidden mortality of monochorionic twin pregnancies. BJOG An Int J Obstet Gynaecol. 1997;104:1203–1207. doi: 10.1111/j.1471-0528.1997.tb10948.x. [DOI] [PubMed] [Google Scholar]
- 29.Chalouhi GE, Stirnemann JJ, Salomon LJ, Essaoui M, Quibel T, Ville Y. Specific complications of monochorionic twin pregnancies: twin-twin transfusion syndrome and twin reversed arterial perfusion sequence. Semin Fetal Neonatal Med. 2010;15:349–356. doi: 10.1016/j.siny.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Reichenberg A, Caspi A, Harrington H, Houts R, Keefe RSE, Murray RM, et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am J Psychiatry. 2010;167:160–169. doi: 10.1176/appi.ajp.2009.09040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López M, García-Oguiza A, Armstrong J, García-Cobaleda I, García-Miñaur S, Santos-Simarro F, et al. Rubinstein-Taybi 2 associated to novel EP300 mutations: deepening the clinical and genetic spectrum. BMC Med Genet. 2018;19:36. doi: 10.1186/s12881-018-0548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JAS, et al. 22q11.2 deletion syndrome. Nat Rev Dis Prim. 2015;1:15071. doi: 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Lily, Boot Erik, Bassett Anne S. Update on the 22q11.2 deletion syndrome and its relevance to schizophrenia. Current Opinion in Psychiatry. 2017;30(3):191–196. doi: 10.1097/YCO.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 35.Cui I, Cui H. Antisense RNAs and epigenetic regulation. Epigenomics [Internet]. Future Medicine. 2010;2:139–150. doi: 10.2217/epi.09.46. [DOI] [PubMed] [Google Scholar]
- 36.Tan XL, Zhai Y, Gao WX, Fan YM, Liu FY, Huang QY, et al. p300 expression is induced by oxygen deficiency and protects neuron cells from damage. Brain Res. 2009;1254:1–9. doi: 10.1016/j.brainres.2008.11.092. [DOI] [PubMed] [Google Scholar]
- 37.Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, et al. Genetic Variations in Tibetan Populations and High-Altitude Adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 38.van Erp TGM, Saleh PA, Rosso IM, Huttunen M, Lönnqvist J, Pirkola T, et al. Contributions of genetic risk and fetal hypoxia to hippocampal volume in patients with schizophrenia or schizoaffective disorder, their unaffected siblings, and healthy unrelated volunteers. Am J Psychiatry. 2002;159:1514–1520. doi: 10.1176/appi.ajp.159.9.1514. [DOI] [PubMed] [Google Scholar]
- 39.Cannon TD, van Erp TGM, Rosso IM, Huttunen M, Lönnqvist J, Pirkola T, et al. Fetal hypoxia and structural brain abnormalities in schizophrenic patients, their siblings, and controls. Arch Gen Psychiatry. 2002;59:35–41. doi: 10.1001/archpsyc.59.1.35. [DOI] [PubMed] [Google Scholar]
- 40.Cannon TD, Yolken R, Buka S, Torrey EF. Decreased neurotrophic response to birth hypoxia in the etiology of schizophrenia. Biol Psychiatry. 2008;64:797–802. doi: 10.1016/j.biopsych.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parets SE, Conneely KN, Kilaru V, Fortunato SJ, Syed TA, Saade G, et al. Fetal DNA methylation associates with early spontaneous preterm birth and gestational age. PLoS One. 2013;8:e67489. doi: 10.1371/journal.pone.0067489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Córdova-Palomera A, Alemany S, Fatjó-Vilas M, Goldberg X, Leza JC, González-Pinto A, et al. Birth weight, working memory and epigenetic signatures in IGF2 and related genes: a MZ twin study. PLoS One. 2014;9:e103639. doi: 10.1371/journal.pone.0103639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Essex MJ, Thomas Boyce W, Hertzman C, Lam LL, Armstrong JM, Neumann SMA, et al. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Dev. 2013;84:58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leenen FAD, Muller CP, Turner JD. DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenetics [Internet]. 2016;8:92. doi: 10.1186/s13148-016-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sago H, Ishii K, Sugibayashi R, Ozawa K, Sumie M, Wada S. Fetoscopic laser photocoagulation for twin–twin transfusion syndrome. J Obstet Gynaecol Res. 2018;44:831–839. doi: 10.1111/jog.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson HP. Sonar measurement of fetal crown-rump length as means of assessing maturity in first trimester of pregnancy. Br Med J. 1973;4:28 LP–28 31. doi: 10.1136/bmj.4.5883.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data. 2016;9:22–24. doi: 10.1016/j.gdata.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morin AM, Gatev E, McEwen LM, MacIsaac JL, Lin DTS, Koen N, et al. Maternal blood contamination of collected cord blood can be identified using DNA methylation at three CpGs. Clin Epigenetics. 2017;9:75. doi: 10.1186/s13148-017-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Development Core Team. R: A Language and environment for statistical computing [Internet]. Team RDC, editor. R Found. Stat. Comput. R Foundation for Statistical Computing; 2011. p. 409. Available from: http://www.r-project.org
- 52.Simpkin AJ, Suderman M, Howe LD. Epigenetic clocks for gestational age: Statistical and study design considerations. Clin Epigenetics. 2017;9:1–2. doi: 10.1186/s13148-017-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mcewen LM, Jones MJ, Tse D, Lin S, Edgar RD, Husquin LT, et al. Systematic evaluation of DNA methylation age estimation with common preprocessing methods and the Infinium MethylationEPIC BeadChip array. Clin Epigenetics. 2018;10:123. doi: 10.1186/s13148-018-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houseman E, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Córdova-Palomera A, Fatjó-Vilas M, Palma-Gudiel H, Blasco-Fontecilla H, Kebir O, Fañanás L. Further evidence of DEPDC7 DNA hypomethylation in depression: a study in adult twins. Eur Psychiatry. 2015;30:715–718. doi: 10.1016/j.eurpsy.2015.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA methylation values for CpG probes used to discard the presence of maternal contamination. (DOCX 29.6 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study together with the associated relevant meta-data are available from the corresponding author upon reasonable request.