Abstract
Carbapenemase-producing Enterobacteriaceae (CPE) are well known to cause many serious infections resulting in increasing mortality rate, treatment cost, and prolonged hospitalization. Among the widely recognized types of carbapenemases, New Delhi β-lactamase (NDM) and Klebsiella pneumoniae carbapenemase (KPC) are the most important enzymes. However, in Vietnam, there are only scattered reports of CPE due to the lack of simple and affordable methods that are suitable to laboratory conditions. This study aims to survey the characteristics of carbapenem-resistant E. coli and K. pneumoniae (CR-E/K) at two hospitals in Southern Vietnam and perform some simple methods to detect the two enzymes. A total of 100 CR-E/K strains were collected from clinical isolates of Gia Dinh People's Hospital and Dong Nai General Hospital, Vietnam, from November 2017 to May 2018. The patient-related information was also included in the analysis. We conducted real-time polymerase chain reaction (PCR), Modified Hodge Test (MHT), and combined disk test (CDT) on all isolates. Carbapenemase-encoding genes were detected in 47 isolates (36 NDM, 10 KPC, and one isolate harboring both genes). The E. coli strain carrying simultaneously these two genes was the first case reported here. Most of isolates were collected from patients in ICU, Infectious Disease Department, and Department of Urologic Surgery. Urine and sputum were two common specimens. The true positive rate (sensitivity, TPR) and specificity (SPC) of the imipenem–EDTA (ethylen diamine tetra acetic acid) for NDM detection and the imipenem–PBA (phenylboronic acid) for KPC detection on E. coli were 93.8%, 97.1% and 66.7%, 95.7%, respectively. Meanwhile, the imipenem–EDTA for NDM detection and the imipenem–PBA for KPC detection among K. pneumonia achieved 90.5%, 100% and 100%, 92.9% TPR and SPC, respectively. However, MHT showed low sensitivity and specificity. Our findings showed that CP-E/K were detected with high prevalence in the two hospitals. We suggest that CDT can be used as a low-priced and accurate method of detection.
1. Introduction
Antibiotic resistance is a tremendous health problem. This includes the resistance to carbapenems which was considered as the last resort for Enterobacteriaceae infections [1]. In 2017, the World Health Organization published a list of superbugs including carbapenem-resistant Enterobacteriaceae. Carbapenemase production and ESBL/AmpC β-lactamase production coupled with porin loss or efflux pump were the common ways that Enterobacteriaceae become resistant to carbapenems [2]. However, the carbapenemase production was more dangerous than the others [2]. E. coli and K. pneumoniae are the most common pathogens in Enterobacteriaceae family. These carbapenemasae-producing bacteria were found in many countries, such as China [3], Parkistan [4], India [5], Turkey [6], Brazil [7], Mexico [8], Peru [9], and Greece [10]. Through this mechanism, New Delhi Metallo-β-lactamase-1 (NDM-1) was found in Asia with the highest frequency; Klebsiella pneumoniae carbapenemase (KPC) was the most popular enzyme causing carbapenem resistance, especially KPC-2 [1, 11, 12]. Furthermore, KPC-producing bacteria have spread all over the world, including Asia [11]. Some gray literature also reported about KPC-producing Enterobacteriaceae in Vietnam [13–15]. In other important views, these two genes harboring bacteria were more dangerous and resistant to carbapenem with a higher level than bacteria that harbored other types of carbapenemase-encoding genes (including OXA-48-like, another popular carbapenemase present in Enterobacteriaceae) [6, 16–18]. For instance, these carbapenemase-producing bacteria result in severe infections with high mortality rate; the infection caused by KPC-producing K. pnuemoniae was from 40% to 56% [16] and up to 88% for NDM-1 producing Enterobacteriaceae [6]. Carbapenem minimum inhibitory concentrations (MICs) for these two types of carbapeenemase producers in recent studies were higher than carbapenem MIC values against OXA-48 type producers [17, 18].
In another aspect, these bacteria have spread rapidly through mobilisable genetic elements [19, 20], for example, plasmid IncX3 [21], IncA/C2 [22]; transposon Tn4401 [23], Tn125 [21], or class I Integron [24]. These elements also play important roles in the existence of multiple genes and transporting various multidrug-resistant genes between bacterial species.
In Vietnam, the prevalence of these CPE is increasingly being reported in the hospital and aquatic environment [25–27]. Data about carbapenemase-producing E. coli and K. pneumoniae were reported notably in Southern and Northern Vietnam. Due to the lack of needed methods for screening and detection, there is an underestimation of CPE in other regions of Vietnam. The requirement for low-priced and efficient methods to screen or confirm carbapenemase-producing bacteria in laboratories, which are likely to be suitable for low-resource settings in Vietnam, is urgent. Today, the combinations of meropenem and varbobactam or imipenem and relebactam are taken into consideration as an option to treat infections caused by KPC-producing organisms [28, 29]. Therefore, it is vital to select an efficient method to detect and characterize these carbapenemase-producing Enterobacteriaceae, particularly in Vietnam.
Among various methods, PCR, real-time PCR, and DNA sequencing are the gold standards for carbapenemase-encoding genes detection, but these methods have not been widely used in Vietnam due to the high cost. Phenotypic tests such as Modified Hodge Test and the combined disk test are suitable because of the lower cost. The combined disk test is based on the synergy between metallo-β-lactamases (MBLs) or KPC inhibitors (EDTA or PBA) and carbapenems. Many studies used these methods but none of them investigated E. coli and K. pneumoniae separately [30, 31]. The purpose of this study was to evaluate the characteristics of NDM/KPC-producing E. coli/K. pneumoniae at the hospitals in the South Vietnam and to assess some simple methods for detecting these bacteria in laboratory conditions.
2. Materials and Methods
2.1. Study Design and Sample Collection
The study was designed as a cross-sectional study, including all clinical isolates from November 2017 to May 2018. The study was conducted at the Molecular Biomedicine Laboratory in the Department of Medical Laboratory Science, University of Medicine and Pharmacy, Ho Chi Minh City. 100 clinical isolate strains (50 E. coli and 50 K. pneumoniae) were collected from the Microbiology Department of Gia Dinh People's Hospital and Dong Nai General Hospital. All of them were nonsusceptible to one of the carbapenems (imipenem, meropenem, or ertapenem) detected by the automatic systems. Patient data such as age, sex, specimen types, and clinical wards were gathered to survey the characteristics of patients. Sample list and relevant information are supplied in Annex 1.
2.2. Bacterial Isolates
Fifty-nine isolates from Gia Dinh People's Hospital were identified and subjected to antimicrobial susceptibility testing by using the Vitek 2 system (bioMérieux Vitek Inc., Hazelwood, MO, USA). Furthermore, ID/AST of 41 isolates from Dong Nai General Hospital was performed by using BD Phoenix™ (USA). From those hospitals, a single colony obtained from pure culture was inoculated into BHI (Heart Infusion Broth, Himedia, India) broth, with 20% glycerol (Xilong medical, China), and stored at -20°C, to reculture for extracting DNA for real-time PCR and performing phenotypic methods.
2.3. Real-Time PCR
2.3.1. DNA Extraction
Bacterial DNA was extracted by heat treatment. One colony form pure isolates incubated overnight on MacConkey agar (Merck KgaA, Darmstadt, Germany) was suspended in 200μL of Tris-EDTA pH 8.0. The suspension was heated at 95°C for 20 minutes, followed by centrifugation at 13,000 revolutions per minute for 10 minutes to collect 150μL supernatant, and stored at -20°C for real-time PCR. DNA quality was evaluated using NanoDrop™ 2000 Spectrophotometer (ThemoFisher Scientific, Wilmington, Delaware, USA) with the acceptable range for the ratio of absorbance at 260 nm/280 nm from 1.6 to 1.8.
2.3.2. Genotypic Detection of blaNDM, blaKPC, and 16S rRNA
The genes were detected by real-time PCR using TaqMan probes for all samples and were confirmed by DNA sequencing of selected positive samples. The bacterial DNA was performed to screen for the presence of blaNDM and blaKPC. The reaction used bacterial 16S rRNA gene as an internal control for DNA extraction and amplification process. The primers and probes used for blaNDM, blaKPC, and 16S rRNA were described by the Center of Disease Control and Prevention [32]. To carry out real-time PCR, 5μL DNA was added to a final volume of 20μL Master Mix, which included 1X PCR buffer, 4 mM MgCl2, 0.2 mM dNTPs, and 5 IU Taq DNA polymerase (Solgent Co., Ltd., South Korea). The concentration for each primers and probes is illustrated in Table 1. The negative control for PCR reaction is sterile DEPC-treated water. DNA from GD018, which is a clinical isolate that carries both blaNDM and blaKPC (confirmed by DNA sequencing), is used as a positive control.
Table 1.
Primers and probes used in this study.
| Oligonucleotide | Nucleotide sequence, 5′–3′ | Origin | Volume of reaction (nM) | Reference |
|
| ||||
| Primers and probes for real-time PCR | ||||
|
| ||||
| NDM-F Primer | GAC CGC CCA GAT CCT CAA | IDT | 300 | |
| NDM-R Primer | CGC GAC CGG CAG GTT | IDT | 300 | |
| NDM-probe | HEX-TG GAT CAA GCA GGA GAT-ZEN/IBFQ | IDT | 200 | |
| KPC-F Primer | GGC CGC CGT GCA ATA C | IDT | 500 | |
| KPC-R Primer | GCC GCC CAA CTC CTT CA | IDT | 500 | [32] |
| KPC- probe | 6FAM-TG ATA ACG CCG CCG CCA ATT TGT-ZEN/IBFQ | IDT | 200 | |
| 16S rRNA-F Primer | TGG AGC ATG TGG TTT AAT TCG A | IDT | 200 | |
| 16S rRNA-R Primer | TGC GGG ACT TAA CCC AAC A | IDT | 200 | |
| 16S rRNA-probe | Cy5-CA CGA GCT GAC GAC AR∗C CAT GCA-IBRQ | IDT | 100 | |
|
| ||||
| Primers for DNA sequencing | ||||
|
| ||||
| NDM-F-seq | AGT CGC TTC CAA CGG TTT | IDT | 300 | This study |
| NDM-R-seq | CAT TGG CAT AAG TCG CAA TCC | IDT | 300 | |
| KPC-F-seq | GGT CAC CCA TCT CGG AAA | IDT | 500 | |
| KPC-R-seq | GGG ATG GCG GAG TTC AG | IDT | 500 | |
The real-time PCR conditions were chosen as follows: Enzyme activation was achieved by primary heating step at 95°C for 15 minutes. This was followed by 40 cycles of 95°C for 15 seconds and 56°C for 20 seconds. Real-time PCR was run on CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, California, USA). Fluorescence signal was detected after each cycle. Positive sample was determined when the automatically normalized fluorescence signal rises above the threshold limit value calculated in each run. A total of 40 cycles takes 70 minutes.
2.3.3. DNA Sequencing
DNA extracts from selected positive samples for blaNDM (GD013, GD002, and GD018) and blaKPC (GD011, GD16, and GD018) were sequenced using the sequencing primers. Initial PCR proceeded in the above-mentioned conditions, and PCR products were sent to 1-BASE (Singapore) for Sanger's sequencing. 16S rRNA genes from two samples GD013 and GD018 were also sequenced to confirm the validity of the internal control primers.
2.4. Modified Hodge Test (MHT)
The MHT was carried out on all isolates, following the Clinical & Laboratory Standards Institute (CLSI) guide on Mueller-Hinton agar (MHA–Becton Diskinson, US) using 10μg meropenem (MEM) disks (Nam Khoa Biotech, Vietnam) [33]. Klebsiella pneumoniae ATCC®BAA–1705 and Klebsiella pneumoniae ATCC®BAA–1706 were used as a positive control and a negative control, respectively.
2.5. Combined Disk Test (PBA/IPM–EDTA/IPM)
Phenylboronic acid solution was prepared by dissolving 2g phenylboronic acid (PBA) (Sigma-Aldrich, Steinheim, Germany) in 100mL solution containing 50mL distilled water and 50mL Dimethyl Sulfoxide (Xilong medical, China). EDTA 0.25M solution prepared by mixing 18.61g disodium ETDA.2H2O in 200 mL of distilled water was adjusted to pH 8.0 by using NaOH. Both solutions were autoclaved at 121°C for 15 minutes. Either 20μL phenylboronic acid solution (400μg PBA) or 10μL EDTA solution (930μg EDTA) was added on a 10μg imipenem disk (IPM) to get a PBA/IPM or EDTA/IPM disk, respectively. These disks were used within 1 hour.
A bacterial suspension with McFarland Equivalence Turbidity Standard 0.5 was streaked on MHA plate using a swab. Three disks, IPM, EDTA/IPM, and PBA/IPM, were placed on the MHA plate with the distance from one disk to another being at least 24 mm. Klebsiella pneumoniae ATCC®BAA–1705 and Klebsiella pneumoniae ATCC®BAA–1706 were used as a positive control and a negative control, respectively. An increase of 7 mm for EDTA/IPM or 5 mm for PBA/IPM in the diameter of the zone of inhibition compared to that of IPM was considered as a positive result for NDM or KPC production, respectively.
2.6. Statistical Analysis
Data were entered using Excel 2010, and all statistical analyses were carried out using SPSS version 22.0 (IBM Corp. 2013). We determined characteristics of patients carrying carbapenemase-producing bacteria with 95% confidence interval (CI); odds ratio (OR) was calculated. Chi-square and Fisher exact tests were used for categorical variables. Independent sample t-test was used for comparing the mean ages of the two groups.
Real-time PCR was considered as a gold standard method. Sensitivity and specificity, negative predictive values (NPV), and positive predictive values (PPV) were also calculated. These values of MHT and CDT were compared using the nonparametric McNemar's test. P-values less than 0.05 were considered as statistical significance.
3. Results and Discussion
3.1. Results
3.1.1. Characteristics of Carbapenemase-Producing E. coli and K. pneumoniae at 2 Hospitals in the Southern Vietnam
In general, of 100 strains from clinical samples (50 isolates of E. coli and 50 isolates of K. pneumoniae), 47 isolates were positive by real-time PCR. In those positive strains, 36 carried blaNDM, 10 carried blaKPC, and one carried both genes. 58.3% (21/36) strains carrying blaNDM and 80% (8/10) strains carrying blaKPC were K. pneumoniae. The one carried both genes was E. coli. CRE were mainly isolated from sputum (37%) and urine (31%). Samples were collected from 13 different sites of ICU (14%), Department of Urologic Surgery (14%), Infectious Disease Department (17%), Geriatrics Department (9%), Respiratory Department (9%), and some other departments (37%). The majority of the study participants were male (55%). The average age was 66.9 (95% CI: 63.6–70.9), the youngest patient was 23, and the oldest one was 94. The proportion of people aged 60 years and older was 69%; the odd ratio of patients in the elder group infected by E. coli or K. pneumoniae having carbapenemase-encoding genes was 3.18 (95% CI: 1.28–7.89) compared to the other group. Furthermore, in Dong Nai General Hospital, the number of the elders infected by NDM/KPC producers was significantly higher than that of the younger ones (p=0.001). The prevalence of pneumonia was 29.0%, the odd of CP-E/K infected patients with pneumonia was 2.32 (95% CI: 0.93–5.78) compared to CP-E/K infected patients without pneumonia; the odd of elders with pneumonia was 9.32 (95% CI: 2.05–42.29) compared to the younger ones.
Moreover, data from our study indicated that the rate of CPE is different between the two hospitals. NDM-producing organisms were the major pathogens in Gia Dinh People's Hospital (45.8%). The prevalence of bacteria carrying blaNDM and blaKPC seems to be the same in Dong Nai General Hospital (22% for blaNDM versus 19.5% for blaKPC) (Figure 1). Other characteristics of CRE infected patients were described in Figures 1 and 2, Tables 2 and 3.
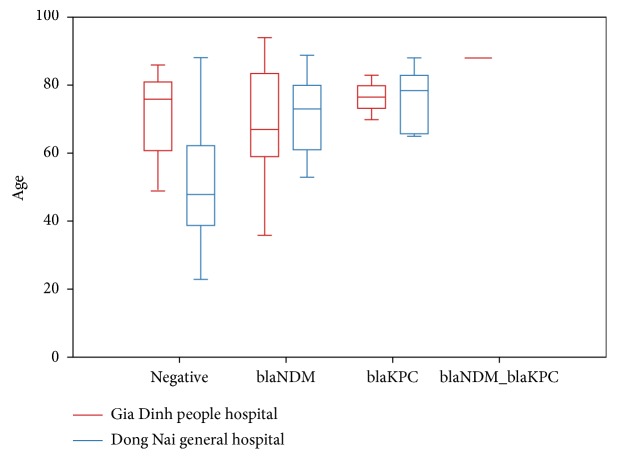
Figure 1.
Different distribution of patients' age following genotype at 2 hospitals.
Figure 2.
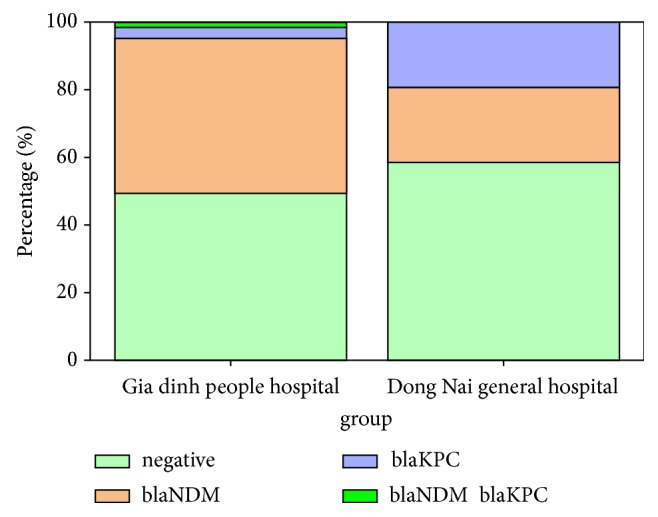
Different distribution of prevalence of CPE at 2 hospitals.
Table 2.
Distribution of diseases.
| Hospital | Disease | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pneumonia | UTI | Wound Infection | Abnormal Infection | Bacteremia | Fever | Surgical Wound Infection | Other | |||
| GD (N=59) |
E | 2 | 14 | 5 | 0 | 5 | 2 | 0 | 1 | 29 |
| K | 13 | 9 | 2 | 1 | 2 | 2 | 1 | 0 | 30 | |
|
| ||||||||||
| DN (N=41) |
E | 3 | 10 | 3 | 0 | 0 | 1 | 2 | 2 | 21 |
| K | 11 | 2 | 3 | 0 | 0 | 0 | 2 | 2 | 20 | |
|
| ||||||||||
| Total | 29 | 35 | 13 | 1 | 7 | 5 | 5 | 5 | 100 | |
∗E: E. coli, K: K. pneumoniae, GD: Gia Dinh People's Hospital, DN: Dong Nai General Hospital, UTI: Urinary Tract Infection.
Table 3.
Particular characteristics of patients at 2 hospitals in this study.
| Characters | Gia Dinh People's Hospital (n=59) | Dong Nai General Hospital (n=41) | P-value |
|---|---|---|---|
| Average age (95% CI) | 71.5 (67.4 – 75.2) | 60.4 (52.2 – 66) | 0.003 |
| Range | 36 – 94 | 23 – 89 | |
| Elder (%) | 76.3 | 58.5 | 0.079 |
| Positive rate (%) | 50.8 | 41.5 | 0.545 |
| Pneumonia rate (%) | 25.4 | 34.1 | |
| Odd of Outcome (Pos/Neg) between Pneumonia and the other | 0.79 | 7.14 | |
| Odd of Pneumonia (Elder/Younger) | 5.88 | 18.91 | |
| Odd of Outcome (Positive/Negative) in Elder group/ the other. | 1.045 | 15 | |
| Therapy departments (%) | ICU (20.3), Geriatrics (15.3), Department of Urologic surgery (10.2), Respiratory Department (8.5), and other departments (45.7) |
Infectious Disease Department (41.4) Department of surgery (19.5), Respiratory Department (9.8), ICU (4.9), and other departments (24.4) |
|
| K. pneumoniae (%) | 50.8 | 48.8 |
3.1.2. Molecular Detection
The conformity of three methods was illustrated in Table 3.
3.1.3. The Modified Hodge Test (MHT)
All isolates were evaluated by using the MHT and real-time PCR. This method showed low sensitivity and specificity compared to real-time PCR, especially in K. pneumoniae. 38% (23/53) of strains were positive with the MHT whereas the real-time PCR results were negative. 8/47 strains were real-time PCR positive but negative for the MHT, and 7 of those 8 strains carried blaNDM. The sensitivity and specificity for E. coli and K. pneumoniae were 72.2%, 68.8% and 89.1%, 38.1%, respectively. Moreover, the PPV and NPV for E. coli and K. pneumoniae were 56.5%, 81.5% and 66.7 %, 72.7%, respectively.
3.1.4. Combined Disk Test
In the same way, all isolates were tested using combined disk test (using EDTA and PBA).
3.2. Discussion
In our study, the prevalence of NDM and KPC-producing bacteria in both hospitals is high. Furthermore, the figures of NDM and KPC-producing E/K among infected patients were different between the two hospitals, which depend on areas and ages of patients (Figure 2). Probably, the high rate of integron and gene cassette carrying E. coli at Gia Dinh People's Hospital (45/65 strains carrying intergron gene and 32 of these 45 intergron harboring strains carrying gene cassette) [34] led to coharboring of the two genes in E. coli strain. In particular, this is the first report of the presence of E. coli strain coharboring both genes in Vietnam.
The average age of patients and the proportion of elder group at Gia Dinh People's Hospital were higher than these figures for Dong Nai General Hospital. In Dong Nai General Hospital, people infected by non-NDM/KPC-producing bacteria were young, most of whom were the group in the range from 40 to 60 (Figure 1); these young people could be infected with ESBL/AmpC producing E. coli/K. pneumoniae. Another hypothesis might be that the isolates from these patients could carry other carbapenemase-encoding genes, such as blaOXA-48-like, but these mechanisms cause a lower level of resistance [2, 17, 18]. These features could play an important role in the difference of the odd of positive and negative outcome of the elder group between these 2 hospitals. The elders who suffered from pneumonia and infection with the two types of carbapenemase producers were more than the younger ones, especially in Dong Nai General Hospital. The reason for the difference between these two hospitals was still unclear. It could be explained that Gia Dinh People's Hospital was established for a long time; the severity of illness was quite different compared with Dong Nai General Hospital.
Regarding the methods, real-time PCR is an accurate and rapid method to detect the presence of carbapenemase-encoding genes. However, the cost associated with the method is one of the major disadvantages that limits its use in common laboratories in Vietnam. Among various phenotypic methods, MHT recommended in CLSI 2017 is a simple method [33]. In this study, SPC of MHT is low (23/53 K. pneumoniae strains were positive, but negative for the real-time PCR). The possible reason for this is that other types of carbapenemase may present in these bacteria. However, it could be explained that AmpC, ESBLs, or strict requirements of MHT process at each step may cause false positive results [35]. Besides, this method shows low sensitivity to NDM-producing Enterobacteriaceae (7/8 strains were found to have a false-negative result) [36, 37]. In our study, the MHT results for K. pneumoniae are consistent with findings in recent studies conducted by Dariush Shokri et al. [38] and Rangneka R aSeeM et al. [39]. This may be the reason why the CLSI 2018 standard excluded MHT in the guideline [40].
In contrast, CDT has been shown to be highly effective. The results of NDM detection by EDTA disk test for K. pneumoniae (Table 4) were in accordance with findings obtained by Paradimitriou–Oliveris et al. [30]. However, it was in contrast to findings reported by Dariush Shokri et al. [38]. This could be explained by the fact that Shokri carried out his study including not only K. pneumoniae, but also A. baumanii and P. aeruginosa. The high rate of blaVIM in P. aeruginosa may be the reason for false-negative results. A study conducted by Ting-ting Qu et al. in 2009 showed that CDT also detected other types of carbapenemase including Verona integron-borne metallo-β-lactamase (VIM), which was the most widespread metallo-β-lactamase in P. aeruginosa [41, 42]. Our results for KPC detection using PBA disk test K. pneumoniae (Table 4) were in accordance with findings obtained by Athanasios Tsakris et al. (using PBA-400μg) [31] and Julio Zúñiga et al. (using 300μg APBA) [43]. The efficiency of PBA disk test in E. coli was low, particularly in sensitivity and PPV because of the sample size of KPC-producing E. coli, only 3 strains with one of them being a false negative. The results in Tables 5 and 6 illustrated the significantly higher specificity of combined disk test compared with MHT.
Table 4.
The agreement of bacteria isolation, real-time PCR, and DNA sequencing results.
| Strains ID | Isolation | Real-time PCR | DNA sequencing |
|---|---|---|---|
| GD013 | K. pneumonia | blaNDM, 16s rRNA | blaNDM, K. pneumoniae |
| GD002 | E. coli | blaNDM | blaNDM |
| GD018 | E. coli | blaNDM, blaKPC, 16S rRNA | blaNDM, blaKPC, E. coli |
| GD011 | K. pneumoniae | blaKPC | blaKPC |
| GD016 | K. pneumoniae | blaKPC | blaKPC |
| DN011 | K. pneumoniae | blaKPC | blaKPC |
Table 5.
Combined disk test results in E. coli and K. pneumonia.
| Combined disk test | Real-time PCR | TPR (%) 95% |
SPC (%) |
PPV (%) |
NPV (%) |
Accuracy (%) |
|||
|---|---|---|---|---|---|---|---|---|---|
| Species | Pos | Ne | |||||||
| NDM detection |
E. coli (n=50) |
Pos | 15 | 1 | 93.8 | 97.1 | 93.8 | 97.1 | 96.0 |
| Neg | 1 | 33 | |||||||
|
K. pneumoniae (n=50) |
Pos | 19 | 0 | 90.5 | 100 | 100 | 93.6 | 96 | |
| Neg | 2 | 29 | |||||||
|
| |||||||||
| KPC detection |
E. coli (n=50) |
Pos | 2 | 2 | 66.7 | 95.7 | 50 | 97.8 | 94.0 |
| Neg | 1 | 45 | |||||||
|
K. pneumoniae (n=50) |
Pos | 8 | 3 | 100 | 92.9 | 72.7 | 100 | 94 | |
| Neg | 0 | 39 | |||||||
∗Pos: positive, Neg: negative.
Table 6.
Comparison between the MHT and combined disk test.
| Bacteria | Index | NDM detection | KPC detection | ||||
|---|---|---|---|---|---|---|---|
| MHT | EDTA | McNemar∗ | MHT | Boronic | McNemar∗ | ||
| E. coli (n=50) | TPR (%) | 72.2 | 93.8 | p=0.25 | 72.2 | 66.7 | p=1.00 |
| SPC (%) | 68.8 | 97.1 | p=0.006 | 68.8 | 95.7 | p<0.001 | |
|
| |||||||
| K. pneumoniae (n=50) | TPR (%) | 89.7 | 90.5 | p=1.00 | 89.7 | 100 | NC |
| SPC (%) | 38.1 | 100 | p<0.001 | 38.1 | 92.9 | p<0.001 | |
∗ means nonparametric McNemar test.
4. Conclusions
The prevalence of CP-E/K in Vietnam is high. The lack of suitable methods to detect and differentiate these bacterial infections has been the main obstacle for clinicians to choosing an appropriate treatment. During this study, we recognized that CDT is reasonable and simple to detect the presence of these important enzymes. These features make it highly applicable to all clinical laboratories in Vietnam.
Acknowledgments
The authors are grateful to Gia Dinh People's Hospital (HCMC) and Dong Nai General Hospital for kindly providing the clinical isolates and also to the staff of Pasteur Institute in Ho Chi Minh City for the comments and suggestions. We would also like to thank Dr. Vu Quang Huy, the Head of the Department of Medical Laboratory Science, University of Medicine and Pharmacy, Ho Chi Minh City, for facilitating the study and thank Pham Thai Binh (Nam Khoa Biotech), Bui The Trung (Children's Hospital no. 2, HCMC), and Nguyen Thi Phuong Thao (Dong Nai Medical College) for providing the ATCC strains of bacteria and some reagents for this study.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Additional Points
Limitation. In our study, other genes such an OXA-48-like, ESBL, and AmpC β-lactamase were not included, so this could affect our study results. The only 10 KPC-producing organisms in this study may have effects on the results of PBA disk test. A lack of genotypic method to identify E.coli and K. pneumoniae was also important.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
List of related information of patients and testing results were detailed in Annex 1. Data analysis results, testing images, reference, and full-text of gray literature, were described in Annexes 2, 3, 4, 5, respectively.
References
- 1.Khan A. U., Maryam L., Zarrilli R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health. BMC Microbiology. 2017;17(1) doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia X., Dai W., Ma W., et al. Corrigendum: carbapenem-resistant e. cloacae in southwest china: molecular analysis of resistance and risk factors for infections caused by ndm-1-producers. Frontiers in Microbiology. 2018;9 doi: 10.3389/fmicb.2018.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Yao X., Luo J., Lv L., Zeng Z., Liu J.-H. Emergence of Escherichia coli coproducing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. Journal of Antimicrobial Chemotherapy. 2018;73(1):252–254. doi: 10.1093/jac/dkx335. [DOI] [PubMed] [Google Scholar]
- 4.Day K. M., Salman M., Kazi B., et al. Prevalence of NDM-1 carbapenemase in patients with diarrhoea in Pakistan and evaluation of two chromogenic culture media. Journal of Applied Microbiology. 2013;114(6):1810–1816. doi: 10.1111/jam.12171. [DOI] [PubMed] [Google Scholar]
- 5.Eshetie S., Unakal C., Gelaw A., Ayelign B., Endris M., Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrobial Resistance and Infection Control. 2015;4(1, article no. 12) doi: 10.1186/s13756-015-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karabay O., Altindis M., Koroglu M., Karatuna O., Aydemir Ö. A., Erdem A. F. The carbapenem-resistant Enterobacteriaceae threat is growing: NDM-1 epidemic at a training hospital in Turkey. Annals of Clinical Microbiology and Antimicrobials. 2016;15(1) doi: 10.1186/s12941-016-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi Gonçalves I., Ferreira M. L., Araujo B. F., et al. Outbreaks of colistin-resistant and colistin-susceptible KPC-producing Klebsiella pneumoniae in a Brazilian intensive care unit. Journal of Hospital Infection. 2016;94(4):322–329. doi: 10.1016/j.jhin.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Torres-González P., Bobadilla-del Valle M., Tovar-Calderón E., et al. Outbreak Caused by Enterobacteriaceae Harboring NDM-1 Metallo-β-Lactamase Carried in an IncFII Plasmid in a Tertiary Care Hospital in Mexico City. Antimicrobial Agents and Chemotherapy. 2015;59(11):7080–7083. doi: 10.1128/AAC.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamariz J., Llanos C., Seas C., et al. Draft genome sequence of the first New Delhi metallo-β-lactamase (NDM-1)-producing Escherichia coli strain isolated in Peru. Genome Announcements. 2018;6(13) doi: 10.1128/genomeA.00199-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maltezou H. C., Giakkoupi P., Maragos A., et al. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece) Infection. 2009;58(3):213–219. doi: 10.1016/j.jinf.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Dortet L., Cuzon G., Ponties V., Nordmann P. Trends in carbapenemase-producing Enterobacteriaceae, France, 2012 to 2014. Eurosurveillance. 2017;22(6) doi: 10.2807/1560-7917.ES.2017.22.6.30461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C.-R., Lee J. H., Park K. S., Kim Y. B., Jeong B. C., Lee S. H. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Frontiers in Microbiology. 2016;7, article no. 895 doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phuong T. N. Klebsiella pneumoniae carbapenemase producing Klebsiella pneumoniae molecular genetic characteristics University of Science. Vietnam National University Ho Chi Minh City; 2016. [Google Scholar]
- 14.Thai P. D., Linh T. D., Thu N. H., Hoang T. H., Son T. H., Phuong T. T. V. Apply Southern-Blotting assay to detect KPC-plasmid of enterobacteriaceae strains from clinical isolates from hospitals in Hanoi. Vietnam . Vietnam Journal of Preventive Medicine. 2017;27(3) [Google Scholar]
- 15.Thu N. H., Linh T. D., Mai N. T. T., Phuong T. V., Yen N. H. L., Duong T. N. Molecular epidemiology of Klebsiella pneumoniae carbapenemase - 2 producing Klebsiella pneumoniae isolated from Saintpaul Hospital. Vietnam Journal of Preventive Medicine. 2016;26(8) [Google Scholar]
- 16.Ramos-Castañeda J. A., Ruano-Ravina A., Barbosa-Lorenzo R., et al. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: Systematic review and meta-analysis: Mortality due to KPC Klebsiella pneumoniae infections. Infection. 2018;76(5):438–448. doi: 10.1016/j.jinf.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Studentova V., Papagiannitsis C. C., Izdebski R., et al. Detection of OXA-48-type carbapenemase-producing Enterobacteriaceae in diagnostic laboratories can be enhanced by addition of bicarbonates to cultivation media or reaction buffers. Folia Microbiologica. 2015;60(2):119–129. doi: 10.1007/s12223-014-0349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fattouh R., Tijet N., McGeer A., Poutanen S. M., Melano R. G., Patel S. N. What is the appropriate meropenem MIC for screening of carbapenemase-producing Enterobacteriaceae in low-prevalence settings? Antimicrobial Agents and Chemotherapy. 2016;60(3):1556–1559. doi: 10.1128/AAC.02304-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer A., Barbour E., Azhar E., et al. Transposable elements in Escherichia coli antimicrobial resistance. Advances in Bioscience and Biotechnology. 2013;04(03):415–423. doi: 10.4236/abb.2013.43A055. [DOI] [Google Scholar]
- 20.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase producing Enterobacteriaceae. Emerging Infectious Diseases. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng J., Qiu Y., Yin Z., et al. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. Journal of Antimicrobial Chemotherapy. 2015;70(11):2987–2991. doi: 10.1093/jac/dkv232. [DOI] [PubMed] [Google Scholar]
- 22.Bosch T., Lutgens S. P. M., Hermans M. H. A., et al. Outbreak of NDM-1-producing klebsiella pneumoniae in a Dutch hospital, with interspecies transfer of the resistance plasmid and unexpected occurrence in unrelated health care centers. Journal of Clinical Microbiology. 2017;55(8):2380–2390. doi: 10.1128/JCM.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiology Reviews. 2017;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 24.Datta S., Mitra S., Chattopadhyay P., Som T., Mukherjee S., Basu S. Spread and exchange of bla NDM-1 in hospitalized neonates: role of mobilizable genetic elements. European Journal of Clinical Microbiology & Infectious Diseases. 2017;36(2):255–265. doi: 10.1007/s10096-016-2794-6. [DOI] [PubMed] [Google Scholar]
- 25.Tada T., Tsuchiya M., Shimada K., et al. Dissemination of Carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of Carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA Methylases (RmtB and RmtC) in Vietnam. BMC Infectious Diseases. 2017;17(1):p. 467. doi: 10.1186/s12879-017-2570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isozumi R., Yoshimatsu K., Yamashiro T., et al. BlaNDM-1-positive Klebsiella pneumoniae from environment, Vietnam. Emerging Infectious Diseases. 2012;18(8):1383–1385. doi: 10.3201/eid1808.111816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang T. H., Wertheim H., Minh N. B., et al. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae strains containing new delhi metallo-beta-lactamase isolated from two patients in Vietnam. Journal of Clinical Microbiology. 2013;51(1):373–374. doi: 10.1128/JCM.02322-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhanel G. G., Lawrence C. K., Adam H., et al. Correction to: Imipenem–Relebactam and Meropenem–Vaborbactam: Two Novel Carbapenem-β-Lactamase Inhibitor Combinations. Drugs. 2018;78(7):p. 787. doi: 10.1007/s40265-018-0910-x. [DOI] [PubMed] [Google Scholar]
- 29.P-WK M., Bm D., Tm J. A, Bc R., Bs A., et al. Relebactam is a Potent Inhibitor of the KPC-2 beta-Lactamase and Restores the Susceptibility of Imipenem Against KPC-Producing Enterobacteriaceae. Antimicrobial Agents and Chemotherapy. 2018 doi: 10.1128/AAC.00174-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christofidou M., Papadimitriou-Olivgeris M., Anastassiou E. D., et al. Performance of four different agar plate methods for rectal swabs, synergy disk tests and metallo-β-lactamase Etest for clinical isolates in detecting carbapenemase-producing Klebsiella pneumoniae. Journal of Medical Microbiology. 2016;65(9):954–961. doi: 10.1099/jmm.0.000318. [DOI] [PubMed] [Google Scholar]
- 31.Tsakris A., Kristo I., Poulou A., et al. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. Journal of Clinical Microbiology. 2009;47(2):362–367. doi: 10.1128/JCM.01922-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Control CfD Prevention. Multiplex real-time PCR detection of Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM-1) genes. Atlanta. 2011;500(6-7) [Google Scholar]
- 33.Institute CLS. Performance standards for antimicrobial susceptibility testing. The Modified Hodge Test for Suspected Carbapenemase Production in Enterobacteriaceae: Clinical and Laboratory Standards Institute. 2017:108–113. [Google Scholar]
- 34.Bich V., Dung D. A., Pha B. N. A., Tuyet N. S. M. Antibiotic resistance of escherichia coli in gia dinh people's hospital. Ho Chi Minh medical Journal. 2009;13(6):253–257. [Google Scholar]
- 35.Jeremiah S. S., Balaji V., Anandan S., Sahni R. D. A possible alternative to the error prone modified Hodge test to correctly identify the carbapenemase producing Gram-negative bacteria. Indian Journal of Medical Microbiology. 2014;32(4):414–418. doi: 10.4103/0255-0857.142258. [DOI] [PubMed] [Google Scholar]
- 36.Kim H.-K., Park J. S., Sung H., Kim M.-N. Further modification of the modified hodge test for detecting metallo-β-lactamase-producing carbapenem-resistant Enterobacteriaceae. Annals of Laboratory Medicine. 2015;35(3):298–305. doi: 10.3343/alm.2015.35.3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sultan B. A., Khan E., Hussain F., Nasir A., Irfan S. Effectiveness of Modified Hodge Test to detect NDM-1 Carbapenemases: An experience from Pakistan. Journal of the Pakistan Medical Association. 2013;63(8):955–960. [Google Scholar]
- 38.Shokri D., Khorasgani M. R., Fatemi S. M., Soleimani-Delfan A. Resistotyping, phenotyping and genotyping of new delhi metallo-β-lactamase (NDM) among gram-negative bacilli from Iranian patients. Journal of Medical Microbiology. 2017;66(4):402–411. doi: 10.1099/jmm.0.000444. [DOI] [PubMed] [Google Scholar]
- 39.Aseem R. Approach to carbapenemase detection in klebsiella pneumoniae in routine diagnostic laboratories. Journal of Clinical and Diagnostic Research. 2016;10(12):DC24–DC7. doi: 10.7860/JCDR/2016/23036.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Institute CLS. Performance standards for antimicrobial susceptibility testing. Modified Carbapenem Inactivation Methods for Suspected Carbapenemase Production in Enterobacteriaceae and P aeruginosa. 2018:112–122. [Google Scholar]
- 41.Qu T., Zhang J., Wang J., et al. Evaluation of phenotypic tests for detection of metallo-β-lactamase-producing pseudomonas aeruginosa strains in china. Journal of Clinical Microbiology. 2009;47(4):1136–1142. doi: 10.1128/JCM.01592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong D. J., Bae I. K., Jang I.-H., Jeong S. H., Kang H.-K., Lee K. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Journal of Infection and Chemotherapy. 2015;47(2):81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Julio Z., Gerardo C., Carlos P., Musharaf T. The combined-disk boronic acid test as an accurate strategy for the detection of KPC carbapenemase in Central America. The Journal of Infection in Developing Countries. 2016;10(3):298–303. doi: 10.3855/jidc.7213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of related information of patients and testing results were detailed in Annex 1. Data analysis results, testing images, reference, and full-text of gray literature, were described in Annexes 2, 3, 4, 5, respectively.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.