Abstract
The aim of the current study was to develop a predictor classifier for response to fluorouracil-based chemotherapy in patients with advanced colorectal cancer (CRC) using microarray gene expression profiles of primary CRC tissues. Using two expression profiles downloaded from the Gene Expression Omnibus database, differentially expressed genes (DEGs) between responders and non-responders to fluorouracil-based chemotherapy were identified. A total of 791 DEGs, including 303 that were upregulated and 488 that were downregulated in responders, were identified. Functional enrichment analysis revealed that the DEGs were primarily involved in ‘cell mitosis’, ‘DNA replication’ and ‘cell cycle’ signaling pathways. Following feature selection using two methods, a random forest classifier for response to fluorouracil-based chemotherapy with 13 DEGs was constructed. The accuracy of the 13-gene classifier was 0.930 in the training set and 0.810 in the validation set. The receiver operating characteristic curve analysis revealed that the area under the curve was 1.000 in the training set and 0.873 in the validation set (P=0.227). The 13-gene-based classifier described in the current study may be used as a potential biomarker to predict the effects of fluorouracil-based chemotherapy in patients with CRC.
Keywords: colorectal cancer, fluorouracil-based chemotherapy, differential expression genes, random forest classifier
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, and it is one of the most common causes of cancer mortality (1). Localized CRCs are amenable to curative surgical resection, however, ~25% of patients present with metastatic disease and ~50% of patients will develop metastases (2). Fluorouracil-based chemotherapy remains the primary treatment for metastatic CRC (3). 5-fluorouracil (5-FU) alone has an objective response rate of ~20% (4). The addition of irinotecan or oxaliplatin to 5-FU increases the objective response rate to ~50% (5). The effects of 5-FU/leucovorin combined with irinotecan (FOLFIRI) or oxaliplatin (FOLFOX) in the first-line treatment of metastatic CRC are comparable (6). In the last decade, the addition of targeted therapies based on these chemotherapy regimens has improved the therapeutic approach and significantly increased progression-free survival and overall survival times (7–9). Fluorouracil-based chemotherapy remains the primary treatment for metastatic CRC. However, ~50% of patients are resistant to fluorouracil-based chemotherapy. In addition, the side effects of systemic chemotherapy, including neurotoxicity, myelotoxicity and gastrointestinal toxicity, may have a major impact on the quality of life of the patients and may lead to life-threatening complications (3). Therefore, identifying effective strategies that predict response to chemotherapy are required. Using these strategies, patients that are predicted to not respond to chemotherapy may receive other potentially effective treatments as early as possible and avoid unnecessary side effects. Gene expression profiling is used to predict the clinical outcome of patients with CRC (10–12). Previous studies have revealed that gene expression profiling may be used to predict cancer response to chemotherapy, including breast cancer and CRC (13–15).
The aim of the present study was to develop a predictor classifier for response to fluorouracil-based chemotherapy in patients with advanced CRC using microarray gene expression profiles of primary CRC tissues.
Materials and methods
Data processing
The raw microarray data (CEL files) of three datasets [GSE52735 (16), GSE62080 (15) and GSE69657 (17)] and corresponding clinical data were downloaded from the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). The microarray data of the 3 datasets were based on the GPL570 Affymetrix Human Genome U133 Plus 2.0 Array platform (Affymetrix; Thermo Fisher Scientific Inc., Waltham, MA, USA). The GSE52735 set contained 37 advanced CRC samples treated with a fluoropyrimidine-based chemotherapy regimen (specific chemotherapy regimens were not available). A total of 23 of the samples were classified as responders and 14 samples were classified as non-responders to the chemotherapy regimen according to Response Evaluation Criteria in Solid Tumors (RECIST) (18). The GSE62080 dataset contained 21 advanced CRC samples treated with the FOLFIRI regimen. A total of 9 samples were classified as responders and 12 samples were classified as non-responders according to the World Health Organization (WHO) criteria (19). The GSE69657 dataset contained 30 advanced CRC samples treated with the FOLFOX4 regimen. However, the raw microarray data was available for only 16 samples. A total of 7 of these samples were classified as responders and 9 samples were classified as non-responders according to RECIST. Two different evaluation criteria used in these three studies due to long time intervals between the studies, Previous studies have revealed that the RECIST criteria are comparable with the WHO criteria in evaluating the response of solid tumors (20–23). Preprocessing and normalization of the raw data were analyzed using the ‘affy’ (version 3.8) package (24) in R (www.r-project.org; version 3.5), using robust multi-array average for background correction and quantiles for normalization. Kernel and nearest neighbor averaging methods were used to impute the missing values using the ‘impute’ package (bioconductor.org/packages/impute; version 3.8) in R. The ComBat function in the ‘sva’ (version 3.8) package (25) was applied to remove batch effects. If one gene matched multiple probes, the average value of the probes was calculated as the expression of the corresponding gene. To build a robust predictive classifier, the GSE52735 and GSE62080 datasets were used as the training set (n=58), while the GSE69657 dataset was used as the validation set (n=16).
Screening of differentially expressed genes (DEGs) and enrichment analysis
Following preprocessing of the raw expression data, the DEGs between responders and non-responders in the training set were screened using the unpaired t-test in the ‘limma’ (version 3.8) package (26) in R. A DEG was defined as |log2 fold change (FC)|≥0.263 and P<0.05. The Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathway enrichment analyses of DEGs were performed using the ‘clusterProfiler’ (version 3.8) package (27) in R with a cut-off of q<0.01.
Principal component analysis (PCA) prior to and following feature selection using the least absolute shrinkage and selection operator (LASSO) method
The expression values of DEGs in each sample were extracted. The LASSO logistic regression model analysis was performed using the ‘glmnet’ package (CRAN.R-project.org/package=glmnet; version 2.0-16) in R. The LASSO method is used to select optimal features in high-dimensional microarray data with a powerful predictive value and a low correlation between each other to prevent over-fitting (28). In the training set, the LASSO logistic regression model was used to select the optimal predictive markers. PCA using the expression profiles of the DEGs was performed prior to feature selection using the LASSO method. PCA was subsequently performed using the expression profiles of the optimal DEGs identified using by the LASSO method. Samples were plotted in two-dimensional plots across the first two principal components.
Feature selection using Boruta and random forest classifier construction
A lower-dimensional model may reduce costs and is more likely to be used by clinicians (29). Following DEGs selection by the LASSO method, a feature selection was performed using the ‘Boruta’ package (www.jstatsoft.org/article/view/v036i11; version 6.0.0) in R. Boruta is a random forest-based feature selection method, which provides an unbiased and stable selection of important and non-important attributes from an information system. A variable importance (VIMP) measure may be calculated and visualized based on Boruta. In the current study, DEGs selected by Boruta were used to develop a gene-based classifier for response to fluorouracil-based chemotherapy in advanced CRCs. The random forest classifier was developed using the ‘randomForest’ package (CRAN.R-project.org/package=randomForest; version 4.6-14) in R. The validation set (GSE69657) was used to confirm the robustness and transferability of the classifier. The performance of the classifier was assessed by accuracy, sensitivity (Se), specificity (Sp), positive predictive value (PPV), negative predictive value (NPV) and receiver operating characteristic (ROC) curves in the training and validation sets. The ROC curves were drawn and compared using the ‘pROC’ (version 1.13.0) package (30) in R.
Results
DEGs in responders and non-responders and enrichment analysis
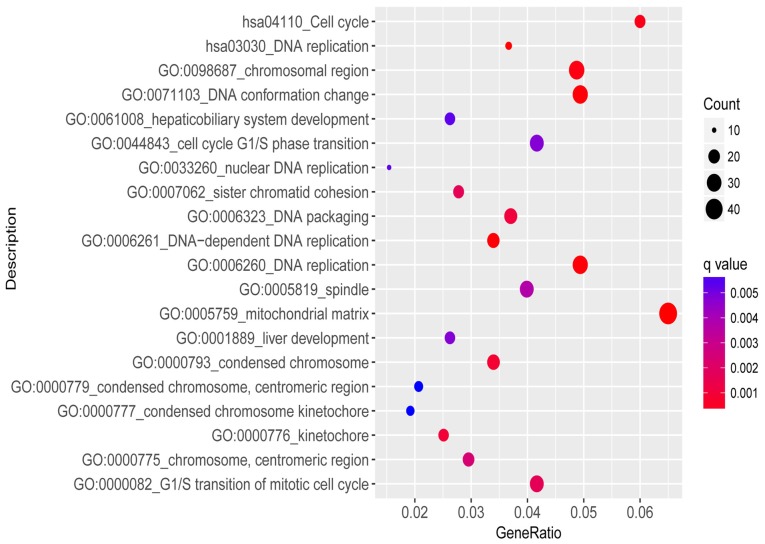
The training set included 32 responders and 26 non-responders. According to the cut-off criteria (|log2FC|≥0.263 and P<0.05), 791 genes were identified as differentially expressed between responders and non-responders. A total of 303 genes were upregulated and 488 genes were downregulated in responders. Functional enrichment analysis revealed that the biological process of DEGs were primarily involved in ‘cell mitosis’, ‘DNA replication’ and ‘cell cycle’ signaling pathways. The results of enrichment analysis are presented in Fig. 1.
Figure 1.
Significantly enriched GO annotation and enriched KEGG pathways of differentially expressed genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
PCA and feature selection using LASSO
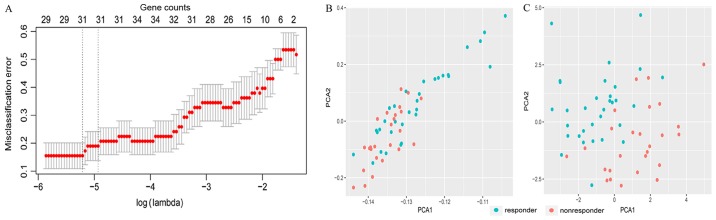
For the first feature selection, LASSO logistic regression was performed using the expression data of DEGs in the training set. The group-wise classifications in 10-fold cross-validations were computed as default. A total of 31 DEGs were identified as optimal genes (Fig. 2A) with non-zero regression coefficients (Table I). Fig. 2B presents the results of PCA prior to feature selection using LASSO and Fig. 2C presents the results of PCA following feature selection using LASSO. As demonstrated in Fig. 2C, responders and non-responders are easily distinguished using the 31 DEGs selected by LASSO.
Figure 2.
LASSO model and principal component analysis. (A) 10-fold cross-validation for tuning parameter selection in the LASSO model. (B) PCA prior to and (C) following LASSO variable reduction. LASSO, least absolute shrinkage and selection operator; PCA, principal component analysis.
Table I.
Overview of the 31 optimal genes.
| Gene | Log2 fold change (Responder/non-responder) | P-value | Coefficients provided by least absolute shrinkage and selection operator | Variable importance provided by Boruta |
|---|---|---|---|---|
| Matrix metallopeptidase 12 | −1.069 | 0.002 | −0.154 | Tentative |
| C-X-C motif chemokine ligand 11 | −1.016 | 0.015 | −0.184 | Rejected |
| Forkhead box P2 | 0.957 | 0.003 | 0.032 | Tentative |
| Small muscle protein X-linked | 0.766 | 0.003 | 0.575 | Confirmed |
| Pleckstrin homology like domain family A member 1 | −0.625 | 0.000 | −0.584 | Confirmed |
| Prostaglandin reductase 2 | −0.602 | 0.000 | −0.792 | Confirmed |
| Chitinase 1 | 0.569 | 0.002 | 0.976 | Confirmed |
| S100 calcium binding protein A2 | −0.541 | 0.039 | −0.091 | Rejected |
| Histone cluster 1 H2B family member c | 0.539 | 0.001 | 0.927 | Confirmed |
| RP1-74M1.3 | −0.515 | 0.005 | −0.023 | Tentative |
| Formin homology 2 domain containing 3 | 0.469 | 0.013 | 0.855 | Confirmed |
| RNA binding motif protein 3 | −0.451 | 0.001 | −0.555 | Tentative |
| Tubulin polymerization promoting protein family member 3 | 0.412 | 0.011 | 0.442 | Rejected |
| Cadherin related family member 2 | 0.411 | 0.047 | 0.913 | Tentative |
| OTUD6B antisense RNA 1 (head to head) | 0.387 | 0.012 | 0.651 | Confirmed |
| Teashirt zinc finger homeobox 1 | 0.384 | 0.004 | 0.347 | Tentative |
| Cholinergic receptor nicotinic β1 subunit | −0.365 | 0.000 | −3.574 | Confirmed |
| Stromal antigen 3-like 4 (pseudogene) | −0.364 | 0.005 | −0.554 | Rejected |
| RPA interacting protein | −0.343 | 0.000 | −0.825 | Confirmed |
| Leucine rich repeat neuronal 1 | −0.334 | 0.017 | −0.331 | Rejected |
| Heparan-α-glucosaminide N-acetyltransferase | 0.334 | 0.006 | 1.374 | Rejected |
| MINDY lysine 48 deubiquitinase 3 | −0.320 | 0.001 | −0.253 | Tentative |
| THAP domain containing 5 | −0.308 | 0.016 | −0.432 | Rejected |
| DNA ligase 4 | 0.298 | 0.002 | 1.692 | Confirmed |
| Zinc finger protein 2 | −0.291 | 0.004 | −1.885 | Tentative |
| ASAP1 intronic transcript 2 | 0.289 | 0.004 | 0.045 | Confirmed |
| Small integral membrane protein 30 | −0.287 | 0.001 | −0.973 | Confirmed |
| c-Maf inducing protein | 0.282 | 0.001 | 0.208 | Confirmed |
| ADAMTS like 2 | 0.278 | 0.005 | 1.088 | Tentative |
| Nucleoporin 133 | −0.273 | 0.011 | −1.718 | Tentative |
| DEAD-box helicase 28 | −0.267 | 0.003 | −0.063 | Tentative |
Features selection using Boruta and construct of the random forest classifier
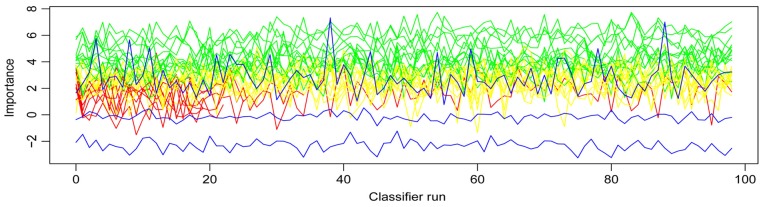
The Boruta function was used to further select features among the 31 DEGs. A total of 13 genes were confirmed as important, 7 genes were rejected and 11 tentative genes remained (Table I)
Fig. 3 presents the variables' importance. These 13 important DEGs included small muscle protein X-linked, pleckstrin homology like domain family A member 1 (PHLDA1), prostaglandin reductase 2 (PTGR2), chitinase 1 (CHIT1), histone cluster 1 H2B family member c, formin homology 2 domain containing 3, OTUD6B antisense RNA 1 (head to head), cholinergic receptor nicotinic β1 subunit (CHRNB1), RPA interacting protein, DNA ligase 4 (LIG4), ASAP1 intronic transcript 2, small integral membrane protein 30 and c-Maf inducing protein. A random forest classifier was constructed using these 13 important DEGs.
Figure 3.
Z score evolution during Boruta run. Green lines correspond to confirmed attributes, yellow to tentative, red to rejected ones; and blue lines correspond to respectively minimal, average and maximal shadow attribute importance.
Performance of the gene-based classifier
The accuracy of the 13-gene classifier was 0.930 in the training set and 0.810 in the validation set. Based on accuracy, Se, Sp, PPV, NPV and area under curve (AUC) values, the sample recognition efficiency of the classifier was high (Table II). ROC curve analysis revealed that the AUC was 1.000 in the training set and 0.873 in the validation set (P=0.227; Fig. 4).
Table II.
Performance of the 13-gene classifier.
| Cohort | Sensitivity | Specificity | Positive predictive value | Negative predictive value | Accuracy | Area under the curve |
|---|---|---|---|---|---|---|
| Training set | 0.970 | 0.960 | 0.910 | 0.960 | 0.930 | 1.000 |
| Validation set | 0.860 | 0.880 | 0.750 | 0.880 | 0.810 | 0.873 |
Figure 4.
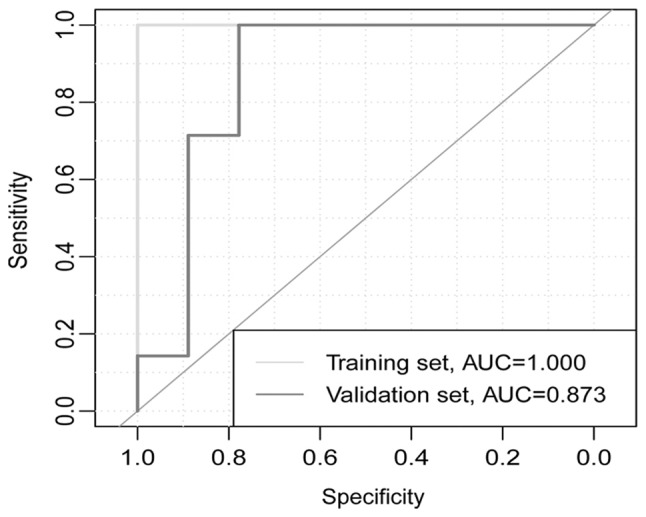
Receiver operating characteristic curves for training and validation sets. AUC, area under the curve.
Discussion
Personalized treatment may improve the treatment outcome of patients with tumors (31). In CRC, the gene expression levels of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) provide the basis for selecting EGFR and VEGF inhibitor combinations (32–36). Monoclonal antibodies against VEGF and EGFR have been approved for treatment of metastatic CRC in combination with 5-FU-based regimens (3). The identification of subsets of patients that respond to specific chemotherapy regimens remains a challenge (3). A previous study demonstrated that tumors with microsatellite instability (MSI) respond well to 5-FU-based therapies; however, further studies are required to substantiate these results (37). Another previously published study suggested that MSI status does not affect the outcome of the treatment (38). Therefore, effective tools for predicting the outcome of chemotherapy are currently lacking. The present study identified 13 genes from 791 DEGs using two feature selection algorithms and developed a 13-gene predictor classifier for response to fluorouracil-based chemotherapy in CRC. The predictor classifier demonstrated high accuracy in the training and validation sets. The training set included two datasets from different centers, and the validation set was from an additional independent center. ROC curve analysis revealed that the AUC was 1.000 in the training set and 0.873 in the validation set, and their difference was not significant (P=0.227). These results suggested that the classifier was robust. The study established a foundation for further research into personalized treatment of CRC.
Previous studies have attempted to identify a single biomarker to predict response to fluorouracil-based chemotherapy in CRC (17,39–42). However, there is currently no single biomarker that is routinely applied in clinical practice. CRC is a heterogeneous disease, which is compounded by changes in the molecular profile of the tumor as it progresses (3). An in vitro study demonstrated that the measurement of multiple, rather than single marker genes, may provide a more accurate assessment of drug response in colon carcinoma (43). Previous studies have been designed to identify a pattern of gene expression capable of predicting response to fluorouracil-based chemotherapy in CRC (15,16). One study identified a set of 14 genes for predicting response to the FOLFIRI regimen based on 21 samples (15), and an expression profile of 7 genes was identified in another study (16). Compared with the two aforementioned studies, the current study performed a comprehensive analysis of more samples (n=58) from two centers and validated the predictor classifier in an independent dataset (n=16). Furthermore, to the best of the authors' knowledge, the current study is the first to construct a random forest classifier to predict response to chemotherapy in CRC. Considering the limited ability of Cox regression analysis to process high-dimensional data (44), it was not performed in the current study. A random forest algorithm was used to construct the classifier, which was subsequently validated with an independent dataset. The results obtained in the current study suggest that the robust classifier developed warrants further investigation.
Functional enrichment analysis revealed that certain DEGs identified in the present study are involved in DNA replication and cell cycle pathways; however, none of the 13 genes were involved in these two signaling pathways. A previous study suggested that PHLDA1 may be associated with CRC progression (45). A previous study demonstrated that PTGR2-knockdown gastric cancer cells rendered them more sensitive to cisplatin and 5-FU compared with the PTGR2-overexpressing cells (46). In addition, two variants of CHIT1, rs61745299 and rs35920428, may increase expression of the gene and have been associated with CRC (47). CHRNB1 may be a biomarker for the detection of relapsed and early relapsed CRC (48). In addition, LIG4 may mediate Wnt signaling-induced radioresistance in CRC (49). With the exception of the aforementioned studies, the association between the 13 genes identified in the current study and CRC or chemotherapy has not been investigated. Therefore, it is not clear whether these genes are causal or merely markers for response to fluorouracil-based chemotherapy in CRC.
Although the current study provides novel insights into the treatment of CRC, it has some limitations. The present study was based on a relatively small sample size; however, it is worth noting that the sample size in our study is relatively large compared with previous studies (15,16). Future studies are required to verify and improve the 13-gene signature in a larger independent cohort of patients.
In conclusion, the current study identified a 13-gene predictor classifier for the response to fluorouracil-based chemotherapy in patients with advanced CRC.
Acknowledgements
Not applicable.
Funding
This research was supported by the Youth Science Foundation of Guangxi Medical University (grant no. GXMUYSF 201716) and the Basic Ability Enhancement Program for Young and Middle-aged Teachers of Guangxi (grant no. 2017KY0120).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
YFL designed the study and revised the manuscript. ZHG and QYZ analyzed the data and wrote the manuscript. YL, ZHX, ZH, and ZCC assisted with analyzing the data and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, ESMO Guidelines Working Group Metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii1–iii9. doi: 10.1093/annonc/mdu260. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.de Gramont A, Bosset JF, Milan C, Rougier P, Bouché O, Etienne PL, Morvan F, Louvet C, Guillot T, François E, Bedenne L. Randomized trial comparing monthly low-dose leucovorin and fluorouracil bolus with bimonthly high-dose leucovorin and fluorouracil bolus plus continuous infusion for advanced colorectal cancer: A French intergroup study. J Clin Oncol. 1997;15:808–815. doi: 10.1200/JCO.1997.15.2.808. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 6.Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: A randomized GERCOR study. J Clin Oncol. 2004;22:229–237. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 7.Yamazaki K, Nagase M, Tamagawa H, Ueda S, Tamura T, Murata K, Eguchi Nakajima T, Baba E, Tsuda M, Moriwaki T, et al. Randomized phase III study of bevacizumab plus FOLFIRI and bevacizumab plus mFOLFOX6 as first-line treatment for patients with metastatic colorectal cancer (WJOG4407G) Ann Oncol. 2016;27:1539–1546. doi: 10.1093/annonc/mdw581.049. [DOI] [PubMed] [Google Scholar]
- 8.Petrelli F, Borgonovo K, Cabiddu M, Ghilardi M, Lonati V, Maspero F, Sauta MG, Beretta GD, Barni S. FOLFIRI-bevacizumab as first-line chemotherapy in 3500 patients with advanced colorectal cancer: A pooled analysis of 29 published trials. Clin Colorectal Cancer. 2013;12:145–151. doi: 10.1016/j.clcc.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Saltz LB, Clarke S, Díaz-Rubio E, Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: A randomized phase III study. J Clin Oncol. 2008;26:2013–2019. doi: 10.1200/JCO.2007.14.9930. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, McLeod HL, Atkins D. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 11.Eschrich S, Yang I, Bloom G, Kwong KY, Boulware D, Cantor A, Coppola D, Kruhøffer M, Aaltonen L, Orntoft TF, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 12.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 13.Wen WH, Bernstein L, Lescallett J, Beazer-Barclay Y, Sullivan-Halley J, White M, Press MF. Comparison of TP53 mutations identified by oligonucleotide microarray and conventional DNA sequence analysis. Cancer Res. 2000;60:2716–2722. [PubMed] [Google Scholar]
- 14.Iwao-Koizumi K, Matoba R, Ueno N, Kim SJ, Ando A, Miyoshi Y, Maeda E, Noguchi S, Kato K. Prediction of docetaxel response in human breast cancer by gene expression profiling. J Clin Oncol. 2005;23:422–431. doi: 10.1200/JCO.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 15.Del Rio M, Molina F, Bascoul-Mollevi C, Copois V, Bibeau F, Chalbos P, Bareil C, Kramar A, Salvetat N, Fraslon C, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 2007;25:773–780. doi: 10.1200/JCO.2006.07.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estevez-Garcia P, Rivera F, Molina-Pinelo S, Benavent M, Gómez J, Limón ML, Pastor MD, Martinez-Perez J, Paz-Ares L, Carnero A, Garcia-Carbonero R. Gene expression profile predictive of response to chemotherapy in metastatic colorectal cancer. Oncotarget. 2015;6:6151–6159. doi: 10.18632/oncotarget.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Lu X, Chi P, Pan J. Identification of HOXB8 and KLK11 expression levels as potential biomarkers to predict the effects of FOLFOX4 chemotherapy. Future Oncol. 2013;9:727–736. doi: 10.2217/fon.13.25. [DOI] [PubMed] [Google Scholar]
- 18.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 19.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–214. doi: 10.1002/1097-0142(19810101)47:1<207::AID-CNCR2820470134>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Aras M, Erdil TY, Dane F, Gungor S, Ones T, Dede F, Inanir S, Turoglu HT. Comparison of WHO RECIST 1.1, EORTC, and PERCIST criteria in the evaluation of treatment response in malignant solid tumors. Nucl Med Commun. 2016;37:9–15. doi: 10.1097/MNM.0000000000000401. [DOI] [PubMed] [Google Scholar]
- 21.Khokher S, Qureshi MU, Chaudhry NA. Comparison of WHO and RECIST criteria for evaluation of clinical response to chemotherapy in patients with advanced breast cancer. Asian Pac J Cancer Prev. 2012;13:3213–3218. doi: 10.7314/APJCP.2012.13.7.3213. [DOI] [PubMed] [Google Scholar]
- 22.Choi JH, Ahn MJ, Rhim HC, Kim JW, Lee GH, Lee YY, Kim IS. Comparison of WHO and RECIST criteria for response in metastatic colorectal carcinoma. Cancer Res Treat. 2005;37:290–293. doi: 10.4143/crt.2005.37.5.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JO, Lee SI, Song SY, Kim K, Kim WS, Jung CW, Park YS, Im YH, Kang WK, Lee MH, et al. Measuring response in solid tumors: Comparison of RECIST and WHO response criteria. Jpn J Clin Oncol. 2003;33:533–537. doi: 10.1093/jjco/hyg093. [DOI] [PubMed] [Google Scholar]
- 24.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 25.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuyle PJ, Prenen H. Current and future biomarkers in the treatment of colorectal cancer. Acta Clin Belg. 2017;72:103–115. doi: 10.1080/17843286.2016.1262996. [DOI] [PubMed] [Google Scholar]
- 30.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stintzing S. Personalized treatment for colorectal carcinomas. Dtsch Med Wochenschr. 2017;142:1652–1659. doi: 10.1055/s-0043-108467. (In German) [DOI] [PubMed] [Google Scholar]
- 32.Gustavsson B, Carlsson G, Machover D, Petrelli N, Roth A, Schmoll HJ, Tveit KM, Gibson F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin Colorectal Cancer. 2015;14:1–10. doi: 10.1016/j.clcc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Iwamoto S, Takahashi T, Tamagawa H, Nakamura M, Munemoto Y, Kato T, Hata T, Denda T, Morita Y, Inukai M, et al. FOLFIRI plus bevacizumab as second-line therapy in patients with metastatic colorectal cancer after first-line bevacizumab plus oxaliplatin-based therapy: The randomized phase III EAGLE study. Ann Oncol. 2015;26:1427–1433. doi: 10.1093/annonc/mdv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bazarbashi S, Aljubran A, Alzahrani A, Mohieldin A, Soudy H, Shoukri M. Phase I/II trial of capecitabine, oxaliplatin, and irinotecan in combination with bevacizumab in first line treatment of metastatic colorectal cancer. Cancer Med. 2015;4:1505–1513. doi: 10.1002/cam4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 36.Stremitzer S, Sebio A, Stintzing S, Lenz HJ. Panitumumab safety for treating colorectal cancer. Expert Opin Drug Saf. 2014;13:843–851. doi: 10.1517/14740338.2014.915024. [DOI] [PubMed] [Google Scholar]
- 37.Saridaki Z, Souglakos J, Georgoulias V. Prognostic and predictive significance of MSI in stages II/III colon cancer. World J Gastroenterol. 2014;20:6809–6814. doi: 10.3748/wjg.v20.i22.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webber EM, Kauffman TL, O'Connor E, Goddard KA. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer. 2015;15:156. doi: 10.1186/s12885-015-1093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi R, Kitao H, Fujinaka Y, Yamashita N, Iimori M, Tokunaga E, Yamashita N, Morita M, Kakeji Y, Maehara Y. FANCJ expression predicts the response to 5-fluorouracil-based chemotherapy in MLH1-proficient colorectal cancer. Ann Surg Oncol. 2012;19:3627–3635. doi: 10.1245/s10434-012-2349-8. [DOI] [PubMed] [Google Scholar]
- 40.Simmer F, Venderbosch S, Dijkstra JR, Vink-Börger EM, Faber C, Mekenkamp LJ, Koopman M, De Haan AF, Punt CJ, Nagtegaal ID. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget. 2015;6:22996–23007. doi: 10.18632/oncotarget.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molina-Pinelo S, Carnero A, Rivera F, Estevez-Garcia P, Bozada JM, Limon ML, Benavent M, Gomez J, Pastor MD, Chaves M, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer. 2014;14:656. doi: 10.1186/1471-2407-14-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Candy PA, Phillips MR, Redfern AD, Colley SM, Davidson JA, Stuart LM, Wood BA, Zeps N, Leedman PJ. Notch-induced transcription factors are predictive of survival and 5-fluorouracil response in colorectal cancer patients. Br J Cancer. 2013;109:1023–1030. doi: 10.1038/bjc.2013.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63:8791–8812. [PubMed] [Google Scholar]
- 44.Kursa MB, Rudnicki WR. Feature selection with the boruta package. J Stat Soft. 2010;36:1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 45.Zhang Z, Huang J. Intestinal stem cells-types and markers. Cell Biol Int. 2013;37:406–414. doi: 10.1002/cbin.10049. [DOI] [PubMed] [Google Scholar]
- 46.Chang EY, Tsai SH, Shun CT, Hee SW, Chang YC, Tsai YC, Tsai JS, Chen HJ, Chou JW, Lin SY, Chuang LM. Prostaglandin reductase 2 modulates ROS-mediated cell death and tumor transformation of gastric cancer cells and is associated with higher mortality in gastric cancer patients. Am J Pathol. 2012;181:1316–1326. doi: 10.1016/j.ajpath.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Li FF, Yan P, Zhao ZX, Liu Z, Song DW, Zhao XW, Wang XS, Wang GY, Liu SL. Polymorphisms in the CHIT1 gene: Associations with colorectal cancer. Oncotarget. 2016;7:39572–39581. doi: 10.18632/oncotarget.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang YT, Yeh YS, Ma CJ, Huang CW, Tsai HL, Huang MY, Cheng TL, Wang JY. Optimization of a multigene biochip for detection of relapsed and early relapsed colorectal cancer. J Surg Res. 2017;220:427–437. doi: 10.1016/j.jss.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 49.Jun S, Jung YS, Suh HN, Wang W, Kim MJ, Oh YS, Lien EM, Shen X, Matsumoto Y, McCrea PD, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994. doi: 10.1038/ncomms10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.