Abstract
Notch signaling is well-known for its role in regulating cell self-renewal and differentiation. Within the cancer research field, it has been identified that dysregulated Notch signaling is involved directly with various types of cancer. Although Notch signaling is generally considered as oncogenic, it sometimes acts as a tumor suppressor, highlighting the complexity of the role of Notch in cancer. A number of studies have associated Notch signaling components with ovarian cancer, but the underlying molecular mechanisms are not well-elucidated. In the present study, the roles of main components of Notch signaling in ovarian cancer were systematically analyzed through large data portals, including Prediction of Clinical Outcomes from Genomic Profiles, Gene Expression across Normal and Tumor tissue, CSIOVDB, Broad Institute Cancer Cell Line Encyclopedia and cBioPortal. Upregulated expression of proteins in the Notch signaling pathway components in ovarian cancer was identified to be generally associated with poor overall and disease-free survival time, and more advanced cancer stages. In addition, Notch components were enriched in ovarian cancer tissues and cell lines. These results led to a proposed neurogenic locus notch homolog protein (NOTCH)2/NOTCH3/Delta-like protein 3/Mastermind-like protein 1/a disintegrin and metalloproteinase domain-containing protein 17 network. Anticancer drugs, developed to target this network, may have high specificity in treating Notch-associated ovarian cancer.
Keywords: neurogenic locus notch homolog protein 2, neurogenic locus notch homolog protein 3, Delta-like protein 3, Mastermind-like protein 1, a disintegrin and metalloproteinase domain-containing protein 17, Notch signaling, ovarian cancer
Introduction
Notch signaling is a highly conserved signaling pathway across numerous species, and is well-known for its role in the regulation of cell self-renewal and differentiation (1,2). The canonical Notch pathway applies a cell-to-cell communication strategy to activate its signaling. In brief, ligands from a neighboring cell bind to the Notch receptors of a target cell, resulting in two rounds of proteolytic cleavage (S2 and S3 cleavage). The S2 cleavage is performed by a disintegrin and metalloproteinase domain-containing (ADAM) family of proteins to break the ligand-receptor bonds. The S3 cleavage is regulated by the γ-secretase complex in order to release the Notch intracellular domain (NICD). The NICD is then translocated into the cell nucleus, where it interacts with the centromere-binding protein 1/suppressor of hairless/DNA-binding protein LAG-1 (recombining binding protein suppressor of hairless in mammals) transcription factor and co-activator Mastermind [Mastermind-like protein (MAML)1–3 in mammals] to activate the downstream targets (Fig. 1) (1,3,4). In mammals, ligands include two protein families, Jagged-1 and −2, and Delta-like proteins (DLL)1, 3 and 4. The Notch receptor has four paralogs, neurogenic locus notch homolog proteins (NOTCH)1-4, the ADAMs include ADAM10 and 17, and the γ-secretase complex contains presenilin (PSEN)1 and 2 and nicastrin (NCSTN) (4,5).
Figure 1.
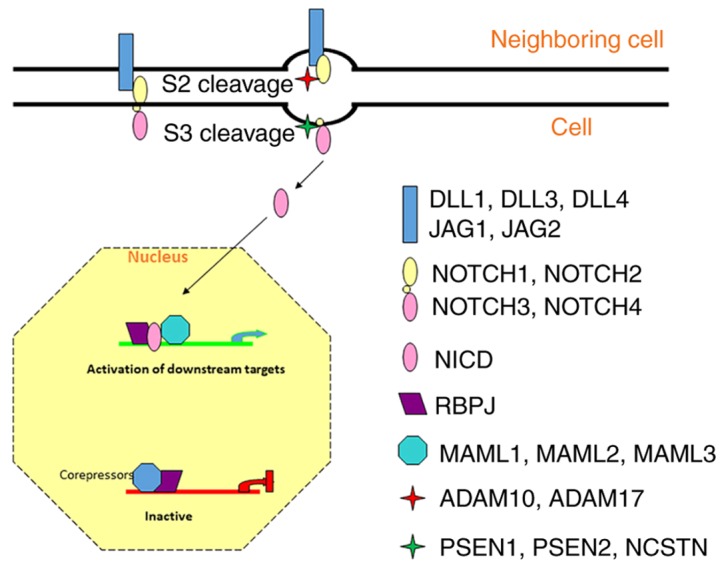
Schematic diagram of Notch signaling and its principal components. DLL, Delta-like protein; JAG, Jagged; NICD, Notch intracellular domain; MAML, Mastermind-like protein; ADAM, a disintegrin and metalloproteinase; PSEN, presenilin; NCSTN, nicastrin; RBPJ, recombination signal binding protein for immunoglobulin kappa J region.
Studies have indicated the involvement of dysregulated Notch signaling in numerous types of cancer. Although Notch signaling has been generally considered as oncogenic, it sometimes acts as a tumor suppressor, highlighting the complexity of the role of Notch in cancer (2,6–8). In addition, Notch signaling has been identified to mediate the immune system, including generation and maintenance of hematopoietic stem cells, lymphocyte development and peripheral T-cell activation (9,10). Notch also enhances the proliferation and angiogenesis of endothelial progenitor cells and mesenchymal stem cells (11). All these types of cells can subsequently influence tumor growth, highlighting the diverse regulatory roles of Notch in cancer development. In ovarian cancer research, a number of studies have reported the critical roles of key Notch components in cancer progression and metastasis. For example, NOTCH1 is associated with tumor differentiation status (12), NOTCH2 is associated with tumor histological subtypes (13), NOTCH3 promotes cancer progression and is associated with poor patient outcome (14,15), DLL4 is overexpressed in tumor samples and acts as an indicator of poor survival (16), and ADAM10 and PSEN1 assist the in vitro nuclear translocation of the L1 cell-adhesion molecule, an indicator of poor prognosis in ovarian cancer (17).
Despite the increasing evidence indicating pivotal roles of Notch signaling components in ovarian cancer, the underlying molecular mechanisms and the functional Notch network remain to be elucidated. In addition, any results obtained from in vitro cell culture experiments may not accurately reflect the clinical reality. In the present study, clinically relevant information was obtained from large data portals, and the roles of main components of the Notch pathway in ovarian cancer were systematically analyzed to demonstrate their association with patient survival time, cancer tissue gene expression enrichment and cancer stages. Finally, a signaling network was proposed for specific drug targeting.
Materials and methods
Prediction of Clinical Outcomes from Genomic Profiles (PRECOG)
The PRECOG portal (precog.stanford.edu) (18) provided processed and integrated data from 166 cancer expression datasets, including mRNA expression levels and clinical outcomes. Particular focus was on the Meta-Z analysis, which offered a Z-score based on large datasets to indicate patient overall survival. Positive and negative values indicated poor and good survival, respectively. Z-scores of the Notch-associated genes were obtained from the PRECOG database (last analysis update, September 22, 2017), and analyzed in Excel 2013 (Microsoft Corporation, Redmond, MA, USA).
Gene Expression across Normal and Tumor tissue (GENT)
The GENT portal (medical-genome.kribb.re.kr/GENT) (19) maintains a collection of 16,400 samples (U133A, 241 datasets), standardizes them and offers gene expression data across diverse tissues. Gene expression data of the Notch-associated genes from normal and ovarian cancer tissues were obtained from the GENT database (last analysis update, September 23, 2017), and analyzed in Excel 2013 and GraphPad Prism (version 7.00; GraphPad Software, Inc., La Jolla, CA, USA).
CSIOVDB
CSIOVDB (csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html), a microarray database of ovarian cancer data (20), provided Kaplan-Meier survival plots comparing the lower and upper quartiles of gene expression. In addition, CSIOVDB was interrogated to investigate the association between Notch-associated genes and ovarian cancer stages. CSIOVDB has an open license to freely use its images and services.
cBioPortal
cBioPortal (www.cbioportal.org) (21,22) helps to visualize and analyze large-scale cancer genomic data. The genes NOTCH2, NOTCH3, DLL3, MAML1 and ADAM17 were inputted into the ovarian serous cystadenocarcinoma dataset (23) to search for genetic alterations and signaling networks. For the signaling network, the inputted genes (referred to as seed nodes with a thick border) were presented in the context of biological interactions derived from a number of public pathway databases (the Human Protein Reference Database hprd.org, the Reactome Pathway Database, reactome.org; the Pathway Interaction Database, this database has been retired; however, its data are now available for the research community via the NDEx database (ndexbio.org); and the Cancer Cell Map, netpath.org). All the genes in the signaling network were color-coded according to multi-dimensional genomic data derived from the ovarian serous cystadenocarcinoma dataset.
Broad Institute Cancer Cell Line Encyclopedia (CCLE)
CCLE (portals.broadinstitute.org/ccle) provides detailed gene expression information of 947 human cancer cell lines, including 65 ovarian cancer cell lines (24). The present study accessed CCLE data to obtain the mRNA levels of Notch signaling components in all the available ovarian cancer cell lines.
Statistical analysis
The bar charts were constructed using Excel 2013, and the box and whisker plots were produced using GraphPad Prism (version 7.00). The P-values for the GENT data were generated using a permutation test (25). The normality assumption suffers when the sample size is small. To overcome this obstacle, a recently developed permutation test was performed. In a two-sample (Ovary-Cancer, Ovary-Normal) scenario, the test worked by first determining the difference between the observed means. The data were then permuted, meaning that the cases were reallocated to the two groups in all possible combinations (with the constraint that the group sizes were held constant at their observed values). The statistic of interest (mean difference) was then calculated for each permutation and a distribution was created. It was further assessed where the original statistic (group-wise mean difference) fell in this distribution. If the original statistic fell in the tails, this indicated that the two samples originated from two different populations. For the CSIOVDB data, Tan et al (20) initially performed a Mann-Whitney test and Spearman's correlation analysis to assess pairwise and binary significance. To control the familywise error rate, the Bonferroni correction (26) was further applied by us for the data in Table SI. Since there are 10 tests in each table, instead of setting the critical P-value for significance to 0.05, a lower critical value of 0.005 was used (0.05/the number of tests). In Table SI, only individual tests with P<0.005 were considered to be significant; these are highlighted in bold.
Results
Upregulated expression of Notch-associated genes in ovarian cancer is associated with poor overall survival time
To systematically investigate the association between the Notch-associated components and patient survival, the PRECOG analysis was performed. PRECOG includes ~18,000 cases in 166 different cancer datasets; however, the ovarian cancer datasets were the main focus of the present study. A number of principal components of the Notch pathway were associated with poor overall survival in ovarian cancer (Fig. 2). Among these, NOTCH2, NOTCH3, DLL1, DLL3, DLL4, MAML1, ADAM17 and NCSTN were associated with poor survival (Z-score cutoff value, 1.00), indicating that these eight genes have a greater probability of participating in ovarian cancer development. Notably, two genes, NOTCH4 and MAML2, were associated with improved survival (Z-score cutoff value, 1.00), highlighting the complexity of Notch signaling. In general, these results demonstrated that the upregulation of Notch-associated genes in ovarian cancer was predominantly associated with poor overall survival time.
Figure 2.
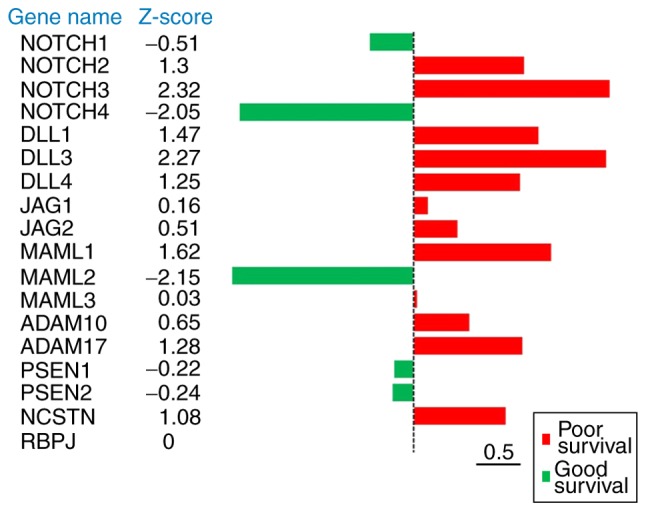
Expression of the principal Notch components in ovarian cancer is associated with poor overall survival. Using the PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) database (precog.stanford.edu), the survival z-scores were obtained for the principal Notch components in ovarian cancer. DLL, Delta-like protein; JAG, Jagged; MAML, Mastermind-like protein; ADAM, a disintegrin and metalloproteinase; PSEN, presenilin; NCSTN, nicastrin; RBPJ, recombination signal binding protein for immunoglobulin kappa J region.
mRNA expression levels of Notch-associated genes are higher in ovarian cancer tissues compared with those in normal ovaries
To further investigate the underlying molecular mechanisms leading to poor survival, the mRNA expression levels of Notch-associated genes in normal ovaries and ovarian cancer tissues were investigated further. The U133A dataset (9,258 cancer tissues and 4,087 normal tissues) from the GENT portal, enabled access to standardized mRNA expression in ovarian tissues, including 9 normal ovaries and 341 ovarian cancer tissues (19). The analysis revealed that higher expression of Notch-associated genes, including NOTCH2, NOTCH3, ADAM17 and NCSTN, was observed in cancer tissues compared with healthy ovaries (Fig. 3). DLL1 and DLL4 data were not available from the GENT dataset.
Figure 3.

mRNA expression levels of Notch-associated genes in normal ovaries and corresponding ovarian cancer tissues. The mRNA expression (log2) data based on the dataset U133A were obtained from the Gene Expression across Normal and Tumor tissue (GENT) portal and were analyzed using GraphPad Prism (version 7.00). P-values are indicated. Ovary-N, normal ovaries; Ovary-C, ovarian cancer; DLL3, Delta-like protein 3; MAML1, Mastermind-like protein 1; ADAM17, a disintegrin and metalloproteinase 17; NCSTN, nicastrin. The box and whisker plots were produced using GraphPad Prism, and the default setting plotted the box from the 10th percentile to the 90th percentile.
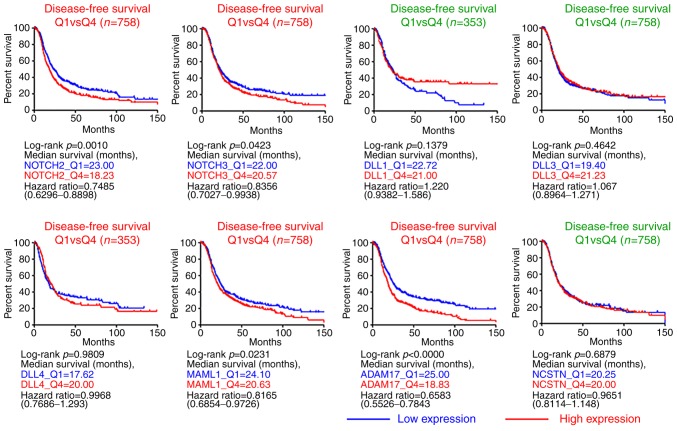
High levels of certain Notch-associated genes in ovarian cancer are associated with poor overall and disease-free survival times
The PRECOG and GENT data analyses indicated the differential expression of Notch-associated factors components in the ovarian cancer tissues, as well their association with poor survival times. A third data portal, CSIOVDB, maintaining a microarray database of ovarian cancer samples (20), was used in order to validate these results. Kaplan-Meier overall survival plots based on the microarray gene expression data indicated that high levels of NOTCH2, NOTCH3, and ADAM17 expression in samples from patients with ovarian cancer were associated with poor overall survival (Fig. 4). In addition, high levels of NOTCH2, NOTCH3, MAML1 and ADAM17 expression were associated with poor disease-free survival (Fig. 5). Together, these results further strengthened the claim that NOTCH2, NOTCH3, MAML1 and ADAM17 serve an important role in ovarian cancer, particularly pertaining to the prognosis of the patient. However, we noticed that high levels of DLL3 were generally associated with poor overall survival, yet were not statistically significant.
Figure 4.
High levels of Notch components in ovarian cancer are generally associated with poor overall survival. Kaplan-Meier overall survival plots of NOTCH2, NOTCH3, DLL1, DLL3, DLL4, MAML1, ADAM17 and NCSTN expression in patients with ovarian cancer were based on the microarray gene expression database CSIOVDB. DLL, Delta-like protein; MAML1, Mastermind-like protein 1; ADAM17, a disintegrin and metalloproteinase 17; NCSTN, nicastrin; Q, quartile. The red text for the plot titles suggested high levels of the genes were generally associated with poor overall survival, while the green text indicated association with either good survival or no clear trend.
Figure 5.
High levels of Notch components in ovarian cancer are generally associated with poor disease-free survival. Kaplan-Meier disease-free survival plots of NOTCH2, NOTCH3, DLL1, DLL3, DLL4, MAML1, ADAM17 and NCSTN expression in patients with ovarian cancer were based on the microarray gene expression database CSIOVDB. DLL, Delta-like protein; MAML1, Mastermind-like protein 1; ADAM17, a disintegrin and metalloproteinase 17; NCSTN, nicastrin; Q, quartile. The red text for the plot titles suggested high levels of the genes were generally associated with poor disease-free survival, while the green text indicated association with either good survival or no clear trend.
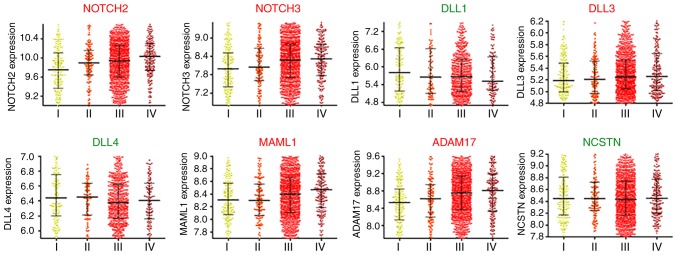
High levels of certain Notch-associated factors in ovarian cancer are associated with advanced cancer stages
In clinical practice, staging is frequently applied to define the extent of the spread of cancer. Ovarian cancer staging is often determined by the system of the International Federation of Gynecology and Obstetrics, which is dependent on the extent of the primary tumor, the metastasis to nearby lymph nodes and the existence of distant metastasis (27–29). Staging is critical for the prognosis of the patient, as different cancer stages are treated differently. From the CSIOVDB data portal, the upregulation of several genes, including NOTCH2, NOTCH3, DLL3, MAML1 and ADAM17, was identified to be associated with increased cancer stages (Fig. 6 and Table SI). In addition, statistical analysis confirmed that the expression levels of NOTCH2, NOTCH3, MAML1 and ADAM17 in advanced stages (stages III and IV) were significantly higher compared with those of stage I (Table SI). These results underscored the significance of these Notch-associated principal components in ovarian cancer progression, and physicians could potentially apply them as molecular parameters during clinical practice.
Figure 6.
High levels of Notch components in ovarian cancer are generally associated with advanced cancer stages. NOTCH2, NOTCH3, DLL1, DLL3, DLL4, MAML1, ADAM17 and NCSTN expression in different cancer stages of ovarian cancer patients were based on the microarray gene expression database CSIOVDB. DLL, Delta-like protein; MAML1, Mastermind-like protein 1; ADAM17, a disintegrin and metalloproteinase 17; NCSTN, nicastrin. The red text for the plot titles suggested high levels of the genes were generally associated with advanced cancer stages, while the green text indicated association with early stages or no clear trend.
NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 network and its associated genes
On the basis of the survival data regarding gene expression levels in tissues and different cancer stages, NOTCH2, NOTCH3, DLL3, MAML1 and ADAM17 were determined to be the top five most relevant genes in Notch-associated ovarian cancer. CCLE data further indicated that the mRNA levels of NOTCH2, NOTCH3, DLL3, MAML1 and ADAM17 are commonly upregulated in ovarian cancer cell lines, including NIHOVCAR3 (Table SII). cBioPortal (21,22) covered the Ovarian Serous Cystadenocarcinoma (The Cancer Genome Atlas) dataset (23), which included ovarian cancer patient data of genetic alterations. In total, 212/489 (43%) sequenced cases/patients had altered mRNA levels in at least one of the five genes NOTCH2, NOTCH3, DLL3, MAML1 and ADAM17 (Fig. S1), indicating the essential roles of these genes in ovarian cancer. To elucidate the signaling factors associated with the NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 axis, a network was created including the five core genes and 50 most frequently altered neighboring genes (Fig. 7 and Table SIII). The most frequently altered neighboring genes are the interacting genes in the same pathway which also have the highest mutation rates. High expression of 17/49 genes (BTC gene was not available in the CSIOVDB database) was generally associated with poor survival, whereas low expression of 9/49 genes was generally associated with poor survival (Table SIV). This network can be utilized in pharmaceutical research to develop more precise drug treatment, targeting the core genes to curb ovarian cancer progression and metastasis.
Figure 7.

NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 network and its associated 50 most frequently altered neighboring genes. Datasets were analyzed using cBioPortal. DLL3, Delta-like protein 3; MAML1, Mastermind-like protein 1; ADAM17, a disintegrin and metalloproteinase 17. For the signaling network, the inputted genes were referred to as seed nodes with a thick border. Higher intensity of the red circles indicates higher mutation rate of the gene. All the genes in the signaling network were color-coded with according to multi-dimensional genomic data derived from the ovarian serous cystadenocarcinoma dataset. Brown color: ‘In Same Component’ indicating connected genes are involved in the same biological component, such as a complex. Blue color: ‘Reacts With’ indicating one gene reacts with another one. Green Color: ‘State Change’ indicating that the gene causes a state change, such as a phosphorylation change, of another gene.
Discussion
The results of the present study confirmed the involvement of Notch signaling in ovarian cancer. The majority of Notch key components were associated with poor survival. However, NOTCH1, NOTCH4 and MAML2 were identified to be associated with good survival. Consistent with the PRECOG data, the mRNA expression levels of NOTCH1 and NOTCH4 were lower in ovarian cancer tissues compared with those in normal ovaries (Fig. S2). In the CSIOVDB data portal, the downregulation of NOTCH1 and NOTCH4 were associated with increased cancer stages (Fig. S3). It has long been a matter of debate whether Notch signaling is oncogenic or tumor-suppressive; the results of the present study indicating opposite effects of NOTCH1–4 further add to the complexity of Notch receptors. The detailed mechanisms of cellular differences are not yet well-understood. One of the main differences among Notch receptors is in the C-terminus of the NICD (2). Since the NICD is the essential functional transcription factor to regulate the expression of downstream targets, we hypothesize that the differences in the termini may yield contrasting signaling outputs. The function complexity of Notch receptors has been associated with the interactions with other signaling pathways (30), and the NICD is known to serve an important role in integrating signals (31). Although a lot is known about the functional differences among NOTCH1-4, MAM1–3 and PSEN1-2, the underlying molecular mechanisms of the transcriptional activation remain unknown. All MAMLs contain an N-terminal basic domain, two middle acidic domains and a C-terminus. Two transcription-activating domain (TAD) regions in the acidic domains are critical for transcriptional activation, and MAML1 possesses a unique p300-binding site in the TAD1 region (32). Although paralogs of NICD and MAMLs are almost structurally identical, the subtle differences may create a totally functionally distinct activator complex, eventually leading to different patient outcomes. We propose a clinical strategy to overexpress the NOTCH4 and MAML2 in patients with cancer, which may inhibit cancer progression and metastasis. However, precautions are required, as an in vitro cell culture study using reverse transcription-polymerase chain reaction analyses identified that high levels of NOTCH4 were observed in cluster of differentiation 24+ ovarian cancer stem cells (33).
In the present study, NOTCH2, NOTCH3, DLL3, MAML1 and ADAM17 were identified to be the top five most relevant genes in Notch-associated ovarian cancer. They were enriched in cancer tissue samples associated with patient survival and cancer stages, indicating that they had pivotal roles in ovarian cancer. Issues such as tumor complexity and heterogeneity may bring confounders to undermine the statistically significant differences identified in the present study. Although the differential expression of these genes was not sufficiently marked, if these genes can be associated with poor patient outcomes across multiple-platform databases, they should be taken into serious consideration. To elucidate the signaling factors associated with the NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 axis, a signaling network was provided to include the 50 most frequently altered neighboring genes. Our proposed genetic network can be an ideal target in pharmaceutical drug development to inhibit ovarian cancer progression and metastasis, thus improving patient survival. A previous study indicated that tarextumab, a NOTCH2/NOTCH3 antagonist, successfully inhibited patient-derived xenograft tumor growth (34), confirming an effective treatment strategy targeting the main components of Notch. In addition, the NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 network can also be used in mathematical models to identify potential drug therapies. For instance, a study in breast cancer utilized the gene nodes from published studies to develop stochastic Boolean network models, which can quantitative assess the vulnerability of each gene node for potential drug targets (35).
We propose that the NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 network is critical in ovarian cancer. It is hoped that the results of the present will shed light on the targeted therapy against Notch activation in ovarian cancer and help to focus future efforts on designing effective drugs.
Supplementary Material
Acknowledgements
The authors are grateful for the research assistance provided by Georgia Southern University, Statesboro, GA, USA. The abstract was presented at the 59th Annual Drosophila Research Conference April 11–15, 2018 in Philadelphia, PA, USA (abstract 620P).
Funding
The present study was supported by Start-Up Funding, and College of Science and Mathematics Research Grant from Georgia Southern University, the Visiting Scholar Foundation of Key Laboratory of Biorheological Science and Technology (Chongqing University), Ministry of Education (grant no. CQKLBST-2016-004) and the Ovarian Cancer Research Fund Ann and Sol Shreiber Mentored Investigator Award.
Availability of data and materials
The data generated during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DJ analyzed and interpreted the data, and wrote the manuscript. JU, QXu and QXie participated in the analysis and contributed in writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 2.Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: molecular basis of cell context dependency. Eur J Cell Biol. 2011;90:572–581. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Jia D, Tamori Y, Pyrowolakis G, Deng WM. Regulation of broad by the Notch pathway affects timing of follicle cell development. Dev Biol. 2014;392:52–61. doi: 10.1016/j.ydbio.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zacharioudaki E, Bray SJ. Tools and methods for studying Notch signaling in Drosophila melanogaster. Methods. 2014;68:173–182. doi: 10.1016/j.ymeth.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaleco AC, Neves H, Hooijberg E, Gameiro P, Clode N, Haury M, Henrique D, Parreira L. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: Switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nature reviews. Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 8.Yuan X, Wu H, Xu H, Xiong H, Chu Q, Yu S, Wu GS, Wu K. Notch signaling: An emerging therapeutic target for cancer treatment. Cancer Lett. 2015;369:20–27. doi: 10.1016/j.canlet.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 9.Yu S, Liu C, Li L, Tian T, Wang M, Hu Y, Yuan C, Zhang L, Ji C, Ma D. Inactivation of Notch signaling reverses the Th17/Treg imbalance in cells from patients with immune thrombocytopenia. Lab Invest. 2015;95:157–167. doi: 10.1038/labinvest.2014.142. [DOI] [PubMed] [Google Scholar]
- 10.Nagase H, Nakayama K. γ-Secretase-regulated signaling typified by Notch signaling in the immune system. Curr Stem Cell Res Ther. 2013;8:341–356. doi: 10.2174/1574888X113089990054. [DOI] [PubMed] [Google Scholar]
- 11.Liang T, Zhu L, Gao W, Gong M, Ren J, Yao H, Wang K, Shi D. Coculture of endothelial progenitor cells and mesenchymal stem cells enhanced their proliferation and angiogenesis through PDGF and Notch signaling. FEBS Open Bio. 2017;7:1722–1736. doi: 10.1002/2211-5463.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Wang J, Wang L, Wu L, Xin X. Notch1 expression correlates with tumor differentiation status in ovarian carcinoma. Med Oncol. 2010;27:1329–1335. doi: 10.1007/s12032-009-9384-8. [DOI] [PubMed] [Google Scholar]
- 13.Galic V, Shawber CJ, Reeves C, Shah M, Murtomaki A, Wright J, Herzog T, Tong GX, Kitajewski J. NOTCH2 expression is decreased in epithelial ovarian cancer and is related to the tumor histological subtype. Pathol Discov. 2013;1:4. doi: 10.7243/2052-7896-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W, Liu T, Ivan C, Sun Y, Huang J, Mangala LS, Miyake T, Dalton HJ, Pradeep S, Rupaimoole R, et al. Notch3 pathway alterations in ovarian cancer. Cancer Res. 2014;74:3282–3293. doi: 10.1158/0008-5472.CAN-13-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung SG, Kwon YD, Song JA, Back MJ, Lee SY, Lee C, Hwang YY, An HJ. Prognostic significance of Notch 3 gene expression in ovarian serous carcinoma. Cancer Sci. 2010;101:1977–1983. doi: 10.1111/j.1349-7006.2010.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W, Lu C, Dong HH, Huang J, Shen DY, Stone RL, Nick AM, Shahzad MM, Mora E, Jennings NB, et al. Biological roles of the Delta family Notch ligand Dll4 in tumor and endothelial cells in ovarian cancer. Cancer Res. 2011;71:6030–6039. doi: 10.1158/0008-5472.CAN-10-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riedle S, Kiefel H, Gast D, Bondong S, Wolterink S, Gutwein P, Altevogt P. Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/gamma-secretase activity. Biochem J. 2009;420:391–402. doi: 10.1042/BJ20081625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: Gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan TZ, Yang H, Ye J, Low J, Choolani M, Tan DS, Thiery JP, Huang RY. CSIOVDB: A microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget. 2015;6:43843–43852. doi: 10.18632/oncotarget.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research N: Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor AB, MacKinnon DP. Four applications of permutation methods to testing a single-mediator model. Behavi Res Methods. 2012;44:806–844. doi: 10.3758/s13428-011-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simes RJ. An improved bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. doi: 10.1093/biomet/73.3.751. [DOI] [Google Scholar]
- 27.Ismail E, Kornovski Y. Surgical staging of ovarian cancer (I FIGO STAGE): IGCS-0018 ovarian cancer. Int J Gynecol Cancer. 2015;25(Suppl 1):S52. doi: 10.1136/00009577-201505001-00041. [DOI] [PubMed] [Google Scholar]
- 28.Ismail E, Kornovski Y. Advanced stage of epithelial ovarian cancer (ii-iv figo stage). The role of lymph node dissection: IGCS-0019 ovarian cancer. Int J Gynecol Cancer. 2015;25(Suppl 1):S53. doi: 10.1097/00009577-201505001-00042. [DOI] [PubMed] [Google Scholar]
- 29.Javadi S, Ganeshan DM, Qayyum A, Iyer RB, Bhosale P. Ovarian cancer, the revised FIGO staging system, and the role of imaging. AJR Am J Roentgenol. 2016;206:1351–1360. doi: 10.2214/AJR.15.15199. [DOI] [PubMed] [Google Scholar]
- 30.Gil-Garcia B, Baladron V. The complex role of NOTCH receptors and their ligands in the development of hepatoblastoma, cholangiocarcinoma and hepatocellular carcinoma. Biol Cell. 2016;108:29–40. doi: 10.1111/boc.201500029. [DOI] [PubMed] [Google Scholar]
- 31.Borggrefe T, Lauth M, Zwijsen A, Huylebroeck D, Oswald F, Giaimo BD. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFbeta/BMP and hypoxia pathways. Biochim Biophys Acta. 2016;1863:303–313. doi: 10.1016/j.bbamcr.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, Griffin JD. Modulation of Notch signaling by mastermind-like (MAML) transcriptional co-activators and their involvement in tumorigenesis. Semin Cancer Biol. 2004;14:348–356. doi: 10.1016/j.semcancer.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29:2672–2680. doi: 10.1038/onc.2010.35. [DOI] [PubMed] [Google Scholar]
- 34.Yen WC, Fischer MM, Axelrod F, Bond C, Cain J, Cancilla B, Henner WR, Meisner R, Sato A, Shah J, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 2015;21:2084–2095. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 35.Zhu PC, Aliabadi HM, Uludag H, Han J. Identification of potential drug targets in cancer signaling pathways using stochastic logical models. Sci Rep Uk. 2016;6:23078. doi: 10.1038/srep23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated during the current study are available from the corresponding author on reasonable request.