Abstract
Liver cancer is a malignant tumor that threatens human health worldwide. It has poor prognosis rates and ineffective therapeutic options. Recently, various miRNAs have been proven to exert promoting or inhibiting functions in different malignancies. However, the definitive mechanisms of miR-99a in liver cancer remain unclear. In the current study, we explored the relationships between the expression of miR-99a and HOXA1 in liver cancer tissues and cells to explore their combined effects on the occurrence and metastasis of liver cancer. The expression of miR-99a and HOXA1 in liver cancer tissue samples and cells was measured by RT-qPCR. Following transfection, transwell assays were conducted to assess the invasion and migration capacities of liver cancer cells. Subsequently, western blots and luciferase reporter assays were performed in liver cancer cells to identify the target of miR-99a. The data indicated that miRNA-99a expression was significantly reduced in both liver cancer tissue samples and cells compared with normal tissues and normal liver cells respectively. By contrast, the HOXA1 expression levels in liver cancer tissues and cells were significantly increased in contrast to the control group. The findings also revealed that the miR-99a expression was negatively correlated with HOXA1 expression in liver cancer tissue samples and miR-99a could suppress cell invasion and migration by targeting HOXA1 in liver cancer.
Keywords: miR-99a, HOXA1, migration, invasion, liver cancer
Introduction
Liver cancer is a leading cause of death worldwide, and its incidence is high in Asia (1). Approximately 745,500 individuals succumb to liver cancer annually (2). Currently, treatment for liver cancer is not satisfactory and generally ineffective (3). As the symptoms of liver cancer become evident at an advanced stage rendering it a disease that is untreatable by surgery, only minority of liver cancer patients are amenable to curative resection. However, due to the high frequency of metastasis and recurrence, the prognosis of the treated liver cancer patients remains poor (4). These challenges necessitate the identification of potential biomarkers and finding of new targets to design more powerful treatments. In the last decade, many studies have been devoted to exploring the etiopathogenesis of liver cancer.
miRNAs can promote or inhibit gene expression by recognizing specific binding sites on the target mRNA 3′UTR playing important roles in various types of cancer (5,6). miRNAs comprise a type of conserved non-coding RNAs (7)by regulating its direct target mRNA, and it acts as a crucial post-transcriptional regulator and plays a critical part in multiple biological processes, including cell formation, growth, apoptosis and metastasis (8). A growing number of studies have revealed that miRNAs may be used as tissue-specific biomarkers for liver cancer. miR-188-5p was confirmed to suppress cancer cell metastasis and proliferation via targeting FGF5 directly in hepatocellular carcinoma (9). miR-345 was reported to inhibit hepatocellular carcinoma metastasis by inhibiting YAP1 (10), and miR-487a can promote proliferation and metastasis in liver cancer (11). Even the significance of different miRNAs in liver cancer has attracted increasing attention recently. However, the regulatory mechanism as well as clinical significances of the majority of miRNAs including miR-99a in liver cancer, remains unclear.
Homeobox A1 (HOXA1), the earliest expressed HOX gene in mammals, affects numerous cell processes (12). HOXA1 is an oncogene and is overexpressed in several tumors, such as gastric cancer (13), small cell lung cancer (14) and oral squamous cell carcinoma (15), and is associated with poor prognosis. Recently, increasing studies showed that HOXA1 expression was associated with invasion and migration, acting as a reliable biomarker of cancer (16). However, the molecular mechanism of HOXA1 in liver cancer is still unclear. TargetScan database analysis showed that HOXA1 was a promising candidate target for miR-99a. The aim of the study was to investigate the expression and clinical significance of HOXA1 in liver cancer and to identify the correlation of miR-99a and the molecular targets regulated by miR-99a in liver cancer.
Materials and methods
Human tissue specimens
We collected liver cancer tissues and matched normal tissue samples from 55 liver cancer patients who underwent surgical treatment in Liaocheng People's Hospital (Liaocheng, China) between 2015 and 2017. All the tissues were snap-frozen in liquid nitrogen immediately and reserved at −80°C. Moreover, all patients involved in this research offered written informed consents and the study was approved by the Ethics Committee of Liaocheng People's Hospital. The clinicopathological characteristics of liver cancer patients were listed in Table I.
Table I.
Association of miR-99a expression with the clinicopathological characteristics of the liver cancer patients.
| miR-99a expression | ||||
|---|---|---|---|---|
| Clinicopathological characteristics | Cases (n=55) | High (n=20) | Low (n=35) | P-value |
| Age (years) | 0.2048 | |||
| >60 | 30 | 11 | 19 | |
| ≤60 | 25 | 9 | 16 | |
| Sex | 0.3257 | |||
| Male | 26 | 8 | 18 | |
| Female | 29 | 12 | 17 | |
| Tumor size (cm) | 0.0824 | |||
| ≥5.0 | 28 | 8 | 20 | |
| <5.0 | 27 | 12 | 15 | |
| TNM stage | 0.0239 | |||
| I–II | 25 | 15 | 10 | |
| III | 30 | 5 | 25 | |
| AFP (ng/ml) | 0.0620 | |||
| ≤20 | 27 | 11 | 16 | |
| >20 | 28 | 9 | 19 | |
| HBV | ||||
| Negative | 22 | 7 | 15 | 0.0658 |
| Positive | 33 | 13 | 20 | |
| Cirrhosis | 0.0587 | |||
| Yes | 33 | 12 | 21 | |
| No | 22 | 8 | 14 | |
| BCLC stage | 0.0116 | |||
| 0-A | 21 | 13 | 8 | |
| B-C | 34 | 7 | 27 | |
TNM, tumor-node-metastasis; AFP, α-fetoprotein; HBV, hepatitis B virus; BCLC, Barcelona Clinic Liver Cancer.
Cell culture
The liver cancer cell lines HepG2 (TCHu72) and Huh7 (TCHu182) (the Cell Bank of the Chinese Academy of Sciences, Shanghai, China) were maintained in DMEM (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified atmosphere at 37°C containing 5% CO2. In addition, the DMEM medium contained 10% FBS.
Cell transfection
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was applied to transfected miR-99a mimics or miR99a inhibitor and the corresponding control into liver cancer cell lines following the manufacturer's protocol. The transfection efficacy was evaluated 48 h after transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNAs of liver cancer tissue samples and cells were extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A miScript Reverse Transcription kit (Qiagen Inc., Valencia, CA, USA) was used strictly following the manufacturer's protocol to reverse transcribe 1 µg RNA of each sample to cDNA. A miScript SYBR-Green PCR kit (Qiagen Inc.) was next used to amplify cDNA. The expression of miR-99a and HOXA1 in tissues was normalized to the expression of U6 and GAPDH, respectively. U6 and GAPDH were used as controls, and the relative expression was calculated as per the 2−∆∆Cq equation (17). The primers used were shown in Table II.
Table II.
Primer sequences for qRT-PCR.
| Primer | Sequence |
|---|---|
| miR-99a | F: 5′-AGTGTGACGTTGACATCCGT-3′ |
| R: 5′-GCAGCTCAGTAACAGTCCGC-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| HOXA1 | F: 5′-TCCTGGAATACCCCATACTTAGC-3′ |
| R: 5′-GCACGACTGGAAAGTTGTAATCC-3′ | |
| GAPDH | F: 5′-GAGTCAACGGATTTGGTCGT-3′ |
| R: 5′-GACAAGCTTCCCGTTCTCAG-3′ |
qRT-PCR, quantitative real-time reverse transcription-PCR; F, forward; R, reverse; U6, small nuclear RNA, snRNA; HOXA1, homeobox A1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Transwell assays
Transwell assays were conducted to assess the invasion and migration abilities of liver cancer cells using 24-well Transwell chambers. Liver cancer cells transfected with miR-99a mimics or inhibitor were collected and resuspended with DMEM. Then, the cell suspensions were added into the upper chamber while medium containing 10% FBS was added into the bottom chamber. After incubation for 12 h, the cells adhering to the upper chamber were removed using cotton swabs completely. The difference between the invasion and migration assays was the top chamber coated with Matrigel for invasion assay. Subsequently, the migrated or invaded cells adherent to the bottom surface were fixed, stained, photographed and counted. The number of migration or invasion cells was counted at a magnification of ×200 from 5 different random fields.
Western blot analysis
HOXA1 protein expression in liver cancer cell lines with the transfections was measured via western blots. The transfected liver cancer cells, HepG2 and Huh7, were lysed using lysis buffer. After centrifugation at 12,000 × g for 20 min at 4°C, a BCA protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to assess the protein concentrations. Proteins were separated with 10% SDS-PAGE and then electrophoretically transferred onto PVDF membrane. After being blocked in 5% non-fat milk 1 h at room temperature, the membrane was then incubated with appropriately diluted primary antibodies [rabbit anti-HOXA1 polyclonal antibody (1:500; cat. no. ab230513; Abcam, Cambridge, UK) and anti-GAPDH polyclonal antibody (1:2,500; cat. no. ab9485; Abcam)] at 4°C overnight. Subsequently, the membranes were incubated with horse-radish peroxidase-linked secondary goat anti-rabbit polyclonal antibody (1:2,000; cat. no. ab6721; Abcam) for 1 h. The enhanced chemiluminescence (ECL-plus, Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA) method was used to detect the proteins.
Luciferase assay
For the luciferase reporter assays, liver cancer cells (HepG2 and Huh7) were co-transfected with miR-99a mimics and HOXA1−3′UTR-WT or corresponding mutant reporter with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Forty-eight hours after the transfections, the cells were collected and the luciferase activities were analyzed by the GloMax fluorescence reader (Promega Corporation, Madison, WI, USA).
Statistical analysis
Data from three separate experiments were presented as mean ± standard error (SE). The statistical analysis was conducted using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 18.0 version (SPSS Inc., Chicago, IL, USA). Correlation between expression levels of miR-99a and HOXA1 was estimated using the Pearson's correlation method. Student's t-test was applied in the present study. P<0.05 was considered to indicate a statistically significant difference.
Results
Low expression of miR-99a and overexpression of HOXA1 in liver cancer
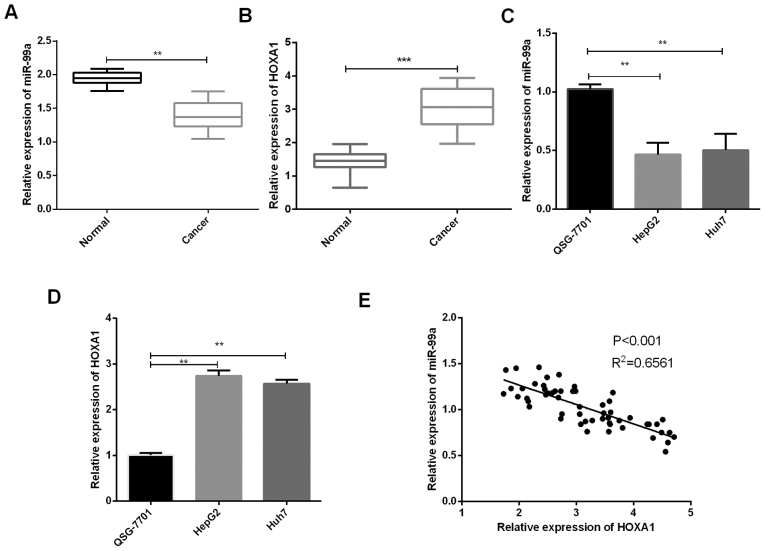
The expression of miR-99a and HOXA1 in liver cancer tissue samples and cell lines were measured using RT-qPCR to determine the function of miR-99a in liver cancer tumorigenesis. Results revealed that miR-99a expression was significantly lower in liver cancer tissues than those in matched benign tissue samples (Fig. 1A). Conversely, the HOXA1 mRNA expression in liver cancer tissue samples was significantly higher than the normal tissue samples (Fig. 1B). We also verified that miR-99a expressions in liver cancer cell lines (HepG2 and Huh7) were decreased in comparison with the normal liver cells (Fig. 1C) while HOXA1 expressions in liver cancer cell lines (HepG2 and Huh7) were increased compared to the normal cells (Fig. 1D). In addition, statistical analysis showed that miR-99a expression was negatively correlated with HOXA1 in liver cancer tissues (Fig. 1Ε). The association between the clinicopathological characteristics of 55 liver cancer patients and miR-99a expression in liver cancer tissue samples is shown in Table I, showing that miR-99a expression was significantly associated with TNM stage (P=0.0239) and BCLC stage (P=0.0116).
Figure 1.
Expression of miR-99a and HOXA1 in liver cancer tissues and cells. (A) RT-qPCR was used to analyze miR-99a expression in liver cancer tissues (n=55) and matched normal tissues (n=55) (**P<0.01). (B) HOXA1 mRNA expression in liver cancer tissues (n=55) and adjacent normal tissues (n=55) was evaluated using RT-qPCR (***P<0.001). (C) miR-99a expressions in liver cancer cells (HepG2 and Huh7) were measured using qRT-PCR (**P<0.01). (D) HOXA1 mRNA expressions were detected using qRT-PCR in liver cancer cells (HepG2 and Huh7) (**P<0.01). (E) Correlation between expression levels of miR-99a and HOXA1.
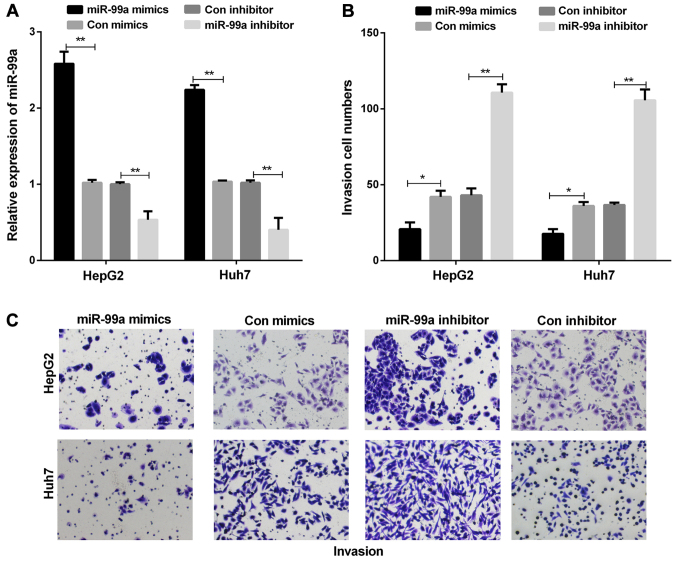
miR-99a suppresses the abilities of cell invasion in liver cancer cell lines
We established miR-99a overexpression models by transiently transfecting miR-99a mimics into liver cancer cells to further investigate the function of miR-99a in liver cancer. The results showed that miR-99a mimics were significantly upregulated in both HepG2 and Huh7 cells (Fig. 2A). Next, we investigated the effect of miR-99a on liver cancer cell invasion. In the cell invasion assays, miR-99a overexpression suppressed liver cancer cell invasion, whereas miR-99a inhibition promoted liver cancer cell invasion (Fig. 2B and C).
Figure 2.
miR-99a suppressed the liver cancer cell invasion abilities. (A) miR-99a expression was measured using RT-qPCR in liver cancer cells transfected with miR-99a mimics or inhibitor, respectively (**P<0.01). (B) The invasion cell numbers of liver cancer cells were counted (*P<0.05, **P<0.01). (C) Cell invasion was observed using the Transwell assay in liver cancer cell lines transfected with miR-99a mimics or inhibitor, respectively
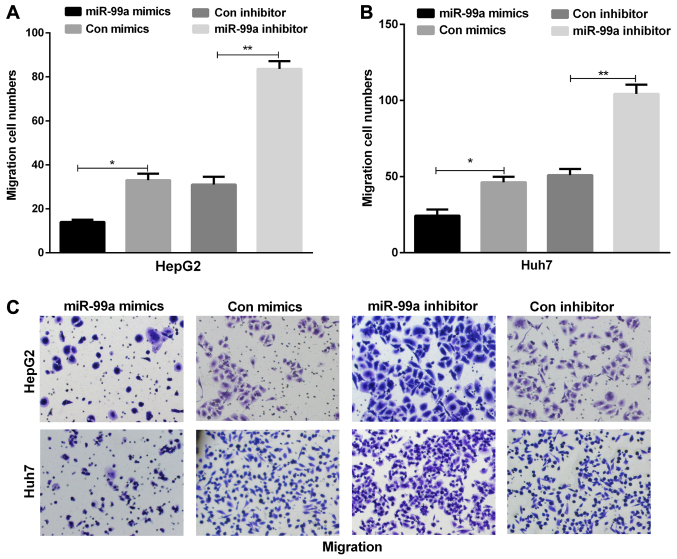
miR-99a inhibits liver cancer cell migration
To explore the metastastic function of miR-99a in liver cancer cells, cell migration assay was performed using Transwell assay. miR-99a overexpression in HepG2 and Huh7 cell lines contributed to a significant reduction in cell migration in contrast to the control group. At the same time, we found that the inhibition of miR-99a increased HepG2 and Huh7 cell migration (Fig. 3A-C).
Figure 3.
miR-99a inhibited liver cancer cell migration. The migration cell numbers of (A) HepG2 and (B) Huh7 were counted (*P<0.05, **P<0.01). (C) Cell migration was detected using the Transwell assays in liver cancer cells transfected with miR-99a mimics or inhibitor.
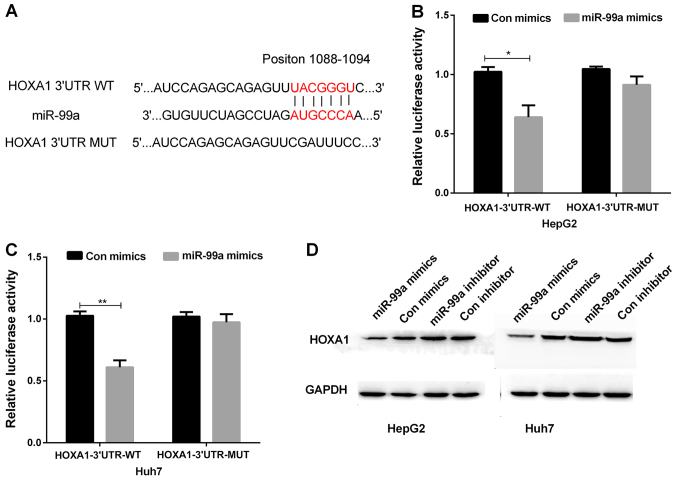
HOXA1 was a direct target for miR-99a
Targetscan was applied to identify the putative target of miR-99a. Among all the genes searched, HOXA1 was selected because of its crucial effect on cell growth. Sequence analysis showed putative binding sites between miR-99a and HOXA1 (Fig. 4A). Luciferase reporter assay was performed on HepG2 and Huh7 cell lines, which were transfected with miR-99a mimics or miR-control. Then, the transfected cells were co-transfected with the luciferase reporters containing WT or MUT of HOXA1 3′UTR, respectively. The results indicated that miR-99a overexpression inhibited the luciferase activity of the HOXA1−3′UTR-WT reporter in both HepG2 and Huh7 cells, not affecting the activity of the HOXA1−3′UTR-MUT reporter (Fig. 4B and C). Moreover, the western blots indicated that HOXA1 protein expression was obviously decreased in both HepG2 and Huh7 cells transfected with miR-99a mimics, whereas the protein expression of HOXA1 was obviously increased in HepG2 and Huh7 cell lines transfected with miR-99a inhibitor compared with their respective control group (Fig. 4D). These results suggested that HOXA1 was a direct target for miR-99a in liver cancer cells.
Figure 4.
miR-99a regulated HOXA1 expression by targeting its 3′UTR. (A) According to the Target Scan, the miR-99a target site in the sequence of HOXA1 was predicted. (B and C) Fluorescence activities of the HOXA1 3′UTR was measured by the luciferase reporter gene assays in liver cancer cells that were co-transfected with miR-99a mimics and wild-type HOXA1 3′UTR or mutational type HOXA1 3′UTR (*P<0.05, **P<0.01). (D) Western blot analysis detected the HOXA1 protein expression in liver cancer cells transfected with miR-99a mimics or inhibitor.
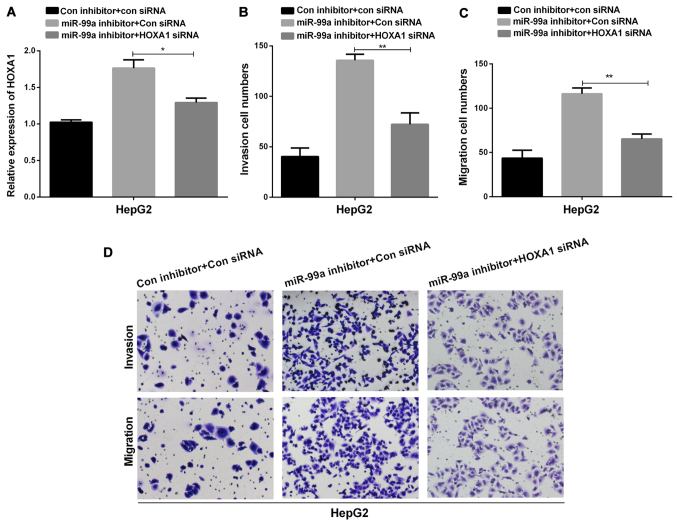
Roles of HOXA1 in regulating the miR-99a ability in liver cancer cell invasion and migration
We also investigated whether HOXA1 was essential in regulating the miR-99a ability in liver cancer cell invasion and migration. HOXA1 siRNAs were transfected into liver cancer cells to knock down endogenous HOXA1 and RT-qPCR was used to determine the HOXA1 mRNA expression. The findings demonstrated that the HOXA1 expression in liver cancer cells transfected with HOXA1 siRNAs was reduced significantly compared to the control group (Fig. 5A). Subsequently, Transwell assays were carried out to observe the migration and invasion capacities of liver cancer cells co-transfected with HOXA1 siRNAs and miR-99a inhibitor. From the results, it was evident that deletion of HOXA1 markedly reversed miR-99a-medicated inhibitory effect on cell invasion and migration in liver cancer cells (Fig. 5B-D).
Figure 5.
HOXA1 may reverse partial function of miR-99a in liver cancer cells. (A) Detection of relative HOXA1 mRNA expression in HepG2 cells after silencing HOXA1 using RT-qPCR (*P<0.05). (B and C) The invasion cell numbers of HepG2 cells co-transfected with miR-99a and HOXA1 siRNA were (**P<0.01). (D) The Transwell assay showed the cell migratory and invasive abilities of HepG2 cells after inhibiting miR-99a alone or inhibiting miR-99a as well as silencing HOXA1.
Discussion
Liver cancer is one of the most lethal cancer types, posing an increasing burden on global health (18). Currently, studies on mechanism have focused primarily on liver cancer. Although liver cancer has been widely researched, the possible mechanisms involved remain to be elucidated because of its complexity. Emerging evidence has shown that the two main biological characteristics of malignant cancers are invasion and metastasis, which affect the prognosis of tumor patients (19). Accommodative dysfunction of miRNA frequently occurs in multifarious human tumors, and the aberrant expression of miRNA often contributes to carcinogenesis by affecting expression levels of multiple genes (20). Thus, it is valuable to fully understand the functions of specific miRNAs in the development of tumor for the diagnosis and treatment of liver cancer.
Recently, as miRNAs have become crucial regulators in the development of tumor, the target therapy of tumor using miRNAs emerges as a fresh diagnostic instrument (21). It has been reported that miR-99a is involved in various tumors, intervening the processes of cell proliferation, apoptosis as well as inflammation (22,23). Yang et al verified that miR-99a exerted a crucial effect on modulating the CSC phenotype of breast carcinoma mediated via targeting the mTOR/HIF-α signaling pathway (24). In addition, miR-99a was frequently downregulated in ATC and miR-99a overexpression could reduce oncogenicity through targeting mTOR (25). Although miR-99a has been identified as a tumor suppressor in various types of cancer, however, the clear functions of miR-99a together with HOXA1 in liver cancer have not been described yet.
HOXA1 is a pivotal transcriptional regulator of early embryonic development (26). A growing number of studies have indicated that the aberrant HOXA1 expression in different cancers is involved in the regulation of biological processes, such as proliferation and apoptosis (27). For instance, overexpression of HOXA1 was found in prostate cancer cells and silencing of HOXA1 suppressed the progression of prostate cancer (16). HOXA1 was found to be a target of miR-10a in pancreatic cancer, and it was involved in tumor cell invasion (28). The current study aimed to determine the relationship between the expression of miR-99a and HOXA1 in liver cancer. The expression of miR-99a and HOXA1 was measured using RT-qPCR, and the results showed that miR-99a was downregulated, whereas HOXA1 was overexpressed in liver cancer. Additionally, we found that miR-99a expression negatively correlated with HOXA1 expression in liver cancer tissues. Moreover, data also showed that miR-99a suppressed cell invasion and migration through HOXA1 in liver cancer.
Collectively, the findings in our study showed that miR-99a exerted a crucial suppressive effect in liver cancer. First, our data revealed that the miR-99a expression was significantly decreased and HOXA1 expression was significantly increased in liver cancer tissues in contrast to the control group. The same result was also found in the liver cancer cell lines. Second, the Transwell assay results revealed that miR-99a could suppress liver cancer cell invasion and migration. In addition, we confirmed that HOXA1 was a direct target of miR-99a. The prevention of tumor progression observed in this study support the further investigation of miRNA therapy for liver cancer prevention and treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CT contributed significantly to data analysis and manuscript preparation. HS helped perform the statistical analysis with constructive discussions. WS contributed to the conception of the study. SL collected, interpreted and analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients involved in this research gave written informed consents and the study was approved by the Liaocheng People's Hospital Ethics Committee (Liaocheng, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ho C, Wang C, Mattu S, Destefanis G, Ladu S, Delogu S, Armbruster J, Fan L, Lee SA, Jiang L, et al. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology. 2012;55:833–845. doi: 10.1002/hep.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 4.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. Circular RNA MTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 5.Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: The potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2:330–334. doi: 10.6026/97320630002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Hoffend NC, Magner WJ, Tomasi TB. The epigenetic regulation of Dicer and microRNA biogenesis by Panobinostat. Epigenetics. 2017;12:105–112. doi: 10.1080/15592294.2016.1267886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H, Yang LY. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63:874–885. doi: 10.1016/j.jhep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Liu H, Bi H. MicroRNA-345 inhibits hepatocellular carcinoma metastasis by inhibiting YAP1. Oncol Rep. 2017;38:843–849. doi: 10.3892/or.2017.5772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2017;23:2593–2604. doi: 10.1158/1078-0432.CCR-16-0851. [DOI] [PubMed] [Google Scholar]
- 12.Makki N, Capecchi MR. Cardiovascular defects in a mouse model of HOXA1 syndrome. Hum Mol Genet. 2012;21:26–31. doi: 10.1093/hmg/ddr434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S, Zhang M, Yu F, Peng Z, Zhou C. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer Res. 2016;35:15. doi: 10.1186/s13046-016-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang J, Yang J, Liao H, Guo L. Downregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100. Eur J Cancer. 2014;50:1541–1554. doi: 10.1016/j.ejca.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Bitu CC, Destro MF, Carrera M, da Silva SD, Graner E, Kowalski LP, Soares FA, Coletta RD. HOXA1 is overexpressed in oral squamous cell carcinomas and its expression is correlated with poor prognosis. BMC Cancer. 2012;12:146. doi: 10.1186/1471-2407-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Liu G, Shen D, Ye H, Huang J, Jiao L, Sun Y. HOXA1 enhances the cell proliferation, invasion and metastasis of prostate cancer cells. Oncol Rep. 2015;34:1203–1210. doi: 10.3892/or.2015.4085. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–424. doi: 10.1038/nrclinonc.2015.103. [DOI] [PubMed] [Google Scholar]
- 19.Gumireddy K, Li A, Cao L, Yan J, Liu L, Xu X, Pazoles C, Huang Q. NOV-002, a glutathione disulfide mimetic, suppresses tumor cell invasion and metastasis. J Carcinog Mutagen. 2013;2013:S7–002. doi: 10.4172/2157-2518.S7-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Hummel R, Hussey DJ, Haier J. MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 22.Tao Z, Zhao H, Wang R, Liu P, Yan F, Zhang C, Ji X, Luo Y. Neuroprotective effect of microRNA-99a against focal cerebral ischemia-reperfusion injury in mice. J Neurol Sci. 2015;355:113–119. doi: 10.1016/j.jns.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Lin KY, Ye H, Han BW, Wang WT, Wei PP, He B, Li XJ, Chen YQ. Genome-wide screen identified let-7c/miR-99a/miR-125b regulating tumor progression and stem-like properties in cholangiocarcinoma. Oncogene. 2016;35:3376–3386. doi: 10.1038/onc.2015.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–595. doi: 10.1111/cpr.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang HG, Luo X, Wu S, Jian B. MiR-99a inhibits cell proliferation and tumorigenesis through targeting mTOR in human anaplastic thyroid cancer. Asian Pac J Cancer Prev. 2015;16:4937–4944. doi: 10.7314/APJCP.2015.16.12.4937. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Zhang X, Li N, Liu Q, Chen D. miR-30b inhibits cancer cell growth, migration, and invasion by targeting homeobox A1 in esophageal cancer. Biochem Biophys Res Commun. 2017;485:506–512. doi: 10.1016/j.bbrc.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 27.López-Romero R, Marrero-Rodríguez D, Romero-Morelos P, Villegas V, Valdivia A, Arreola H, Huerta-Padilla V, Salcedo M. The role of developmental HOX genes in cervical cancer. Rev Med Inst Mex Seguro Soc. 2015;53(Suppl 2):S188–S193. (In Spanish) [PubMed] [Google Scholar]
- 28.Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, Toma H, Nakamura M, Nagai E, Hashizume M, et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. 2012;19:2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.