Abstract
Epidermal growth factor receptor (EGFR) is the most important driver gene of non-small cell lung cancer (NSCLC) as EGFR mutations determine the efficacy of EGFR tyrosine kinase inhibitor (EGFR-TKI) therapy. In the present study, the comprehensive ability of widely used polymerase chain reaction (PCR) methods to detect EGFR mutations was determined. Among the 35 EGFR mutations detected via the direct sequencing of 73 patients with NSCLC, 11 types were identified in exons 18, 19 and 21. Among the 11 mutation types, all exon 18 and 21 mutations were identified by 2 widely used PCR methods, namely, Scorpion-Amplification Refractory Mutation System and cobas v2. However, among the 9 different exon 19 deletions, 3 types were not identified by the 2 methods. In addition, 25 samples with EGFR mutations were analyzed by the 2 methods, including a sample from a patient with an unidentified exon 19 deletion, the T751_I759 deletion and insertion S; this patient had long-term disease control as a result of EGFR-TKI therapy. The 2 methods could not detect this unidentified deletion, whereas sizing capillary electrophoresis for the comprehensive detection of exon 19 deletions detected this deletion. It is generally thought that patients with exon 19 mutations have higher response rates to EGFR-TKI therapy than patients with exon 21 mutations. The present study confirmed the EGFR mutation status by comparing the mutations with the Catalog Of Somatic Mutations In Cancer, which is the world's largest and most comprehensive resource for analyzing the effects of somatic mutations in human cancers. The predicted frequency of EGFR mutations identified by the 2 methods was 85%. The frequency of mutations detectable by the 2 methods was less for exon 19 than exon 21. Therefore, the results of the present study suggest that decreasing false-negative detection of exon 19 deletions is crucial for the clinical testing of EGFR mutations.
Keywords: non-small cell lung cancer, epidermal growth factor receptor, tyrosine kinase inhibitor, PCR, exon 19 deletion
Introduction
Epidermal growth factor receptor (EGFR) mutations are important factors in non-small lung cancer (NSCLC), as EGFR tyrosine kinase inhibitor (EGFR-TKI) treatment has exhibited high efficacy in NSCLC patients with EGFR mutations (1–6). Polymerase chain reaction (PCR) methods are used clinically for the detection of EGFR mutations. Two widely used PCR methods are approved as in vitro diagnostic (IVD) methods, namely, the Scorpion Amplification Refractory Mutation System (ARMS; QIAGEN therascreen® EGFR; Qiagen, Inc., Valencia, CA, USA) and the cobas® EGFR Mutation Test v2 (Roche Diagnostics, Indianapolis, IN, USA) (7,8). These 2 methods are a real-time PCR test for the qualitative detection of defined mutations of the EGFR gene in DNA derived from formalin-fixed paraffin-embedded (FFPE) tumor tissue from NSCLC patients. The test is intended to aid in identifying patients with NSCLC whose tumors have defined EGFR mutations and for whom safety and efficacy of EGFR-TKI have been established. The first EGFR-TKI is gefitinib, which was approved from July 2002 in Japan. Erlotinib, afatinib, dacomitinib and osimertinib are subsequently approved as EGFR-TKIs. Dacomitinib is a second-generation, irreversible EGFR-TKI. In NSCLC patients with EGFR mutations detected by Scorpion-ARMS method, dacomitinib significantly improved progression-free survival over gefitinib in first-line treatment (5). Osimertinib is a third-generation, irreversible EGFR-TKI. In the first-line treatment of EGFR mutation-positive advanced NSCLC identified by cobas v2, osimertinib showed efficacy superior to that of gefitinib or erlotinib with a similar safety profile and lower rates of serious adverse events (6). In addition, cobas v2 can be used with plasma samples, as a companion diagnostic for NSCLC therapy. The Scorpion-ARMS and the cobas v2 are useful, rapid and cost-effective methods as a companion diagnostic. However, they can only identify a small proportion of the different types of mutation, including common exon 19 deletions and exon 21 L858R. The present study therefore analyzed the frequency of detectable EGFR mutations and the clinical significance of mutations that are not detected by these 2 methods.
Materials and methods
Patients
The present study included a cohort of 73 Japanese patients with NSCLC, from whom written informed consent was obtained for the use of their samples in this research. These patients presented with recurrent disease following surgery between 1992 and 2004. The response of patients with EGFR mutations to EGFR-TKI treatment, which was a daily dose of gefitinib (250 mg) administered between April 2002 and October 2005, was evaluated. During this period, only gefitinib was approved as an EGFR-TKI therapy for NSCLC patients in Japan (Table I). The present study received ethics approval from the Institutional Review Board of Tokyo Medical University (Tokyo, Japan).
Table I.
Background information of the 73 patients with NSCLC.
| Variable | Total n, (%) |
|---|---|
| Age, years, range (median) | 30–92 (65.6) |
| Sex: Male/Female | 45 (61.6)/28 (38.4) |
| Histology: ADC/SCC/La/Pleomorphic | 59 (80.8)/9 (12.3)/4 (5.5)/1 (1.4) |
| Smoking: Never/Former and Current | 29 (39.7)/44 (60.3) |
| EGFR mutation | 35 (47.9) |
NSCLC, non-small cell lung cancer; ADC, adenocarcinoma; SCC, squamous cell carcinoma; La, large cell carcinoma; Pleomorphic, pleomorphic carcinoma; EGFR, epidermal growth factor receptor.
DNA extraction and direct sequencing analysis
All 73 surgical samples were FFPE; analysis of EGFR mutations was performed. Tumor cells were microdissected under stereoscopic microscopy, and DNA was extracted from tumor cells with a DNeasy Tissue kit (Qiagen, Inc.). PCR was performed using a primer set flanking exons 18 to 21. PCR products were run on a gel, and the appropriate bands were cut, and then DNA was extracted from the gel with the QIAquick Gel Extraction kit (Qiagen, Inc.). Sequencing analysis was performed using an ABI Prism 3100-Avant genetic analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol.
Determination of the efficacy of EGFR-TKI therapy
Definition of the efficacy of EGFR-TKI therapy was applied according to the Response Evaluation Criteria in Solid Tumors (9). In the case of a partial response (PR) or complete response (CR), changes in tumor measurements were confirmed by repeated assessments that were performed no less than 4 weeks following the fulfillment of the response criteria. Stable disease (SD) was defined as fulfilling the SD criteria for at least 8 weeks following the start of EGFR-TKI therapy. Patients receiving >4 weeks of treatment were considered assessable, and patients who had received EGFR-TKI therapy for <4 weeks were considered not assessable. Response rate (RR) was defined as the percentage of PR and CR divided by the total of the assessable patients, and disease control rate (DCR) was the percentage of response and SD divided by the total of the assessable patients (Table II).
Table II.
Information on 35 NSCLC patients with EGFR mutations receiving EGFR- tyrosine kinase inhibitor therapy.
| Variable | Total n |
|---|---|
| Assessable patients/patients with EGFR mutations | 30/35 |
| Mutations of exon 18/19/21 | 1/23/11 |
| Mutation types in 30 assessable patients | |
| Mutations of exon 18/19/21 | 1/19/10 |
| EGFR mutation RR | 36.7% (11/30) |
| EGFR mutation DCR | 97% (29/30) |
| Exon mutations | |
| Exon 18 mutation RR | 100% (1/1) |
| Exon 19 mutations RR | 42.1% (8/19) |
| Exon 21 mutations RR | 20% (2/10) |
NSCLC, non-small cell lung cancer; EGFR, epidermal growth factor receptor; RR, response rate; DCR, disease control rate.
Frequency of detectable mutations by the 2 IVD PCR methods
Two widely used IVD PCR methods (Scorpion ARMS and cobas v2) were used in the present study. Scorpion-ARMS can detect 29 types of EGFR nucleotide mutations, and cobas v2 can detect 42 types of EGFR nucleotide mutations (Table III) (7,8). The two methods of detection have previously identified common mutations such as specific exon 19 deletions and specific exon 21 mutations including L858R. Using the 2 methods, patients who harbored mutations that could not be identified were confirmed, and the frequency of detectable EGFR mutations was calculated. The results from the 2 methods were compared with the predicted frequency of detectable mutations from the catalog of somatic mutations in cancer (COSMIC) version 84 database. COSMIC is the world's largest and most comprehensive resource for analyzing the effects of somatic mutations in human cancers. EGFR consists of 564 nucleotides from exon 18 to exon 21 (codons 688–875), and EGFR mutations include substitutions, deletions, insertions, and combinations of these (9–12). The COSMIC v84 database was used in the present study to count the types of EGFR mutations (released on February 13, 2018).
Table III.
Types of EGFR mutations identified by the 2 IVD PCR methods.
| PCR method | Exon 18 | Exon 19 | Exon 20 | Exon 21 | Total n |
|---|---|---|---|---|---|
| Scorpion-ARMS | 3 | 19 | 5 | 2 | 29 |
| Cobas v2 | 3 | 29 | 7 | 3 | 42 |
EGFR, epidermal growth factor receptor; IVD, in vitro diagnostics; PCR, polymerase chain reaction; Scorpion-ARMS, Scorpion Amplification Refractory Mutation System.
Detection of EGFR mutations by the 2 IVD PCR methods
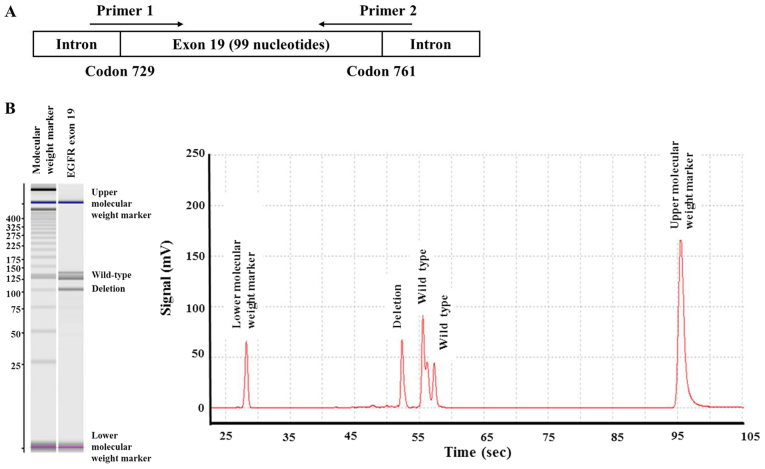
Owing to the large number of clinical samples, DNA was extracted from FFPE tumor specimens without microdissection, for analysis by the 2 IVD PCR methods. The DNA was subjected to Scorpion-ARMS at SRL Inc. (Tokyo, Japan) and cobas v2 at BML Inc. (Tokyo, Japan), along with sizing capillary electrophoresis using MCE-202 MultiNA (Shimazu Corporation, Kyoto, Japan) at BML Inc. for the comprehensive detection of exon 19 deletions. Sizing capillary electrophoresis measures the length of DNA to distinguish between wild-type exons and exons with deletions, and covers all exon 19 deletions along the 99 nucleotides from codon 729 to 761 (Fig. 1A). This was performed to detect exon 19 deletions that were not identified by the 2 IVD PCR methods.
Figure 1.
(A) Diagram presenting the primer and amino acid positions of exon 19, from codon 729 to 761, in sizing capillary electrophoresis. (B) Sizing capillary electrophoresis was able to detect 1 exon 19 deletion in a patient with adenocarcinoma and long stable disease (43.4 months) who received EGFR-tyrosine kinase inhibitor therapy, namely, the T751_I759 deletion and insertion S, which were undetected by Scorpion-Amplification Refractory Mutation System and cobas v2. EGFR, epidermal growth factor receptor.
Results
EGFR mutation status and response to EGFR-TKI therapy
EGFR mutations were detected by direct sequencing in 35 patients (47.9% of clinical samples), including, 1 exon 18 mutation, 23 exon 19 mutations and 11 exon 21 mutations. Among the 35 patients with EGFR mutations who received EGFR-TKI therapy, 30 patients were assessable, which included 1 patient with an exon 18 mutation, 19 patients with exon 19 mutations and 10 patients with exon 21 mutations. The RR of the EGFR mutations was 36.7% and the DCR was 97%. In exons 19 and 21, including common mutations, the RR of exon 19 mutations was 42.1% and the RR of exon 21 mutations was 20% (Table II).
Frequency of mutations identified by the 2 IVD PCR methods
In all 73 cases, there were 11 types of EGFR mutations among the 35 patients with an EGFR mutation, including 1 type of exon 18 mutation, 9 types of exon 19 mutations and 1 type of exon 21 mutation, as detected by direct sequencing. The exon 18 mutation was G719S, the exon 19 mutations were all deletions (9 types), including deletion and insertion compound mutations, and the exon 21 mutation was L858R. All exon 18 and 21 mutations were identified by Scorpion ARMS and cobas v2. However, among the 9 types of exon 19 deletions, 3 types were not identified by the 2 methods. These unidentified deletions were the E746_S752 deletion, the T751_I759 deletion with insertion S and the K757_L760 deletion with insertion N. Two patients with unidentified deletions had achieved disease control (CR and SD) via EGFR-TKI therapy (Table IV). Of the 35 patients with EGFR mutations, the frequency of detectable EGFR mutations by the 2 methods was 91.4% (32/35); that of exon 19 mutations was 87.0% (20/23) and that of exon 21 mutations was 100% (11/11).
Table IV.
Information on 35 EGFR mutations detected by direct sequencing and the EGFR-TKI response of the patients.
| Amino acid/deletion type | Nucleotide | No. | Histology ADC/SCC | Response CR/PR/SD/PD/NA |
|---|---|---|---|---|
| Exon 18 | ||||
| G719S | G2155A | 1 | 2/0 | 0/1/0/0/0 |
| Exon 19 | 23 | 20/3 | 2/6/10/1/4 | |
| E746-A750 del | 2,235-2,249 | – | – | – |
| 2,236-2,250 | 13 | 11/2 | 0/3/7/1/2 | |
| E746-S752 dela | 2,236-2,256 | 1 | 1/0 | 1/0/0/0/0 |
| L747-E749 del | 2,239-2,247 | 1 | 1/0 | 1/0/0/0/0 |
| L747-L751 del | 2,239-2,253 | – | – | – |
| 2,240-2,254 | 3 | 3/0 | 0/2/0/0/1 | |
| L747-P753 ins S | 2,240-2,257 | 3 | 3/0 | 0/1/2/0/0 |
| T751-I759 ins Sa | 2,252-2,275 T2276G | 1 | 1/0 | 0/0/1/0/0 |
| K757-L760 ins Na | 2,271-2,279 | 1 | 0/1 | 0/0/0/0/1 |
| Exon 21 | ||||
| L858R | T2573G | 11 | 11/0 | 0/2/8/0/1 |
| Total EGFR mutations | – | 35 | 32/3 | 2/9/18/1/5 |
Exon 19 deletion that was not identified by Scorpion-ARMS and cobas v2. EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; ADC, adenocarcinoma; SCC, squamous cell carcinoma; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; NA, no assessable patients; del, deletion; ins, insertion.
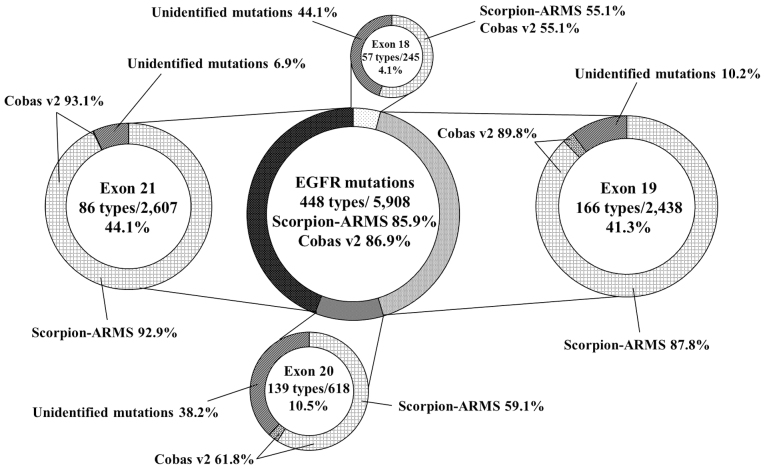
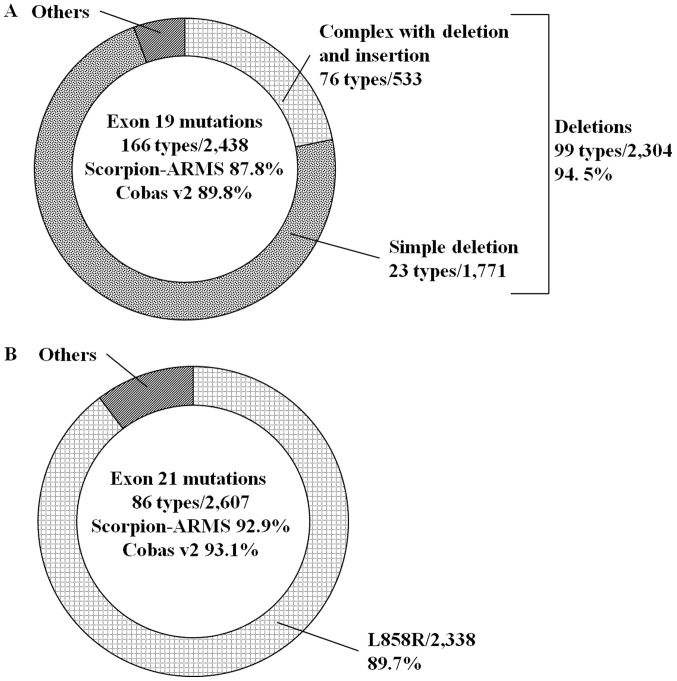
The COSMIC v84 database contains a total of 5,908 EGFR mutations comprising 448 types. The distribution of the mutations was 4.1% in exon 18, 41.3% in exon 19, 10.5% in exon 20 and 44.1% in exon 21. In the COSMIC v84 database, the predicted frequencies of detectable EGFR mutations by Scorpion-ARMS and cobas v2 were 85.9 and 86.9%, respectively (Fig. 2). Regarding exon 19 mutations, the distribution of deletions, including deletion and insertion compound mutations, was 94.5%, and the predicted frequencies of these mutations by the two methods were 87.8 and 89.8%, respectively (Fig. 3A). Regarding exon 21 mutations, the distribution of L858R was 89.7%, and the predicted frequencies of the two methods were 92.9 and 93.1%, respectively (Fig. 3B).
Figure 2.
Distribution of EGFR mutations and predicted frequencies of mutations from the Catalog Of Somatic Mutations In Cancer v84 database that were detectable by the 2 polymerase chain reaction methods. EGFR, epidermal growth factor receptor; Scorpion-ARMS, Scorpion-Amplification Refractory Mutation System.
Figure 3.
Percentage of deletions among (A) exon 19 mutations and L858R among (B) exon 21 mutations in the Catalog Of Somatic Mutations In Cancer v84 database. Scorpion-ARMS, Scorpion-Amplification Refractory Mutation System.
Detection of EGFR mutations by the 2 IVD PCR methods
Scorpion-ARMS and cobas v2 analyses were performed on 25 samples among the 35 patients who had EGFR mutations that were detected by direct sequencing, which included 1 exon 18 mutation (G719S), 18 exon 19 deletions comprised of 6 different types (including 1 exon 19 deletion not identified by the 2 methods), and 6 exon 21 mutations (L858R). All exon 18 and 21 mutations were detected by the two methods; however, both methods failed to detect exon 19 deletions in 4 patients (2 squamous cell carcinoma and 2 adenocarcinoma patients). The undetected deletions of exon 19 from 3 samples were actually the common E746-A750 mutation, which was expected to be identified by the two methods. One undetected complex with a deletion and insertion of exon 19 was T751_I759 and insertion S (COSMIC ID 1667027), which were not identified by either of the methods. The deletion was identified in a patient with adenocarcinoma, with long SD (43.4 months) by EGFR-TKI therapy. Subsequent sizing capillary electrophoresis was able to detect the deletion (Fig. 1B).
Discussion
Gefitinib and erlotinib were approved as a first-generation EGFR-TKI. Subsequently, afatinib and dacomitinib as a second-generation and osimertinib as a third-generation were approved. EGFR mutations are the major factors determining the efficacy of EGFR-TKI therapy in patients with NSCLC (1–6). EGFR consists of 564 nucleotides from exons 18 to 21, and EGFR mutations include substitutions, deletions, insertions, and a combination of any those (10–13). Therefore, different types of mutations are continuously being identified. In the COSMIC v84 database, exon 19 and 21 mutations account for the majority of EGFR mutations, and the distribution of these mutations among each exon is similar to previous studies (14,15) Scorpion-ARMS and cobas v2 can detect a proportion of the EGFR mutations, including common exon 19 deletions and the exon 21 L858R mutation. The present results revealed that the 2 methods did not identify 3 exon 19 deletions, in which 2 deletions were adenocarcinomas and 1 deletion was squamous cell carcinoma. In squamous cell carcinoma with EGFR mutations, a patient with a common exon 19 deletion (E746-A750) had achieved disease control via EGFR TKI therapy. Regardless of the histological type of lung cancer, exon 19 deletions are important for EGFR TKI therapy.
In the present study and COSMIC v84 database, the frequencies of detectable EGFR mutations by the 2 methods were similar, despite the different number of mutations identified by the 2 methods. In particular, in regard to exon 19 mutations, Scorpion-ARMS and cobas v2 identified 19 and 29 deletions, respectively. Mutations identified by the two methods include common mutations (exon 19 E746-A750 deletion and exon 21 L858R mutation); however, there are many other types of mutations, for example there are 99 different types of exon 19 deletions listed in the COSMIC database v84. In a recent clinical study, the efficacy of EGFR-TKI therapy in NSCLC patients with common exon 19 deletions and the exon 21 L858R mutation were analyzed (4–6). Methodological studies tend to use methods with higher sensitivity for liquid biopsy. Scorpion-ARMS and cobas v2 are highly sensitive PCR methods for the detection of common mutations. Cobas v2 is a recently established reliable PCR method, which is the companion diagnostic for osimertinib (6). However, the frequency of detectable EGFR mutations and the clinical significance of unidentified mutations by the 2 methods are unclear. Previous studies have demonstrated that NSCLC patients with tumors harboring exon 19 deletions have a longer survival rate when compared with those harboring the L858R point mutation of exon 21, following treatment with EGFR-TKIs, which was reported between 2006 and 2007 prior to the approval of cobas v2 as an IVD (16–18). Afatinib treatment resulted in a significant improvement in the overall survival of NSCLC patients with exon 19 deletions, compared with cytotoxic chemotherapy (4). In the present study, the RR of patients with exon 19 deletions was higher than that of exon 21 L858R. Among patients with exon 19 deletions, that were not identified by Scorpion-ARMS and cobas v2, there were patients who responded to EGFR-TKI therapy.
Sizing capillary electrophoresis not only detected the exon 19 deletions identified by Scorpion-ARMS and cobas v2, but also detected the exon 19 deletion that was not identified by the 2 methods. This was observed in a patient with adenocarcinoma for whom EGFR-TKI therapy was effective. The samples of 3 patients with the exon 19 E746–750 deletion (common mutation), which was detected by sequencing were concluded to be wild-type by the 2 methods. The reason for this was that the tumors are heterogeneous, comprised of a mixture of wild-type cells and cells with mutations, and as there was a time lag between performing sequencing and the 2 methods, the DNA may have been damaged during this delay.
Sequencing has the ability to analyze the whole exome. EGFR contains 564 nucleotides in exons 18 to 21 (codons 688–875), and for the majority of NSCLC patients, sequencing of the whole exome of EGFR is difficult due to the high cost and long time required (19,20). The frequency of false negatives is an important factor in screening prior to treatment, as patients may lose the opportunity to receive EGFR-TKI therapy. Exon 19 mutations in particular had 10% false negatives in Scorpion-ARMS and cobas v2, and were expected to achieve improved overall survival via EGFR TKIs therapy when compared with other exon mutations. In addition, different types of exon 19 deletions have continuously increased in the COSMIC database between v84 and v87 over the 10 month time period between versions, therefore the number of false negatives is increasing. In conclusion, exon 19 deletions are the most important EGFR mutations in exons 18 to 21 for EGFR-TKI therapy, and the frequencies of exon 19 deletions detected by Scorpion-ARMS and cobas v2 were less than those of exon 21 mutations with L858R. Therefore, the results of the present study suggest that decreasing the rate of the false negatives of exon 19 deletions via widely used PCR methods may be important for the clinical testing of EGFR mutations.
Acknowledgements
The authors would like to thank the Department of International Medical Communications of Tokyo Medical University (Tokyo, Japan) for their assistance with the manuscript.
Glossary
Abbreviations
- EGFR
epidermal growth factor receptor
- NSCLC
non-small cell lung cancer
- TKI
tyrosine kinase inhibitor
- PCR
polymerase chain reaction
- IVD
in vitro diagnostics
- Scorpion-ARMS
Scorpion Amplification Refractory Mutation System
- FFPE
formalin-fixed and paraffin-embedded
- COSMIC
the catalog of somatic mutations in cancer
- PR
partial response
- CR
complete response
- SD
stable disease
- RR
response rate
- DCR
disease control rate
Funding
No funding was received.
Availability of data and materials
The datasets regarding the clinical samples used and analyzed during this study are available from the corresponding author on reasonable request. The datasets analyzed in this study are available in the COSMIC database repository (cancer.sanger.ac.uk/cosmic/).
Authors' contributions
EN and MS conceived and designed the study. KF obtained the clinical samples from Tokyo Medical University Ibaraki Medical Center (Ibaraki, Japan). HT and OU obtained the clinical samples from Tokyo Medical University Hachioji Medical Center (Tokyo, Japan). YK, TO and NI obtained the clinical samples from Tokyo Medical University Hospital (Tokyo, Japan). EN, MS and TO processed and analyzed the samples. TO, NI and FRH interpreted the data. JM and WAF analyzed and interpreted the pathological samples. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study received ethics approval from the Institutional Review Board of Tokyo Medical University (Tokyo, Japan), and written informed consent to participate was obtained from all of the patients.
Patient consent for publication
The present study obtained consent for publication from each patient.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. Gefitinib or chemotherapy for non-small cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 5.Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa K, Niho S, Tsuji F, Linke R, Rosell R, Corral J, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18:1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 6.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 7.Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, Nishio K. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 8.Kimura H, Ohira T, Uchida O, Matsubayashi J, Shimizu S, Nagao T, Ikeda N, Nishio K. Analytical performance of the cobas EGFR mutation assay for Japanese non-small lung cancer. Lung Cancer. 2014;83:329–333. doi: 10.1016/j.lungcan.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 10.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: Biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 11.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 12.Sonobe M, Manabe T, Wada H, Tanaka F. Mutations in the epidermal growth factor receptor gene are linked to smoking-independent, lung adenocarcinoma. Br J Cancer. 2005;93:355–363. doi: 10.1038/sj.bjc.6602707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 14.Mitsudomi T, Kosaka T, Yatabe Y. Biological and clinical implications of EGFR mutations in lung cancer. Int J Clin Oncol. 2006;11:190–198. doi: 10.1007/s10147-006-0583-4. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi Y, Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 2016;107:1179–1186. doi: 10.1111/cas.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, Zakowski MF, Kris MG, Ladanyi M, Miller VA. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 17.Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, Bell DW, Huberman MS, Halmos B, Rabin MS, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch FR, Varella-Garcia M, Cappuzzo F, McCoy J, Bemis L, Xavier AC, Dziadziuszko R, Gumerlock P, Chansky K, West H, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18:752–760. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 19.Lin MT, Mosier SL, Thiess M, Beierl KF, Debeljak M, Tseng LH, Chen G, Yegnasubramanian S, Ho H, Cope L, et al. Clinical validation of KRAS BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141:856–866. doi: 10.1309/AJCPMWGWGO34EGOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Yang Y, Li H, Chen Z, Jiang G, Fei K. Assessment of the clinical application of detecting EGFR, KRAS, PIK3CA and BRAF mutations in patients with non-small cell lung cancer using next-generation sequencing. Scand J Clin Lab Invest. 2016;76:386–392. doi: 10.1080/00365513.2016.1183813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets regarding the clinical samples used and analyzed during this study are available from the corresponding author on reasonable request. The datasets analyzed in this study are available in the COSMIC database repository (cancer.sanger.ac.uk/cosmic/).