Abstract
Background
Interferon gamma (IFNγ) plays an important role in the development of chronic lung diseases via the production of inflammatory mediators, although the exact mechanism remains unclear. The present study aimed at investigating the potential mechanisms by which IFNγ induced over-production of interleukins through the interaction between carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) pathway.
Methods
IFN-γ induced over-production of interleukin (IL) 6 and IL8, and RNA expression of CEACAM1 and its subtypes or PI3K and its subtypes in human bronchial epithelial cells (HBE). The production of IL6 and IL8 or cell proliferation and movement were also evaluated in cellCEACAM1− or cellCEACAM1+ after the induction of IFN-γ. Roles of PI3K subtype proteins, e.g. PI3Kp110α/δ, Akt, p110α/γ/δ/β/mTOR, PI3Kp110α/δ/β, PI3Kp110δ, or pan-PI3K in IFN-γ-induced CEACAM1 subtype alterations were furthermore validated using those proteins of PI3K subtypes.
Results
CEACAM1, especially CEACAM1-S isoforms, was significantly up-regulated in HBE cells after treatment with IFN-γ. CEACAM1 played roles in expression of IL-6 and IL-8, and facilitated cellular proliferation and migration. IFN-γ up-regulated the expression of CEACAM1 in airway epithelial cells, especially CEACAM1-S isoforms, promoting cellular proliferation, migration, and the production of inflammatory factors. PI3K (p110δ)/Akt/mTOR pathway was involved in the process of IFN-γ-upregulated CEACAM1, especially CEACAM1-S. On the other hand, CEACAM1 could promote the activation of PI3K/Akt/mTOR pathway.
Conclusion
IFN-γ could induce inflammatory responses, cellular growth and proliferation through the interaction of CEACAM1 (especially CEACAM1-S isoforms) and PI3K(p110δ)/Akt/mTOR in airway epithelial cells, which might be new alternative of future therapies against epithelial transition from inflammation to cancer.
Electronic supplementary material
The online version of this article (10.1186/s12967-019-1894-3) contains supplementary material, which is available to authorized users.
Keywords: COPD, Lung cancer, CEACAM1, PI3K/Akt pathway, IFN-γ
Background
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) belongs to immunoglobulin superfamily, presents in various cells, and participates in cellular proliferation, apoptosis, angiogenesis, tumor metastasis, and pathogen recognition. CEACAM1 mediates transcellular transcytosis, suppresses immune cell activities and inflammatory responses, and plays a critical role in the development of lung cancer and acute exacerbation of chronic obstructive pulmonary diseases (AECOPD). We emphasized the importance to understand the interaction and network of driver molecules and develop new biomarkers and therapeutics during the transformation from chronic lung inflammation to cancer [1]. There is a clear correlation between COPD and lung cancer, although molecular mechanisms of COPD-lung cancer transits remain unclear. The chronic inflammation in COPD might be associated with increased DNA errors during tissue remodeling after injury, while acute exacerbations of COPD might worsen the condition, and finally resulting in amplification of carcinogenic effects [2–7]. CEACAM1 can be one of potential target candidates to play a critical role in the COPD-lung cancer transits. The human CEACAM1 gene has 12 different isoforms via different splicing, of which isoforms (4L, 3L, 4S, 3S, 3, 4C1, 3C2) are involved in immune regulation or inflammation [8]. Lipopolysaccharide (LPS) and interferon gamma (IFN-γ) as key intermediates contribute to the initiation of inflammation probably through signal pathways of PI3K and NF-κB [9, 10]. PI3K activation plays a central role in the development of airway inflammation and tissue remodeling [11]. CEACAM1 was also a key factor in inflammation, fibrosis and tissue remodeling [12, 13]. PI3K inhibitor could induce apoptosis in high CEACAM1 expressed cells, rather than CEACAM1 knockdown cells [14]. These studies suggested that the regulations of PI3K signaling on inflammation, tissue remodeling and apoptosis might more dependent on CEACAM1.
We previously evaluated the dynamic gene expression profiling of peripheral blood mononuclear cells (PBMCs) from patients with AECOPD on day 1, 3 and 10 after hospitalization, and selected up-regulated CEACAM1 as one of AECOPD-specific biomarkers [15]. The role of CEACAM1 on regulation of adhesion and function of B cells is in a PI3K-dependent manner [16–18]. IFN-γ was found to induce the malignant transformation of healthy lactating bovine mammary epithelial cells, closely associated with the promotion of cell growth and even the carcinogenesis of breast cancer [19]. The present study aims at evaluating the expression of CEACAM1 in lung diseases and the profiling of CEACAM1 isoforms and PI3K subtypes after IFN-γ treatment, and investigating the role of PI3K pathway in IFN-γ-induced CEACAM1 expression in airway epithelial cells. In order to determine the role of CEACAM1 in alternations of IFN-γ-induced cell function, gene expression of inflammatory factors and proliferation and migration of cellCEACAM1− were furthermore determined.
Materials and methods
Cell lines and reagents
Human bronchial epithelial cell line HBE cells were cultured in RPMI 1640 supplemented with penicillin (100 U/ml), streptomycin (100 mg/ml), and 10% heat inactivated fetal bovine serum (FBS). Lipopolysaccharide(LPS, Escherichia coli, 055:B5)was purchased from Sigma (Missouri, USA). Interferon-γ was purchased from R&D Systems (Minnesota, USA). PI3K/Akt/mTOR inhibitors (GDC0941, GSK690693, BEZ235, LY294002 and CAL101) were purchased from Biovision (California, USA). SHBM1009 (a new PI3K/mTOR inhibitor) was synthesized by Fudan University. CEACAM1 and GAPDH antibodies for western blot were purchased from Abcam (Hong Kong, China). SYBR Premix Ex Taq was from TaKaRa (Shiga, Japan). Lipofectamine™ 2000 Transfection Reagent was from Invitrogen (Grand Island, NY, USA).
Small interfering RNA transfection
Small interfering RNA (siRNA) transfections were performed, according to the manufacturer’s protocol, in 6-well plates or 24-well plates, using Lipofectamine™ 2000 with siRNA duplexes (Table 1) targeting CEACAM1 and a double-stranded RNA negative control (GenePharma, Shanghai, China). As for 6-well plates, 4 μl of Lipofectamine™ 2000 and 160 pmol of each siRNA were transfected; as for 24-well plates, 1 μl of Lipofectamine™ 2000 and 40 pmol of each siRNA were transfected, each group in triplicate. Cells were then treated with IFN-γ for an additional 24 h and then prepared for quantitative RT-PCR, western blot, and other measurements 24 h after transfection.
Table 1.
Sequences mentioned in the article
| Name | Sequences | |
|---|---|---|
| Forward primer | Reverse primer | |
| CEACAM1 | 5′-GCTGGCATTGTGATTGGAGTA-3′ | 5′-TTAGGTGGGTCATTGGAGTG-3′ |
| CEACAM1-4S | 5′-AAGACGATCATAGTCACTGAGCT-3′ | 5′-ATTGGAGTGGTCCTGAGCTG-3′ |
| CEACAM1-4L | 5′-AAGACGATCATAGTCACTGAGCT-3′ | 5′-GGAGACTGAGGGTTTGTGCT-3′ |
| CEACAM1-3S | 5′-TCATAGTCACTGATAATGCTCTACC-3′ | 5′-ATTGGAGTGGTCCTGAGCTG-3′ |
| CEACAM1-3L | 5′-TCATAGTCACTGATAATGCTCTACC-3′ | 5′-GGAGACTGAGGGTTTGTGCT-3′ |
| CEACAM1-4C1 | 5′-AAGACGATCATAGTCACTGAGCT-3′ | 5′-TTGCACACCATTGACAGAGT-3′ |
| CEACAM1-3 | 5′-CAGTGACCCAGTCACCTTGA-3′ | 5′-TGGACTTGTTTGTGCCTGTTG-3′ |
| CEACAM1-3C2 | 5′-CAAGACGATCATAGTCACTGAGTC-3′ | 5′-AGAGGGACATATAGGAAGGGGT-3′ |
| PIK3CA | 5′-GGGATGATTTACGGCAAGATA-3′ | 5′-CCACACAGTCACCGATTGA-3′ |
| PIK3CB | 5′-TGCGACAGATGAGTGATGAA-3′ | 5′-TCCTCCGATTACCAAGTGCT-3′ |
| PIK3CD | 5′-GCCAACATCCAACTCAACAA-3′ | 5′-CCACACAATAGCCAGCACAG-3′ |
| PIK3CG | 5′-TAGACCACCGTTTCCTCCTG-3′ | 5′-CGTGAGTTTGTCAGCATTGA-3′ |
| PIK3C2A | 5′-GATTACCTGGGCCTTCCAC-3′ | 5′-AGTGGGCATTCTTGGATTGA-3′ |
| PIK3C2B | 5′-TCCACCTTGAACTACCTCGTC-3′ | 5′-AAGTCTCCATCAGCCAGCAG-3′ |
| PIK3C2G | 5′-CTCCTGGCATCAAGTTAGCA-3′ | 5′-TCTGGAATCATCAGCACCAT-3′ |
| PIK3C3 | 5′-ATCCCGTTGCCTTTAGAACC-3′ | 5′-TGCCTCCATCTTCCGTCTTA-3′ |
| PIK3R1 | 5′-GGACGGCGAAGTAAAGCAT-3′ | 5′-TGACATTGAGGGAGTCGTTG-3′ |
| PIK3R2 | 5′-GATGGGCACTATGGCTTCTC-3′ | 5′-TGCTGGTATTTGGACACAGG-3′ |
| PIK3R3 | 5′-AGCACAACGACTCCCTCAAC-3′ | 5′-AAATGCCAGAGAACCACCTC-3′ |
| PIK3R4 | 5′-TGGCATTTGTGTCCCTTTGT-3′ | 5′-TGCTGGATGAGTTGCTGAAG-3′ |
| PIK3R5 | 5′-TACACCACACTTCCCACCAG-3′ | 5′-CTCAGCCCTCACCAGTCTCT-3′ |
| PIK3R6 | 5′-TTCCCACTTCTCCCACTGTC-3′ | 5′-TGCTGTTCCTTGCTTCCAAT-3′ |
| IL-6 | 5′-GACAGCCACTCACCTCTTCAG-3′ | 5′-CATCCATCTTTTTCAGCCATC-3′ |
| IL-8 | 5′-TTGCCAAGGAGTGCTAAAGAA-3′ | 5′-GCCCTCTTCAAAAACTTCTCC-3′ |
| MCP-1 | 5′-AGGAACCGAGAGGCTGAGA-3′ | 5′-GGAATGAAGGTGGCTGCTAT-3′ |
| TGF-β | 5′-GCCAGAGTGGTTATCTTTTGATG-3′ | 5′-AGTGTGTTATCCCTGCTGTCAC-3′ |
| VEGF | 5′-AGGGCAGAATCATCACGAAGT-3′ | 5′-AGGGTCTCGATTGGATGGCA-3′ |
| GAPDH | 5′-CCACCCATGGCAAATTCCATGGCA-3′ | 5′-TCTACACGGCAGGTCAGGTCCACC-3′ |
| siRNA-CEACAM1 | 5′-CACCUUGAAUGUCACCUAUTT-3′ | 5′-AUAGGUGACAUUCAAGGUGTT-3′ |
| siRNA-NC | 5′-UUCUCCGAACGUGUCACGUTT-3′ | 5′-ACGUGACACGUUCGGAGAATT-3′ |
siRNA-NC is a negative control siRNA with the same nucleotide composition but which lacks significant sequence homology to CEACAM1
Measurement of gene expression
Total RNA was isolated using a guanidinium isothiocyanate/chloroform based technique (TRIZOL, Invitrogen, USA) and measured with OD 260 nm. RNA was subsequently reversed and transcribed to cDNA by using PrimeScript RT reagent Kit (TaKaRa, China). Real-time PCR was carried out using a CFX96 Touch Real-time PCR instrument (Bio-Rad, USA) with the two-stage program parameters, as follows: 1 min at 95 °C, and then 40 cycles of 5 s at 95 °C and 30 s at 60 °C. The sequences of the primer sets used for this analysis are listed in Table 1. Specificity of the produced amplification product was confirmed by examination of dissociation reaction plots. Each sample was tested in triplicate with quantitative RT-PCR, and each group had six wells.
Western blot
Protein samples were mixed with one-fourth volume of SDS sample buffer, boiled for 5 min, and then separated through 10% SDS-PAGE gels. After electrophoresis, proteins were transferred to PVDF membranes through electrophoretic transfer. Membranes were blocked in 5% bovine serum albumin for 2 h, rinsed and incubated with primary antibodies directed against CEACAM1 (rabbit monoclonal, 1:5000; Abcam) or GAPDH (mouse monoclonal, 1:1000; Beyotime) at 4 °C overnight, then washed in TBS-tween thrice and treated with secondary HRP-conjugated anti-rabbit (1:1000) or anti-mouse (1:1000) for 2 h at room temperature. Secondary antibody-bound protein was detected using the ECL kit (Beyotime). Data were analyzed using the Image J software.
Wound-healing assay
HBE cells transfected with siRNA-CEACAM1 or siRNA-NC for 24 h were then treated with IFN-γ in 24-well plates. After another 24 h, a sterilized pin was used to wipe off the adherent cells on two intersecting lines, to create the wounds in each well. The migration of HBE was assessed using an inverted light microscope at the original magnification 40× and migrated distances of HBE were measured at 0, 12, 24, 48 or 72 h.
Cell proliferation assay
The effect of CEACAM1 knockdown on HBE cell proliferation was detected by Cell counting kit-8 (CCK-8, Beyotime, Shanghai, China). HBE cells transfected with siRNA-CEACAM1 or siRNA-NC were plated to 96-well plates. After IFN-γ treatment for 0, 24, 48, 72 h, CCK-8 was added (10 μl) to each well, and incubated in 37 °C. The absorbance at 450 nm was measured 2 h later. Each group has six wells.
Statistical analysis
Data was presented as mean ± standard errors. Statistical analysis was performed using SPSS software (SPSS 20.0; SPSS Inc; Chicago, IL, USA). Data were evaluated using ANOVA with LSD test for multiple comparisons and Students t test between two groups. Increased rates of total cell number and differentiation were calculated as the following: Rate (%) = (value at each time point-value of primary seeding cells)/value of primary seeding cells × 100. P-values less than 0.05 were considered to be statistically significant.
Results
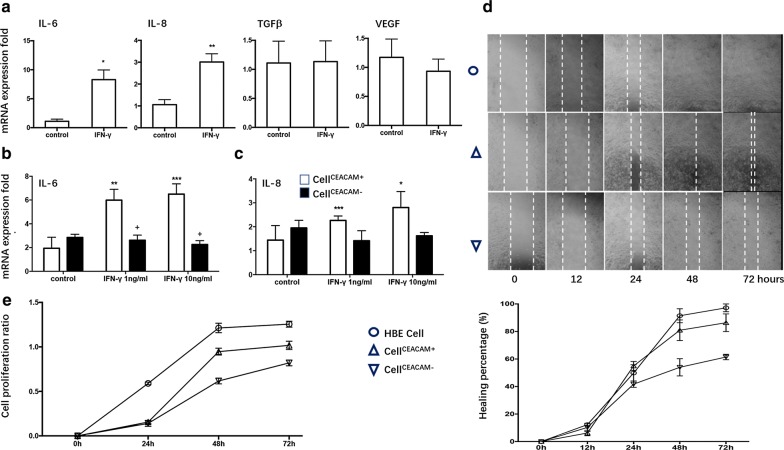
We evaluated mRNA expression of CEACAM1 in HBE cells stimulated by LPS or IFN-γ, respectively, and found no significant difference of CEACAM1 expression 24 h after LPS stimulation at the concentration of 0.1 μg/ml or 1 μg/ml (Additional file 1: Figure S1), while mRNA (Fig. 1a) and protein (Fig. 1g) expression of CEACAM1 significantly up-regulated after IFN-γ stimulation at 1 or 10 ng/ml in a concentration-dependent pattern. CEACAM1 mRNA expression significantly increased from 3 h and on after IFN-γ stimulation at 10 ng/ml and reached the peak at 24 h (Fig. 1h). Of CEACAM-1 subtypes, CEACAM1-4S and -3S isoforms are dominant in HBE cells, and mRNA of CEACAM1-4S, 4L, 3S, and 3 isoforms increased significantly after IFN-γ stimulation (Fig. 1b–f). Of interleukin (IL)-6, IL-8, transforming growth factor-β (TGF-β), vascular endothelial growth factor (VEGF), and monocyte chemoattractant protein-1 (MCP-1), we found that IFN-γ stimulation increased expression of IL-6 and IL-8 in HBE cells (Fig. 2a), while not in cellCEACAM1− (Fig. 2b, c). Delayed cell migration (Fig. 2d) and decreased cell proliferation (Fig. 2e) were noticed in cellCEACAM1−, as compared with HBE cells or cellCEACAM1+, respectively.
Fig. 1.
Expression of CEACAM1 after IFN-γ stimulation in HBE cells. mRNA expression of CEACAM1 (a), CEACAM1-4L (b), CEACAM1-4S (c), CEACAM1-3L (d), CEACAM1-3S (e), CEACAM1-3 (f), as well as protein levels of CEACAM1 protein (g) were measured in HBE cells treated with vehicle (control) or with IFN-γ at concentration of 0, 1, 10 ng/ml for 24 h. Dynamic expression of CEACAM1 gene in HBE cells 0, 3, 6, 12, 24, 48, 72 h after treatment with vehicle (control) or with IFN-γ at 10 ng/ml (h). *, **, ***p-values less than 0.05, 0.01 and 0.005, as compared to control
Fig. 2.
Role of CEACAM1 in HBE cells with IFN-γ treatment. mRNA expression of IL-6, IL-8, TGFβ, and VEGF in HBE cells treated with vehicle (control) or with IFN-γ at 10 ng/ml for 24 h (a). mRNA expression of IL6 (b) and IL8 (c) in cellCEACAM1− or cellCEACAM1+ treated with vehicle (control) or with IFN-γ at 1.0 or 10 ng/ml. Cell movement measured by wound healing assay (d) and cell proliferation measured by CCK8 0, 12, 24, 48, or 72 h after treatment with vehicle (HBE cell) and treated with IFN-γ. Healing percentage (%) = (S0 − St)/S0 × 100% (S0: wound area at 0 h; St: wound area at specific time point). Cell proliferation rate = (ODt-OD0)/OD0 (OD0: OD value at 450 nm at 0 h after treated with IFN-γ; ODt: OD value at 450 nm at t hours after treated with IFN-γ). *, **, ***p-values less than 0.05, 0.01 and 0.005, as compared to control
To define roles of CEACAM1 in PI3Ks, we evaluated the mRNA expression profiling of ten PI3K subtypes between HBE cellCEACAM1+ and cellCEACAM1−, and found the expression of PIK3CA, PIK3CB, PIK3C2A, PIK3C3, PIK3R3, and PIK3R4 up-regulated in cellCEACAM1−, as compared to those in cellCEACAM1+ (Fig. 3). The low concentration (1 ng/ml) of IFNγ increased the expression of PIK3CB, PIK3C2B, PIK3R1, and PIK3R2, but not the high concentration (10 ng/ml). This indicates a clear correlation of biological functions between CEACAM1 and PI3K subunits and the regulatory role of CEACAM1 in expression of PI3K subunits, of which some were regulated by the certain concentration of IFNγ. However, all these results were without statistical significance.
Fig. 3.
Effect of CEACAM1 on PI3K subtype gene expression. mRNA expression of 10 PI3K subunits (PIK3CA, PIK3CB, PIK3CD, PIK3C2A, PIK3C2B, PIK3C3, PIK3R1, PIK3R2, PIK3R3, PIK3R4) in cellCEACAM1− or cellCEACAM1+ treated with vehicle (control) or with IFN-γ at 1.0 or 10 ng/ml. mRNA levels of PIK3CG, PIK3C2G, PIK3R5, PIK3R6 expression could not be detected
HBE cells were pretreated with different PI3K inhibitors, including GDC0941, GSK690693, BEZ235, LY294002, CAL101 and SHBM1009, and then stimulated by IFN-γ for 24 h. Of those inhibitors, GDC0941 (Fig. 4a), BEZ235 (Fig. 4b), GSK 690693 (Fig. 4c), LY294002 (Fig. 4d) and CAL 101 (Fig. 4e) showed inhibitory effects on CEACAM1-4L, -4S, -3L, 3S mRNA levels, as compared with cells stimulated with IFN-γ and treated with vehicle, while levels of those mRNA expression were still higher than in cells stimulated and treated with vehicle (p < 0.05 or less, respectively). BEZ235 showed most strong inhibitory effect on both –S and –L isoforms. Levels of CEACAM1-4L and -3L were higher in cells treated with SHBM1009, while CEACAM-4S and -3S were lower in cells with SHBM1009 at high dose (Fig. 4f, p < 0.05 or less, respectively). Levels of CEACAM1 protein increased significantly 24 h after IFNγ stimulation (p < 0.01, vs control, Fig. 4g), which was down-regulated by treatment with GDC0941, GSK690693, BEZ235, LY294002, CAL101, or SHBM1009 (p < 0.01, vs IFNγ with vehicle, respectively). Levels of CEACAM1 protein in cells stimulated with IFNγ and treated with various PI3K inhibitors were still significantly higher than in those stimulated and treated with vehicle (p < 0.01, respectively, Fig. 5a).
Fig. 4.
Effect of PI3K/Akt pathway inhibitors on IFN-γ induced CEACAM1 and its isoform expression. mRNA expression of CEACAM1-4L, -4S, -3L, and -3L in HBE cells treated with p110α/δ inhibitor (GDC0941) at 0.05 or 0.5 ng/ml (a), Akt inhibitor (GSK690693) at 0.1 or 1.0 ng/ml (b), p110α/γ/δ/β/mTOR inhibitor (BEZ235) at 0.2 or 2.0 ng/ml (c), p110α/δ/β inhibitor (LY294002) at 2.0 or 20 ng/ml (d), p110δ inhibitor (CAL101) at 0.5 or 5.0 ng/ml (E), pan-PI3K inhibitor (SHBM1009) at 0.1 or 1.0 ng/ml (F) 24 h after stimulation and treatment with vehicle (Vehicle + Vehicle), IFN-γ with vehicle, IFN-γ with various inhibitors. *, **, ***p-values less than 0.05, 0.01 and 0.005, as compared to HBE control; +, ++, +++p-values less than 0.05, 0.01 and 0.005, as compared to IFN-γ with vehicle, respectively
Fig. 5.
Interaction between CEACAM1 and PI3K. a Protein levels of CEACAM1 in HBE cells pretreated with GDC0941 (GDC), BEZ235 (BEZ), GSK690693 (GSK), LY294002 (LY), CAL101 (CAL), and SHBM1009 (SHBM) 24 h after stimulation with IFN-γ for 24 h, as compared with control group with vehicle (CON). *, **, ***p-values less than 0.05, 0.01 and 0.005, as compared to HBE control, or +, ++, +++p-values less than 0.05, 0.01 and 0.005, as compared to IFN-γ with vehicle, respectively. b Levels of phosphorylated Akt protein (p-AKT) in cellCEACAM1− or cellCEACAM1+ treated with vehicle (control) or with IFN-γ at 10 ng/ml for 0 and 10 min. *p-values less than 0.05, as compared to control, +p-values less than 0.05, as compared to 0 min group, respectively
Figure 5b demonstrated that levels of phosphatized Atk were lower in cellCEACAM−, and decreased even more significantly 10 min after the culture (p < 0.05, vs cellCEACAM− at 0 min or cellCEACAM+, respectively).
Discussion
CEACAM1 is a member of CECAM family in various cells derived from normal tissues or malignant tumors. CEACAM1 primary transcript generates 12 isoforms in human through alternative splicing, which are mainly allocated to long (L) and short (S) tails of intracellular domains. L tails contain two immunoreceptor tyrosine-based inhibitory motifs (ITIMs), coordinating inhibitory signaling, which S tails lack. CEACAM1-L mediates negative signaling, rather than CEACAM1-S [20]. CEACAM1 was involved in the processes of inflammation or tumor genesis, during which levels of CEACAM1 isoforms, or CEACAM1-S/CEACAM1-L changed in the pathological conditions. The present study found that the expression level of CEACAM1 was upregulated in AECOPD patients, and CEACAM1 could increase production of IL-6 and IL-8, which induced inflammation in the bronchial epithelial cells. CEACAM1 could regulate LPS driven production of IL-6 and IL-1β, mediating the appearance of clinical phenomes, e.g. fever response, through the ITIM receptor [21].
CEACAM1 as an important factor in carcinogenesis has dual roles of different isoforms [22]. For example, CEACAM-S isoform and CEACAM1-S/CEACAM1-L ratio were upregulated in non-small-cell lung cancer tissues [23], while CEACAM1-L isoforms in melanoma were upregulated [24]. Although the function of CEACAM1-L and CEACAM1-S in carcinogenesis seems varied among diseases, there is no doubt that CEACAM1 and its isoforms ratio is pivotal in carcinogenesis. The association between lung cancer and COPD has been emphasized for a while, even though the mechanism remains unclear [25]. The chronic inflammation, genetic susceptibility, DNA damage and repair are critical factors responsible for the occurrence of carcinogenesis [26–29]. Our data demonstrated that CEACAM1 was closely related to the severity of inflammation, cellular repair and migration which are critical in carcinogenesis. It indicates that CEACAM1 may be a key factor linking COPD and lung cancer, and the regulation of CEACAM1-S and -L isoforms may be a therapeutic target for COPD-lung cancer transit.
IFN-γ as a cytokine with a crucial immunological function against viral infections and mycobacteria, has also been suggested for treatment of malignant tumors [30], while recent study demonstrated that IFN-γ could promote cell malignant growth and even carcinogenesis of cancer after long-term application of IFN-γ [19], although the exact mechanism remains unclear. IFN-γ up regulates CEACAM1 expression in various epithelial cells, including human normal bronchial epithelial cells [31, 32]. The present study demonstrated that IFN-γ could stimulate airway epithelial cell proliferation and induce up-regulation of CEACAM1 expression, mainly CEACAM1-S isoforms.
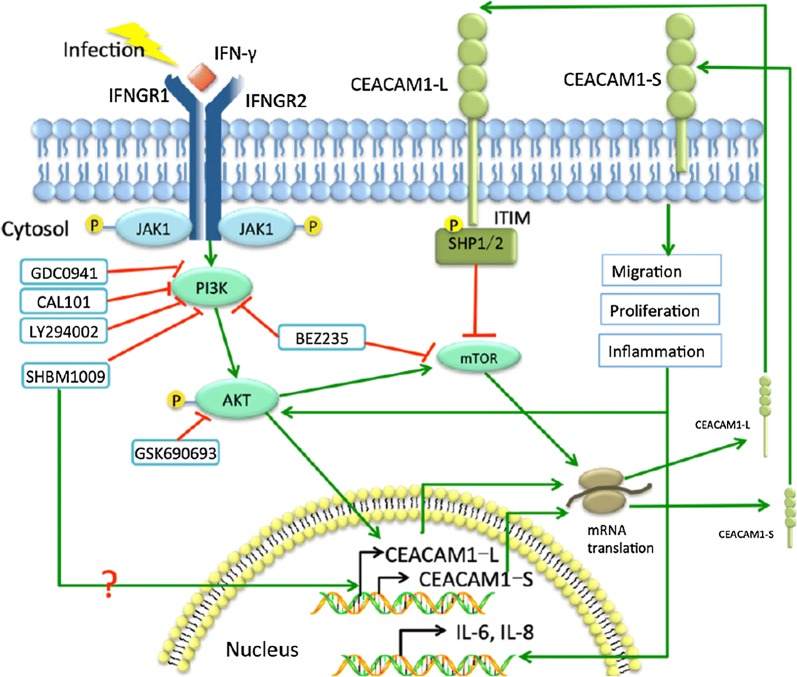
PI3K plays an important role in cell growth, proliferation, differentiation, apoptosis, cytoskeleton, and intracellular trafficking through the function of four classes (Class I, II, III and IV) [33, 34]. In the present study, we chose 6 different PI3K inhibitors targeting various subunits of PI3K and investigated effects of those PI3K inhibitors on the signaling of downstream molecules (e.g. PI3Kp110α/δ, Akt, p110α/γ/δ/β/mTOR, PI3Kp110α/δ/β, PI3Kp110δ, or pan-PI3K), in order to understand the function of specific PI3K subunits on CEACAM1 and its isoforms’ expression. The present study showed that different kinds of PI3K pathway inhibitors could suppress the expression of CEACAM1 to various degrees, especially the CEACAM1-S isoforms. Our data indicate that Akt, PI3K/mTOR, and p110δ subunit are more involved in the regulation of CEACAM1-S and CEACAM1-L isoform functions, while p110α/δ and p110α/δ/β were much weaker. Comparing p110α/δ and p110α/δ/β, the p110δ subunit may play a more important role in IFN-γ-upregulated CEACAM1 expression. PI3K(p110δ)/Akt/mTOR may contribute more to the regulation of signaling pathway between IFN-γ and CEACAM1-S or CEACAM1-L isoforms in airway epithelial cells, as explained in Fig. 6. Our data firstly described that SHBM1009 up-regulated function of CEACAM1-L, while down-regulated CEACAM1-S, on basis of previous study [35]. However, the detailed target of SHBM1009 is unclear and the mechanism by which SHBM1009 regulates CEACAM1 isoforms needs to be further investigated. On the other hand, we also noticed that CEACAM1 also plays critical roles in IFN-γ-induced alterations of PI3K subunit expression and functions. The present study firstly explores the existence of intercommunication between CEACAM1 and PI3K at isoform/subunit levels, although such trade-off needs to be defined and validated in more precise targeting ways and in the in vivo system.
Fig. 6.
Potential mechanism of CEACAM1 and PI3K interactions. In the infectious environment during acute exacerbation of COPD, the increased IFN-γ up-regulated the expression of CEACAM1, especially the -S isoforms, which further promotes cellular migration, proliferation and induces the production of inflammatory factors such as IL-6 and IL-8. The -L isoforms of CEACAM1 contains two ITIMs in the cytoplasmic tail, which negatively regulates down-stream signal transduction pathways. PI3K/Akt pathway inhibitors (GDC0941, CAL101, LY294002, SHBM1009, BEZ235 and GSK69063) inhibited CAECAM1 expression, which suggested that PI3K/Akt pathway induces the up-regulation of CEACAM1 after IFN-γ stimulation. Different from other PI3K/Akt pathway inhibitors, SHBM1009 could specifically up regulate CEACAM1-L isoforms. As a positive feedback, CEACAM1 promoted the activation of PI3K/Akt pathway
Our data evidenced that CEACAM1 plays a decisive role in inflammatory responses of epithelial cells through the production of inflammatory mediators, which is at least one of critical effects. We also found IFN-γ induced the overproduction of IL-6 and IL-8 through CEACAM1, which may take part in the pathogenesis of AECOPD or the transition from inflammation to cancer [36, 37]. We furthermore found that CEACAM1 promoted the migration and regeneration of airway epithelial cells, which promoted the malignancy in tumors. Studies on malignant tumors showed that the effect of CEACAM1 on cellular proliferation depends on the ratio of L/S isoforms, e.g. CEACAM1-S promotes proliferation while CEACAM-L has a totally opposite effect [38]. Our results suggested that CEACAM1-4S and CEACAM1-3S were more involved in IFN-γ-induced epithelial cell proliferation and migration, and might be the promoters to bridge between chronic inflammations and cancer.
Conclusion
In conclusion, IFN-γ up-regulated the expression of CEACAM1 in airway epithelial cells, especially CEACAM1-S isoforms, promoting cellular proliferation, migration, and the production of inflammatory factors. PI3K (p110δ)/Akt/mTOR pathway was involved in the process of IFN-γ-upregulated CEACAM1, especially CEACAM1-S. On the other hand, CEACAM1 could promote the activation of PI3K/Akt/mTOR pathway. Thus, we believe that the signal pathway between CEACAM1-S and PI3K (p110δ)/Akt/mTOR can be new alternative of future therapies against epithelial transition from inflammation to cancer.
Additional file
Additional file 1: Figure S1. Expression of CEACAM1 after LPS stimulation in HBE cells. CEACAM1 gene expression in HBE cells treated with LPS at concentration of 0, 0.1, 1 μg/ml for 24 h, no significant change of CEACAM1 was shown.
Authors’ contributions
Conceived and designed the study: XDW; performed the biological experiments: YCZ; statistical analysis: YCZ and DLS. Wrote the paper: YCZ and DLS. All authors read and approved the final manuscript.
Acknowledgements
The work was supported by Zhongshan Distinguished Professor Grant (XDW), The National Natural Science Foundation of China (81600027, 81500025, 81800034, 81600014, 91230204, 81270099, 81320108001, 81270131, 81300010, 81700008, 81873409), The Shanghai Committee of Science and Technology (12JC1402200, 12431900207, 11410708600, 14431905100), Operation funding of Shanghai Institute of Clinical Bioinformatics, Ministry of Education for Academic Special Science and Research Foundation for PhD Education (20130071110043), and National Key Research and Development Program (2016YFC0902400, 2017YFSF090207).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CEACAM1
carcinoembryonic antigen-related cell adhesion molecule 1
- AECOPD
acute exacerbation of chronic obstructive pulmonary disease
- IFN-γ
interferon-γ
- IL-6
interleukin 6
- IL-8
interleukin 8
- CCK-8
cell counting kit-8
- PI3K
phosphatidylinositide 3-kinase
- COPD
chronic obstructive pulmonary disease
- PBMC
peripheral blood mononuclear cell
- LPS
lipopolysaccharide
- siRNA
small interfering RNA
- TGF-β
transforming growth factor-β
- VEGF
vascular endothelial growth factor
- MCP-1
monocyte chemoattractant protein-1
References
- 1.Wang X. New biomarkers and therapeutics can be discovered during COPD-lung cancer transition. Cell Biol Toxicol. 2016;32:359–361. doi: 10.1007/s10565-016-9350-0. [DOI] [PubMed] [Google Scholar]
- 2.Shi L, Zhu B, Xu M, Wang X. Selection of AECOPD-specific immunomodulatory biomarkers by integrating genomics and proteomics with clinical informatics. Cell Biol Toxicol. 2018;34:109–123. doi: 10.1007/s10565-017-9405-x. [DOI] [PubMed] [Google Scholar]
- 3.Zhu HX, Shi L, Zhang Y, Zhu YC, Bai CX, Wang XD, Zhou JB. Myocyte enhancer factor 2D provides a cross-talk between chronic inflammation and lung cancer. J Transl Med. 2017;15:65. doi: 10.1186/s12967-017-1168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang DC, Wang W, Zhu B, Wang X. Lung cancer heterogeneity and new strategies for drug therapy. Annu Rev Pharmacol Toxicol. 2018;58:531–546. doi: 10.1146/annurev-pharmtox-010716-104523. [DOI] [PubMed] [Google Scholar]
- 5.Wang DC, Wang X. Genome dimensions control biological and toxicological functions; myth or reality? Cell Biol Toxicol. 2018;34:333–336. doi: 10.1007/s10565-018-9440-2. [DOI] [PubMed] [Google Scholar]
- 6.Lv J, Gao D, Zhang Y, Wu D, Shen L, Wang X. Heterogeneity of lipidomic profiles among lung cancer subtypes of patients. J Cell Mol Med. 2018;22:5155–5159. doi: 10.1111/jcmm.13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunnumakkara AB, Sailo BL, Banik K, Harsha C, Prasad S, Gupta SC, Bharti AC, Aggarwal BB. Chronic diseases, inflammation, and spices: how are they linked? J Transl Med. 2018;16:14. doi: 10.1186/s12967-018-1381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 9.Shi L, Dong N, Ji D, Huang X, Ying Z, Wang X, Chen C. Lipopolysaccharide-induced CCN1 production enhances interleukin-6 secretion in bronchial epithelial cells. Cell Biol Toxicol. 2018;34:39–49. doi: 10.1007/s10565-017-9401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii T, Hosoki K, Nikura Y, Yamashita N, Nagase T, Yamashita N. IFN regulatory factor 3 potentiates emphysematous aggravation by lipopolysaccharide. J Immunol. 2017;198:3637–3649. doi: 10.4049/jimmunol.1601069. [DOI] [PubMed] [Google Scholar]
- 11.Campa CC, Silva RL, Margaria JP, Pirali T, Mattos MS, Kraemer LR, Reis DC, Grosa G, Copperi F, Dalmarco EM, et al. Inhalation of the prodrug PI3K inhibitor CL27c improves lung function in asthma and fibrosis. Nat Commun. 2018;9:5232. doi: 10.1038/s41467-018-07698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Chen Y, Yan Y, Li X, Chen G, He N, Shen S, Chen G, Zhang C, Liao W, et al. Loss of CEACAM1, a tumor-associated factor, attenuates post-infarction cardiac remodeling by inhibiting apoptosis. Sci Rep. 2016;6:21972. doi: 10.1038/srep21972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Culver SA, Quadri S, Ledford KL, Al-Share QY, Ghadieh HE, Najjar SM, Siragy HM. High-fat diet amplifies renal renin angiotensin system expression, blood pressure elevation, and renal dysfunction caused by Ceacam1 null deletion. Am J Physiol Endocrinol Metab. 2015;309:E802–E810. doi: 10.1152/ajpendo.00158.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobo EO, Zhang Z, Shively JE. Pivotal advance: CEACAM1 is a negative co-receptor for the B cell receptor and promotes CD19-mediated adhesion of B cells in a PI3K-dependent manner. J Leukoc Biol. 2009;86:205–218. doi: 10.1189/jlb.0109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Sun X, Chen C, Bai C, Wang X. Dynamic gene expressions of peripheral blood mononuclear cells in patients with acute exacerbation of chronic obstructive pulmonary disease: a preliminary study. Crit Care. 2014;18:508. doi: 10.1186/s13054-014-0508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Ledford KJ, Pitkin WB, Russo L, Najjar SM, Siragy HM. Targeted deletion of murine CEACAM 1 activates PI3K-Akt signaling and contributes to the expression of (Pro)renin receptor via CREB family and NF-kappaB transcription factors. Hypertension. 2013;62:317–323. doi: 10.1161/HYPERTENSIONAHA.113.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HX, Yang JJ, Zhang SA, Zhang SM, Wang JX, Xu ZY, Lin RY. HIF-1alpha promotes inflammatory response of chronic obstructive pulmonary disease by activating EGFR/PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:6077–6084. doi: 10.26355/eurrev_201809_15946. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Li R, Zhong R. Extracellular matrix promotes proliferation, migration and adhesion of airway smooth muscle cells in a rat model of chronic obstructive pulmonary disease via upregulation of the PI3K/AKT signaling pathway. Mol Med Rep. 2018;18:3143–3152. doi: 10.3892/mmr.2018.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren WB, Xia XJ, Huang J, Guo WF, Che YY, Huang TH, Lei LC. Interferon-gamma regulates cell malignant growth via the c-Abl/HDAC2 signaling pathway in mammary epithelial cells. J Zhejiang Univ Sci B. 2019;20:39–48. doi: 10.1631/jzus.B1800211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueshima C, Kataoka TR, Takei Y, Hirata M, Sugimoto A, Hirokawa M, Okayama Y, Blumberg RS, Haga H. CEACAM1 long isoform has opposite effects on the growth of human mastocytosis and medullary thyroid carcinoma cells. Cancer Med. 2017;6:845–856. doi: 10.1002/cam4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, La Placa D, Nguyen T, Kujawski M, Le K, Li L, Shively JE. CEACAM1 regulates the IL-6 mediated fever response to LPS through the RP105 receptor in murine monocytes. BMC Immunol. 2019;20:7. doi: 10.1186/s12865-019-0287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calinescu A, Turcu G, Nedelcu RI, Brinzea A, Hodorogea A, Antohe M, Diaconu C, Bleotu C, Pirici D, Jilaveanu LB, et al. On the dual role of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in human malignancies. J Immunol Res. 2018;2018:7169081. doi: 10.1155/2018/7169081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou MQ, Du Y, Liu YW, Wang YZ, He YQ, Yang CX, Wang WJ, Gao F. Clinical and experimental studies regarding the expression and diagnostic value of carcinoembryonic antigen-related cell adhesion molecule 1 in non-small-cell lung cancer. BMC Cancer. 2013;13:359. doi: 10.1186/1471-2407-13-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullrich N, Heinemann A, Nilewski E, Scheffrahn I, Klode J, Scherag A, Schadendorf D, Singer BB, Helfrich I. CEACAM1-3S drives melanoma cells into NK cell-mediated cytolysis and enhances patient survival. Cancer Res. 2015;75:1897–1907. doi: 10.1158/0008-5472.CAN-14-1752. [DOI] [PubMed] [Google Scholar]
- 25.Seijo LM, Zulueta JJ. Understanding the links between lung cancer, COPD, and emphysema: a key to more effective treatment and screening. Oncology (Williston Park) 2017;31:93–102. [PubMed] [Google Scholar]
- 26.Wu D, Cheng Y, Wang X. Definition of clinical gene tests. Cell Biol Toxicol. 2019;35:83–87. doi: 10.1007/s10565-019-09464-w. [DOI] [PubMed] [Google Scholar]
- 27.Song D, Yang D, Powell CA, Wang X. Cell-cell communication: old mystery and new opportunity. Cell Biol Toxicol. 2019;35:89–93. doi: 10.1007/s10565-019-09470-y. [DOI] [PubMed] [Google Scholar]
- 28.Lv J, Wang L, Shen H, Wang X. Regulatory roles of OASL in lung cancer cell sensitivity to Actinidia chinensis Planch root extract (acRoots) Cell Biol Toxicol. 2018;34:207–218. doi: 10.1007/s10565-018-9422-4. [DOI] [PubMed] [Google Scholar]
- 29.Ghufran MS, Soni P, Kanade SR. Aflatoxin-induced upregulation of protein arginine methyltransferase 5 is mediated by protein kinase C and extracellular signal-regulated kinase. Cell Biol Toxicol. 2019;35:67–80. doi: 10.1007/s10565-018-9439-8. [DOI] [PubMed] [Google Scholar]
- 30.Razaghi A, Owens L, Heimann K. Review of the recombinant human interferon gamma as an immunotherapeutic: impacts of production platforms and glycosylation. J Biotechnol. 2016;240:48–60. doi: 10.1016/j.jbiotec.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Liu X, Duan Y, Liu Y, Wang H, Lian S, Zhuang G, Fan Y. Combined blockade of T cell immunoglobulin and mucin domain 3 and carcinoembryonic antigen-related cell adhesion molecule 1 results in durable therapeutic efficacy in mice with intracranial gliomas. Med Sci Monit. 2017;23:3593–3602. doi: 10.12659/MSM.903098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dery KJ, Kujawski M, Grunert D, Wu X, Ngyuen T, Cheung C, Yim JH, Shively JE. IRF-1 regulates alternative mRNA splicing of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in breast epithelial cells generating an immunoreceptor tyrosine-based inhibition motif (ITIM) containing isoform. Mol Cancer. 2014;13:64. doi: 10.1186/1476-4598-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein RC, Waterfield MD. PI3-kinase inhibition: a target for drug development? Mol Med Today. 2000;6:347–357. doi: 10.1016/S1357-4310(00)01770-6. [DOI] [PubMed] [Google Scholar]
- 34.Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. 2014;127:923–928. doi: 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang X, Li K, Tao X, Chen C, Wang X, Wang L, Wang DC, Zhang Y, Bai C, Wang X. Effects of phosphoinositide 3-kinase on protease-induced acute and chronic lung inflammation, remodeling, and emphysema in rats. Chest. 2013;143:1025–1035. doi: 10.1378/chest.12-1040. [DOI] [PubMed] [Google Scholar]
- 36.Jones SA, Jenkins BJ. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol. 2018;18:773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 37.Peng S, Chen Y, Gong Y, Li Z, Xie R, Lin Y, Zou B, Li J, Zeng L. Predictive value of intratumour inflammatory cytokine mRNA levels of hepatocellular carcinoma patients and activation of two distinct pathways govern IL-8 induced epithelial-mesenchymal transition in human hepatic cancer cell lines. Cytokine. 2019;119:81–89. doi: 10.1016/j.cyto.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Singer BB, Scheffrahn I, Obrink B. The tumor growth-inhibiting cell adhesion molecule CEACAM1 (C-CAM) is differently expressed in proliferating and quiescent epithelial cells and regulates cell proliferation. Cancer Res. 2000;60:1236–1244. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Expression of CEACAM1 after LPS stimulation in HBE cells. CEACAM1 gene expression in HBE cells treated with LPS at concentration of 0, 0.1, 1 μg/ml for 24 h, no significant change of CEACAM1 was shown.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.