Abstract
Epidermal growth factor receptor (EGFR)-positive non-small cell lung cancer (non-SCLC) benefits from first-line treatment with first generation tyrosine kinase inhibitors (TKIs). However, drug resistance is inevitable through different mechanisms and is dominated by the acquisition of the T790M mutation within EGFR, which occurs in ~50% of cases. The present study reports the case of a patient originally diagnosed with stage IA lung adenocarcinoma, with a recurrent tumor lesion in each side of the lungs following the surgical removal of the primary tumor. Erlotinib treatment of the recurrent tumors eliminated the tumor on the right side of the lung and resulted in the histological transformation of the tumor on the left side to SCLC following 6 years of treatment. Genetic profiling of the SCLC lesions using targeted next-generation sequencing identified different genetic alterations from the primary tumor, characterized by the newly acquired copy number loss of tumor protein p53 and transcriptional coreceptor 1, and the copy number gain of SRY-box 2. Continuation of treatment with chemotherapy and erlotinib demonstrated moderate disease control for ~1 year prior to the outbreak of a new lung lesion. Liquid biopsy profiling of circulating tumor DNA revealed the acquisition of KRAS proto-oncogene, GTPase (KRAS) p.G12C mutation, indicating the occurrence of another resistance mechanism to erlotinib. During erlotinib treatment, the lung adenocarcinoma progressed through two atypical mechanisms, notably from the transformation to SCLC and the acquisition of the KRAS mutation to surpass EGFR inhibition. However, the combinational and interchanging usage of chemotherapy and TKI resulted in persistent and effective disease control.
Keywords: non-small cell lung cancer, epidermal growth factor receptor, erlotinib, small cell lung cancer, KRAS mutation, next-generation sequencing
Introduction
Lung cancer is the leading cause of cancer-related mortality globally. Lung cancer is classified into two main histological subtypes, non-small cell lung cancer (NSCLC) and SCLC, with substantially different treatment strategies (1,2). In patients with epidermal growth factor receptor (EGFR)-activating mutations in NSCLC, the most frequently occurring mutations are deletion of exon 19 and a leucine to arginine change at position 858 (p.L858R) according to 2018 National Comprehensive Cancer Network Practice Guidelines in Oncology for NSCLC (3). EGFR tyrosine kinase inhibitors (EGFR-TKIs) including gefitinib and erlotinib are generally used as a first line treatment, and confer an improved progression-free survival (PFS) time compared with traditional chemotherapy (4,5). The standard treatment for SCLC is chemotherapy or radiotherapy, due to the low frequency of druggable mutations in tumors (6). The median PFS time for EGFR-TKI treatment is ~10 months, and drug resistance is inevitable through various mechanisms, including the acquisition of a secondary EGFR mutation (p.T790M), inactivation of tumor protein p53 (TP53), bypassed activation of downstream effectors, including AKT, PIK3CA, KRAS, alterations of alternative pathways, including MET and ERBB2, epithelial-to-mesenchymal transition, and more rarely, histological transformation from NSCLC to SCLC (7–9). The present study reports the case of a patient with stage IA lung adenocarcinoma (ADC), who displayed severe adverse effects to chemotherapy and underwent 6 years of erlotinib maintenance.
Case report
A 65-year-old female patient with a history of smoking (smoking index, 60 pack/year) was admitted to Peking University 3rd Hospital (Beijing, China) in April 2008 and diagnosed with stage IA3 (T1cN0M0) peripheral lung ADC in the right upper lobe according to the 8th edition of UICC TNM classification (10,11). Radical resection by video-assisted thoracoscopic surgery was performed immediately. The excised tumor was confirmed as poorly differentiated ADC, which is a high-risk tumor requiring postoperative adjuvant chemotherapy according to the 2008 NCCN Practice Guidelines in Oncology for NSCLC (11).
Immunohistochemistry was performed to determine expression of pan-cytokeratin (CKpan), Ki-67 proliferation factor (Ki-67), cytokeratin-7 (CK7), thyroid transcription factor-1 (TTF-1), napsin A, CK5/6, p40, p63, neural cell adhesion molecule (CD56), chromogranin A (CgA) and synaptophysin (Syn). The tissue sample was fixed in 10% formalin for 24–48 h at room temperature, embedded in paraffin, and cut into 4-µm thick sections. Immunohistochemistry was performed by blocking with goat serum for 30 min at room temperature and staining with the following primary antibodies at 1:1,000 for 1 h at room temperature: CK7, CK5/6, Ki-67, TTF-1 and p63 (cat. nos. GM701807, GM723714, GT209407, GM357514, GM724714; Gene Tech Biotechnology Co., Ltd, Shanghai, China); CKpan and CgA (cat. nos. GA053, M0869; Dakocytomation, Glostrup, Denmark); Syn (cat. no. 1870-1; Epitomics; Abcam, Cambridge, MA, USA); and p40, napsin A and CD56 (cat. nos. ZM-0472, ZM-0057; ZSGB-BIO Technologies, Inc., Beijing, China). Sections were then incubated in the following horseradish peroxidase-conjugated secondary antibodies at 1:1,000 for 20 min at room temperature (MaxVision TM HRP-Polymer anti-Mouse/Rabbit IHC Kit; cat. no. KIT-5020; MXB Biotechnologies Inc., Fuzhou, China). The sections were imaged using a Olympus Vanox microscope at ×400 magnification. All tests were performed in the clinical testing lab of Peking University 3rd Hospital.
Immunohistochemistry revealed positive expression of ADC markers CK7, TTF-1 and napsin A, with no expression of histological markers of SCLC (CD56, CgA and Syn; Table I). The primary tumor was sent to a commercial sequencing center (Geneseeq Technology Inc., Nanjing, China) for targeted next generation sequencing (NGS) with a customized panel (Geneseeq One™) covering 382 cancer-related genes as previously described (12). In brief, tumor DNA was extracted with DNeasy Blood & Tissue kit (cat. no. 69506; Qiagen China Co., Ltd., Shanghai, China) and fragmented into 350 bp DNA fragments using the Covaris M220 instrument (Covaris, Woburn, MA, USA). The DNA library was prepared using KAPA Hyper Prep kit (cat. no. KK8504; KAPA Biosystems; Roche Diagnostics, Basel, Switzerland). Library hybridization was performed with NimbleGen SeqCap EZ Hybridization and Wash Kit (cat. no. 5634253001; Roche Diagnostics) and Dynabeads M-270 (cat. no. 65305; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the probes for enrichment were customized and synthesized by Integrated DNA Technologies (Coralville, IA, USA). The prepared library was sequenced on Illumina HiSeq4000 (Illumina, Inc., San Diego, CA, USA). A loss-of-function mutation in TP53, p.G244C and two point mutations in the GNAS gene with unknown significance were identified (Table II). Gene copy number variations (CNVs) were detected with ADTEx 2.0 software (http://adtex.sourceforge.net) using default parameters (13).
Table I.
Summary of immunohistochemistry results for diagnostic markers of ADC and SCLC.
| Samples | |||
|---|---|---|---|
| Markers | Histological Type | Primary lesion, surgical resection | EBUS + IBNA, biopsy |
| CKpan | Carcinoma | + | + |
| Ki-67 | Proliferation | 5% + | 90% |
| CK7 | ADC | + | Na |
| TTF-1 | ADC | Partially + | Partially + |
| Napsin A | ADC | + | Na |
| CK5/6 | SCC | − | Na |
| p40 | SCC | − | − |
| p63 | SCC | − | Na |
| CD56 | SCLC | − | Partially + |
| CgA | SCLC | − | − |
| Syn | SCLC | − | Partially + |
-, negative staining; +, positive staining; na, not tested; ADC, adenocarcinoma; SCC, squamous cell carcinoma; SCLC, small-cell lung cancer; EBUS + TBNA, endobronchial ultrasound-guided trans-bronchial needle aspiration; CKpan, pan-cytokeratin; CK, cytokeratin; TTF-1, thyroid transcription factor-1; CD56, neural cell adhesion molecule; CgA, chromogranin A; P, tumor protein; Syn, synaptophysin.
Table II.
Mutation profiling by targeted next generation sequencing.
| Right lung: ADC | Left lung: SCLC | Peripheral Blood | |||
|---|---|---|---|---|---|
| Genes | Variations | Primary lesion, surgical resection | Hilar lymph node, biopsy | Bronchial neoplasia, biopsy | Plasma ctDNA |
| GNAS | p.P438T | 8.5% | |||
| p.S252R | 5.9% | − | − | ||
| TP53 | p.G244C | 12.6% | − | − | |
| p.G245V | − | 83% | 89% | − | |
| CNV | − | 0.5-fold | 0.5-fold | ||
| RB1 | p.S81fs | − | 65% | 60% | |
| CNV | 0.5-fold | 0.5-fold | − | ||
| SOX2 | CNV | − | 4-fold | 3.2-fold | − |
| PKHD1 | c.8546delG | − | 45% | 31% | − |
| KRAS | p.G12C | − | − | − | 1% |
Mutant allele frequency and CNV fold-change are provided for each variation calculated with the methods as previous described (11). -, not detected; del, deletion; ADC, adenocarcinoma; SCLC, small cell lung cancer; CNV, copy number variation; ctDNA, circulating tumor DNA; GNAS, GNAS complex locus; TP53, tumor protein p53; RB1, retinoblastoma 1; SOX2, superoxide dismutase 2; PKHD1, polycystic kidney and hepatic disease 1; KRAS, KRAS proto-oncogene.
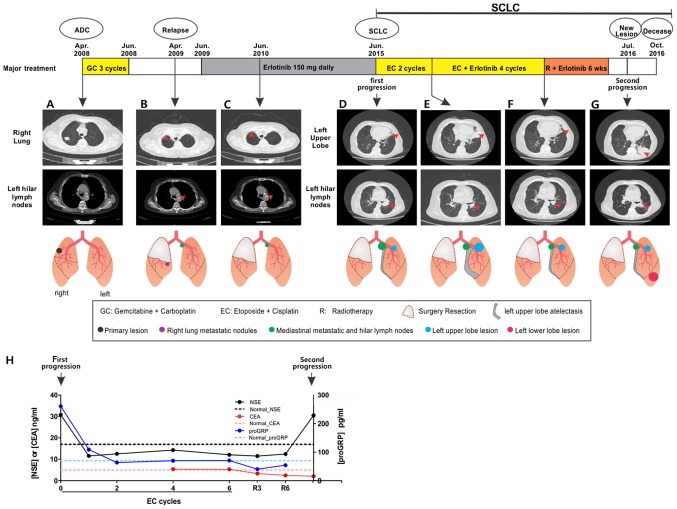
The patient quit smoking following the diagnosis of ADC and was treated with gemcitabine at a dose of 1,000 mg/m2 body surface area (BSA) on day 1 and 8, and carboplatin (area under the curve=5, d1, every 21 days, calculated by Calvert Formula) as adjuvant chemotherapy for only 3 cycles due to severe adverse effects. A relapse occurred with a non-productive cough in April 2009. Computed tomography (CT) scans revealed a metastatic nodule in the right middle lobe and infiltration into the bilateral mediastinal and hilar lymph nodes (Fig. 1B). Serum progastrin-releasing peptide (ProGRP), carcinoembryonic antigen (CEA) and neuron-specific enolase (NSE) levels were normal. Chemiluminescent testing kits for these protein markers were obtained from Roche Diagnostics (cat. nos. 3404883, 3404885, 3404881), and the experiments were performed according to the manufacturer's protocols in the clinical testing lab of Peking University 3rd Hospital. As the patient was not tolerant to either surgery or chemotherapy, erlotinib treatment was commenced at a dose of 150 mg/day in June 2009 and continued thereafter for 6 years until June 2015. The EGFR inhibitor was used as erlotinib is recommended for treating NSCLC following first-line or second-line chemotherapy regardless of the EGFR mutation status according to the BR.21 clinical trial in 2005 (14). Unexpectedly, following treatment with erlotinib at the aforementioned dose, the non-productive cough eased and the metastatic nodule in the right middle lobe fully responded and disappeared, while the left hilar lymph nodes remained unchanged with 1 year of erlotinib therapy (Fig. 1C). However, due to the hard-to-access position of the lymph nodes, no tumor tissues were acquired for histological and genetic analysis.
Figure 1.
Clinical tracking of disease progression in the patient with non-SCLC. The CT images and the schematic diagrams represent the different multiple lesions over the course of the disease, with the time scale above and arrows indicating the different time points at which the images were obtained. Tumor sites of interest in the CT scans are labeled using red arrows. (A) At diagnosis and prior to surgery. (B) At 1 year post-surgery. (C) After 1 year of erlotinib treatment. (D) After 6 years of erlotinib treatment. (E) After 2 cycles of EC chemotherapy without erlotinib. (F) After an additional 4 cycles of EC chemotherapy along with erlotinib. (G) Second progression with new lesion. (H) Serum levels of NSE, CEA and proGRP were plotted under the treatment timeline. R3, radiotherapy for 3 weeks; R6, radiotherapy for 6 weeks. wks, weeks; CT, computed tomography; EC, etoposide and cisplatin; NSE, neuron-specific enolase; CEA, carcinoembryonic antigen; proGRP, pro-gastrin-releasing peptide; GC, gemcitabine and carboplatin; ADC, adenocarcinoma; SCLC, small cell lung cancer.
Following 6 years of erlotinib treatment at the aforementioned dose until June 2015, the patient developed a cough and CT revealed that the left hilar lymph node was markedly enlarged, which caused left upper lobe atelectasis (Fig. 1D). Endobronchial ultrasound-guided trans-bronchial needle aspiration (EBUS + TBNA) revealed that the left hilar lymph nodes and the left upper lobe bronchial neoplasms were enlarged. Biopsies at these two sites confirmed SCLC, with positive IHC staining for the SCLC markers CD56, Syn, TTF-1 and CKpan (Table I). Ki-67 staining revealed a high proliferation rate in >90% of cells, which is an additional feature of SCLC. Targeted NGS identified completely different genetic profiles in the two SCLC samples from the primary lesion (Table II). The two SCLC biopsies had acquired loss-of-function mutations in TP53 (p.G245V) and retinoblastoma 1 (RB1, p.S81fs), as well as reduced copy number in TP53 and RB1, and increased copy number in the superoxide dismutase 2 (SOX2) gene. This mutation profile is consistent with the SCLC genetic signature, which is characterized by a high prevalence of TP53 and RB1 inactivation, and a frequent SOX2 copy number gain (15). Staging examination by brain MRI, bone scan and abdomen ultrasound revealed no distant metastasis to the brain, bone or abdominal cavity. The levels of serum SCLC markers, including Pro-GRP and NSE, were markedly elevated (Fig. 1H).
Following the histological transformation of ADC to SCLC, treatment with erlotinib was replaced with chemotherapy using etoposide (100 mg/m2 BSA on days 1–3) and cisplatin (administered at the standard rate of 75 mg/m2) every 3 weeks. Partial remission was achieved with the shrinkage of the left hilar lymph node, and the serum levels of Pro-GRP and NSE returned to normal (Pro-GRP, <70 pg/ml; NSE, 0–17 ng/ml) after 2 cycles of treatment (Fig. 1E and H). However, the left upper lobe atelectasis progressed rapidly (Fig. 1E), thus erlotinib (150 mg/day) was re-prescribed along with chemotherapy for another 4 cycles of 3 weeks of treatment. At the end of the treatment, atelectasis was improved and the left upper lobe lesion was markedly reduced, suggesting that it may contain a mutation sensitive to EGFR-TKI (Fig. 1F). However, positron emission tomography (PET)-CT with IV injection of 18F-fluorodeoxyglucose (10–20 mCi) immediately after these 4 cycles of combination therapy, revealed no improvement in the left hilar lymph node. Therefore, intensity-modulated radiotherapy was administrated to the mediastinum and bilateral hilum for 6 weeks (95% planning target volume, 50.4 Gy/1.8 Gy/28f), with oral erlotinib (150 mg/day). During the treatment, serum Pro-GRP, NSE and CEA levels were decreased and/or within a normal range (Fig. 1H).
In July 2016, the patient developed an aggravated cough with sputum and hematemesis, and a CT scan revealed a new lesion in the left lower lobe, with no changes in all the former lesions (Fig. 1G). The serum NSE level was markedly elevated, while Pro-GRP and CEA remained normal (Fig. 1H). The patient refused another biopsy; therefore, peripheral blood samples were collected for a liquid biopsy to test the plasma circulating tumor DNA (ctDNA) instead (Table II). Targeted NGS on plasma ctDNA using the aforementioned sequencing panel, revealed a KRAS-activating mutation (p.G12C), which may impact the downstream signaling of EGFR and therefore cause resistance to EGFR-TKI treatment (16). The patient refused further treatment and succumbed to the disease in October 2016.
Discussion
In the present study, although the initial genetic sequencing of the surgical resection sample did not identify any detectable EGFR activation mutations, erlotinib treatment resulted in an overall survival time of 7 years. Retrospectively reviewing the present study, we hypothesize that the patient may have harbored an EGFR-activating mutation in the right lung metastatic nodule, as this nodule began to shrink one month after erlotinib treatment and was undetectable after one year. However, a tumor biopsy sample was not acquired at the time for confirmation.
Notably, the recurrent lesions had distinct responses to EGFR-TKI, in the presence/absence of chemotherapy and radiotherapy treatments, which may be ascribed to their diverse genetic profiles. The long-term treatment of erlotinib prompted the formation of two SCLC lesions in the mediastinal metastatic and hilar lymph nodes, and subsequently, a lesion in the left upper lobe. The diagnosis of SCLC was supported by the positive histological staining of protein biomarkers, including CD56 and Syn, and the detection of the SCLC genetic signatures, including the copy number loss in RB1 and the copy number gain in SOX2. The initial lesion in the right upper lobe responded to chemotherapy, while the second lesion, in the right middle lobe, responded to erlotinib treatment. The transformed SCLC normally carries the same EGFR mutation as the primary NSCLC (9). In the present study, no shared somatic mutations were identified from the ADC tissue and SCLC biopsies, suggesting that SCLC lesions in this case may represent a novel and independently developed disease, and the loss of RB1 is essential for the development of SCLC (9).
Following the formation of a novel lesion in the left lower lobe, the KRAS p.G12C mutation was detected in the liquid biopsy, while other SCLC genetic signatures, including frameshift mutation and copy number loss in RB1 and copy number gain in SOX2 (17) were not detected, suggesting the acquisition of a new resistance mechanism to erlotinib. As a known oncogenic driver in NSCLC, KRAS is not markedly mutated in SCLC (9), but its mutations have been reported as a primary resistance mechanism to EGFR-TKI treatments in NSCLC (4).
In conclusion, the present study reports a complex and diversified tumor evolution under erlotinib treatment, by phenotypic transformation from ADC to SCLC with the acquisition of a KRAS activating mutation. With the assistance of NGS-based genetic testing and biomarker monitoring, the treatment strategy was adjusted and achieved long-term disease control.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data analyzed during this study are included in the published article.
Authors' contributions
LL designed the study, revised the manuscript and provided project supervision. YG, XZ and BC aided in the collection of clinical information, NGS performed data analysis and interpretation and writing the manuscript. XW, XT and YWS performed DNA extraction, NGS undertook library preparation and bioinformatics analysis.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of Peking University 3rd Hospital. The specimens of this study were obtained from Biological Sample Bank of Peking University 3rd Hospital, Beijing, China.
Patient consent for publication
The patient provided written informed consent for research and publication of their clinical details.
Competing interests
YWS, XW and XT are shareholders or employees of Geneseeq Technology Inc.
References
- 1.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 2.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378:1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 3.www.nccn.org NCCN practice guidelines in oncology-non-small cell lung cancer, V.2.2018. Available at. [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 6.Wakuda K, Kenmotsu H, Serizawa M, Koh Y, Isaka M, Takahashi S, Ono A, Taira T, Naito T, Murakami H, et al. Molecular profiling of small cell lung cancer in a Japanese cohort. Lung Cancer. 2014;84:139–144. doi: 10.1016/j.lungcan.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, Capelli L, Casanova C, De Luigi N, Mariotti M, et al. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res. 2017;23:2195–2202. doi: 10.1158/1078-0432.CCR-16-0966. [DOI] [PubMed] [Google Scholar]
- 8.Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res. 2015;4:67–81. doi: 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: Molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–e172. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 8th. Wiley-Blackwell; Jan, 2017. [Google Scholar]
- 11.www.nccn.org NCCN Practice Guidelines in Oncology-Non-small Cell Lung Cancer, V.2.2008. Available at. [Google Scholar]
- 12.Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao Y, Chen X, Sun J, Wang Z, Hong Z, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7:583. doi: 10.1038/s41598-017-00520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amarasinghe KC, Li J, Hunter SM, Ryland GL, Cowin PA, Campbell IG, Halgamuge SK. Inferring copy number and genotype in tumour exome data. BMC Genomics. 2014;15:732. doi: 10.1186/1471-2164-15-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 15.Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 17.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed during this study are included in the published article.